Abstract
BACKGROUND:
Ventilator-associated pneumonia (VAP) remains a common complication in critically ill surgical patients, and its diagnosis remains problematic. Exhaled breath contains aerosolized droplets that reflect the lung microbiota. We hypothesized that exhaled breath condensate fluid (EBCF) in hygroscopic condenser humidifier/heat and moisture exchanger (HCH/HME) filters would contain bacterial DNA that qualitatively and quantitatively correlate with pathogens isolated from quantitative BAL samples obtained for clinical suspicion of pneumonia.
METHODS:
Forty-eight adult patients who were mechanically ventilated and undergoing quantitative BAL (n = 51) for suspected pneumonia in the surgical ICU were enrolled. Per protocol, patients fulfilling VAP clinical criteria undergo quantitative BAL bacterial culture. Immediately prior to BAL, time-matched HCH/HME filters were collected for study of EBCF by real-time polymerase chain reaction. Additionally, convenience samples of serially collected filters in patients with BAL-diagnosed VAP were analyzed.
RESULTS:
Forty-nine of 51 time-matched EBCF/BAL fluid samples were fully concordant (concordance > 95% by κ statistic) relative to identified pathogens and strongly correlated with clinical cultures. Regression analysis of quantitative bacterial DNA in paired samples revealed a statistically significant positive correlation (r = 0.85). In a convenience sample, qualitative and quantitative polymerase chain reaction analysis of serial HCH/HME samples for bacterial DNA demonstrated an increase in load that preceded the suspicion of pneumonia.
CONCLUSIONS:
Bacterial DNA within EBCF demonstrates a high correlation with BAL fluid and clinical cultures. Bacterial DNA within EBCF increases prior to the suspicion of pneumonia. Further study of this novel approach may allow development of a noninvasive tool for the early diagnosis of VAP.
Ventilator-associated pneumonia (VAP) is among the most common health-care-associated infections in severely ill and injured patients, accounting for substantial morbidity, increased length of ICU and hospital stay, and excess cost and mortality.1‐3 Critically ill and injured patients are particularly prone to VAP, due to several factors that both increase the risk and confound the diagnosis of pneumonia, making accurate and timely diagnosis of pneumonia problematic.1,4‐9 Atelectasis, pulmonary contusions, acute lung injury, aspiration pneumonitis, and the systemic inflammatory response syndrome are all common following major operative interventions or severe trauma and can mimic pulmonary infections. The standard clinical criteria for the diagnosis of VAP overestimates the rate twofold when compared with quantitative culture using BAL or protected brush specimens.10‐13 Quantitative scoring systems of clinical and radiographic findings have failed to increase the diagnostic accuracy when compared to quantitative cultures.14‐18 Quantitative culture techniques increase the specificity of the diagnosis and may improve overall outcomes.11 However, these techniques mandate the development of clinical symptoms to introduce clinical suspicion, requiring approximately three additional days for culture and sensitivity results, necessitating empirical antibiotic coverage while results are pending. Thus, existing diagnostic strategies delay therapy until the infection is well established and require significant empirical therapy, contributing to unnecessary antibiotic exposure.19
While the diagnosis of pneumonia is difficult to establish in a rapid or specific fashion, significant evidence supports reduced morbidity and mortality when appropriate antibiotic therapy is initiated early in patients with VAP.20‐23 Inadequate empirical antibiotic coverage for VAP is associated with a twofold increase in mortality, and retrospective studies suggest that delays as short as 30 min from the onset of fever in infected patients may increase mortality.20‐24 However, unnecessary antibiotic exposure is associated with increased risk for subsequent infectious complications, colonization, and infection with resistant pathogens, and increased hospital costs.21,25‐31 Thus, early identification of patients with pneumonia is necessary to improve outcomes in this population.
The goal of our research is to develop tools that can improve the timeliness, specificity, and sensitivity of VAP diagnosis in critically ill patients. This would enable clinicians to diagnose pneumonia earlier in its course, select more-specific antimicrobial therapies, and more accurately monitor response to treatments. Exhaled breath contains aerosolized droplets of widely varying size (majority between 5 μm and 100 μm) that carry bacteria, as first described by Flugge in 1897.32,33 These droplets reflect the pathogens in the lower respiratory tree, transmitting them to the environment.32‐35 Thus, we hypothesized that bacteria within these aerosolized breath droplets would collect within the hygroscopic condenser humidifier/heat and moisture exchanger (HCH/HME) filters between the endotracheal tubes and ventilator circuit and provide a quantitative assessment of pulmonary bacterial growth. The purpose of this current study is to examine if polymerase chain reaction (PCR) analysis of exhaled breath condensate fluid (EBCF) would correlate quantitatively and qualitatively with fluid samples from time-matched, semiquantitative BAL fluid (BALF) obtained for the clinical suspicion of pneumonia, thus providing qualitative and quantitative results in hours rather than the 3 days required by current techniques.
Materials and Methods
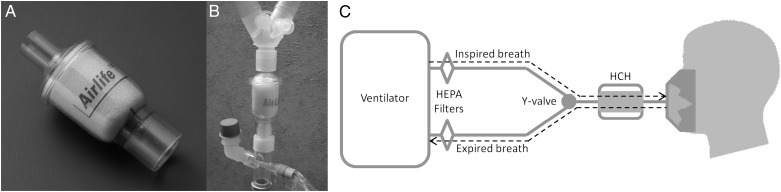
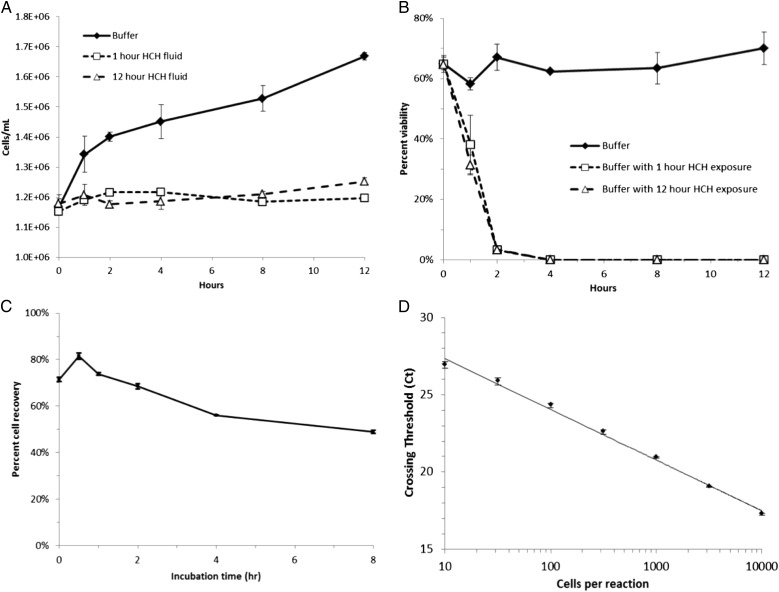
We previously reported that HCH/HME filters (Fig 1) serve as a reservoir for pathogens carried in exhaled breath condensate but do not allow bacterial proliferation, since they are bacteriocidal. As a result, bacterial cells remain intact and can be reliably quantified by real-time PCR (RT-PCR) (Fig 2).36 Additionally, we have demonstrated that bacterial recovery is independent of species and that these isolated bacteria can be uniformly lysed and quantified without bias or loss of specific species. Finally, we have demonstrated that samples can be normalized by surfactant content via surfactant-associated protein B (SP-B) and dipalmitoylphosphatidylcholine (DPPC) measurements.
Figure 1 –
An HCH/heat and moisture exchanger filter unit in isolation and as part of the ventilator circuit. HCH = hygroscopic condenser humidifier; HEPA = high-efficiency particulate air.
Figure 2 –
Extraction of information from HCH/heat and moisture exchanger (HCH/HME) filters is reliable. A, B, Filter environment does not allow proliferation of Escherichia coli and is bacteriocidal. C, A large, representative, and reproducible fraction of bacteria inoculated can be recovered from filters through quantitative assay of nonviable E coli by Coulter counter. D, Linear regression line (r2 = 0.995) of E coli inoculum vs quantitation by real-time polymerase chain reaction, demonstrating the sensitivity and accuracy of polymerase chain reaction analysis of HCH/HME fluid. See Figure 1 legend for expansion of other abbreviation.
Patient Recruitment and Sample Collection
The study was approved by the Vanderbilt University Medical Center (VUMC) institutional review board (No. 101185) and funded by the Vanderbilt Institute for Clinical and Translational Research (CTSA 1 UL1 RR024975). The current standard of care for critically ill patients who were mechanically ventilated in the Vanderbilt surgical ICU (SICU) with suspected pneumonia is to undergo protocol directed, semiquantitative BAL for standard microbiologic culture and empirical antibiotic therapy until BAL results are available. Antibiotics are discontinued for BAL culture results < 104 colony-forming unit (CFU) of bacteria and deescalated to cover only pathogens > 104 CFU.
Between April and August of 2011, adult patients who were ventilated in the SICU, with a clinical suspicion of pneumonia, were enrolled. Two to 4 h prior to BAL, the existing HCH/HME filter unit was exchanged for a new “study” unit. Immediately prior to BAL, the study filter was removed and placed in a biohazard bag. Following the BAL, 2 mL excess BALF was placed in a sterile specimen vial for PCR analysis and residual BALF was sent to the clinical microbiology laboratory, per standard of care. The BALF sample and the time-matched EBCF collected from the study filter were processed separately in identical fashion, with positive and negative control samples. PCR findings from matched BALF and EBCF samples were compared retrospectively with clinical microbiology results of the corresponding BALF sample.37
To assess quantitative changes in bacterial DNA over time within the EBCF in patients who were ventilated, serial HCH/HME filters were collected from a small number of patients who developed VAP during mechanical ventilation. Stored samples and clinical course were analyzed retrospectively after positive BAL cultures were identified.
Sample Processing
DNA Extraction:
EBCF samples from HCH/HME filters and BALF samples were prepared as previously described.36 Briefly, cells were concentrated (independently) on 13-mm-diameter, 0.45 micron Durapore membranes (Millipore Corp). Total DNA was purified from the membranes using the DNeasy Blood and Tissue Kit (Qiagen NV) with the following modifications. Lysis buffer consisted of 20 mM tris(hydroxymethyl)aminomethane, 136 mM sodium chloride, pH 7.4, plus 2 mg/mL lysozyme (Sigma-Aldrich Co LLC), 5 mM ethylenediaminetetraacetic acid, and 10 units/mL lysostaphin (Sigma-Aldrich Co LLC). Each concentrated cell sample was resuspended in 200 uL lysis buffer and incubated for 2 h at 37°C with 200 rpm shaking. Then, 20 uL proteinase K solution and 200 uL buffer AL from the DNeasy kit was added, and incubated at 56°C for 30 min.
Quantitative PCR:
Sample analysis was performed as described previously.36,38 Eight genomic targets, seven bacterial species-specific genes, and a species-independent bacterial 16S rDNA were used.38‐44 These seven species represent 70% of SICU pathogens causing VAP, as reported to the Centers for Disease Control and Prevention. Primer details are given in Table 1. All reactions were conducted in triplicate at 10 uL total volume, consisting of 5 uL SsoFast EvaGreen Supermix (BioRad Laboratories Inc), 1 uL DNA template, 1 uL primer mix, and 3 uL nuclease-free distilled water. Reactions were run on a CFX96 Real-Time PCR Detection System (BioRad Laboratories Inc). The cycling parameters were as follows: an initial incubation of 2 min at 98°C, then 40 cycles of 5 s at 98°C and 10 s at 60°C with real-time fluorescence measurement. Purified DNA samples were first analyzed by universal 16S quantitative PCR to conserve time and reagents by not further studying negative samples. Positive samples were then subjected to the full panel of species-specific probes. Absolute quantification was achieved using four concentrations (100, 10,000, 100,000, and 1 million copies) of each amplified sequence that had been ligated into TOPO-TA vectors, expressed in transformed Escherichia coli, extracted and purified, then quantified by fluorescent NanoDrop (Thermo Fisher Scientific Inc) using SYBR Green II (Life Technologies Corp) as a dye and a DNA standard.
TABLE 1 ] .
Pathogen-Specific Targets for Polymerase Chain Reaction Analysis
| Organisma | Gene Target | Amplicon Size |
| Acinetobacter baumannii | Citrate synthase | 722 |
| Escherichia coli | β-Glucuronidase | 486 |
| Enterococcus faecalis | 16s rDNA | 138 |
| Enterococcus faecium | d-ala:d-ala ligase | 550 |
| Klebsiella pneumoniae | 16s-23s rDNA IS | 260 |
| Pseudomonas aeruginosa | OM lipoprotein | 504 |
| Staphylococcus aureus | Thermonuclease | 106 |
| Universal | 16s rDNA | 470 |
Selected pathogens account for > 70% of surgical ICU ventilator-associated pneumonia pathogens.
Surfactant Assay:
To aid in quality control and standardization, SP-B concentrations from EBCF samples, BALF samples, or both were determined as reported by Krämer et al45 with the following modifications: Infasurf (ONY, Inc) was diluted in phosphate-buffered saline to create standards. Polyclonal rabbit anti-SP-B (Millipore Inc) was used at a 1:5,000 dilution in phosphate-buffered saline, 0.1% bovine serum albumin. Plates were developed with the Amplex ELISA Development Kit for Rabbit IgG (Invitrogen Corp) according to manufacturer’s directions and read at 590 nm with 530 nm excitation on a Spectramax M5 (Molecular Devices LLC) in the VUMC Center Molecular Cell Biology Resource Core. DPPC concentrations in all samples were determined as previously described by Ivanova et al46 using 200 ng/mL D62-DPPC (Avanti Polar Lipids Inc) as an internal standard and analyzed on a TSQ Quantum quadrupole mass spectrometer (Thermo Scientific Fisher Inc) in the VUMC Mass Spectrometry Core.
Statistical Methods:
The binary outcome data (target amplified or not relative to controls) were categorized using a contingency table with EBCF results in the rows. The CIs for proportions, such as percent agreement, sensitivity, and specificity, were constructed using the Wilson score method with continuity correction.47 CIs for the correlation coefficients were constructed using the Fisher Z transform method. The concordance coefficient κ, and an accompanying CI, was calculated from the matched sample data according to the description by Hanley48 by considering the sample space to consist of all 27 = 128 possible outcomes from performing seven distinct tests with binary response.
Results
Forty-eight subjects with 51 episodes of clinically suspected VAP who underwent BAL prior to the initiation of empirical antibiotic therapy were enrolled. The matched EBCF/BALF pairs were analyzed by RT-PCR for both qualitative and quantitative agreement of pathogens and compared with quantitative clinical cultures reported by the clinical microbiology laboratory.
Clinical Culture Results
Of 51 episodes of suspected VAP, 20 patients had confirmed VAP by quantitative BAL (cultures with > 104 CFU/mL) and continued deescalated antibiotic therapy for a full therapeutic course. RT-PCR of matched EBCF detected the pathogens in 100% of patients. In 31 episodes of suspected VAP, quantitative BAL did not confirm pneumonia (< 104 CFU/mL) and empirical antibiotics were discontinued. In 24 of these 31 episodes, BAL revealed no growth. RT-PCR detected low copies of bacterial DNA from both EBCF and BALF in eight cases when BAL clinical cultures did not.
Qualitative Agreement of PCR Detection of Bacterial Pathogens in EBCF and BALF
Fifty-one time-matched pairs of EBCF and BALF samples were collected from 48 patients. Three patients had repeat BAL procedures performed on different days. The qualitative results from the 51 time-matched pairs are summarized in Table 2, grouped by pathogen results in the 24 patterns observed. The qualitative PCR results for the matched (EBCF/BALF) pairs are in near-perfect agreement, with two minor exceptions (Table 2). In the first exception, < 100 copies/mL of Entercoccus faecalis DNA was found in the BALF sample but not the EBCF sample, and no growth occurred in the standard microbiologic culture sample. In the second exception, < 200 copies/mL of Staphylococcus aureus DNA was detected in the BALF sample but not in the EBCF sample, with no growth in microbiologic culture sample. In two matched pairs (Table 2), the clinical microbiologic cultures detected < 103 CFU/mL of Klebsiella pneumoniae not detected by PCR in either EBCF or BALF.
TABLE 2 ] .
Summary Results of 51 Matched Pairs Grouped by Pattern of Occurrence
| No. | EBCF Bacteria by PCR | BALF Bacteria by PCR | BALF Bacteria by Culture | No. | EBCF Bacteria by PCR | BALF Bacteria by PCR | BALF Bacteria by Culture |
| 1 | Nonea | Sa | None | 1 | Pa | Pa | None |
| 2 | Ec | Ec | None | 1 | Sa | Sa | None |
| 1 | Ef | Ef | None | 1 | None | None | Kpb |
| 2 | Kp | Kp | None | 15 | None | None | None |
| 1 | Efs | Efs | Efs | 1 | Ab | Ab | Ab |
| Pa | Pa | Pa | |||||
| 1 | Pa | Pa | Pa | 1 | Ef | Ef | Ef |
| Sa | Sa | Sa | |||||
| 1 | Kp | Kp | Kp | 3 | Kp | Kp | Kp |
| Pa | Pa | Pa | |||||
| 1 | Ec | Ec | Ec | 2 | Pa | Pa | Pa |
| Sa | Sa | Sa | |||||
| 1 | Ef | Ef | Ef | 9 | Sa | Sa | Sa |
| Sa | Sa | Sa | |||||
| 1 | Kp | Kp | Kp | 1 | Ec | Ec | Ec |
| Sa | Sa | a | Efs | ||||
| 1 | Ab | Ab | Ab | 1 | Sa | Sa | Sa |
| Pa | Pa | ||||||
| Sa | Sa | Sa | Kpb | ||||
| 1 | Ab | Ab | Ab | 1 | Efs | Efs | None |
| Efs | Efs | ||||||
| Pa | Pa | Sa | Sa | ||||
| Sa | Sa | Sa |
Ab = Acinetobacter baumannii; BALF = BAL fluid; CFU = colony-forming unit; Ec = Escherichia coli; EBCF = exhaled breath condensate fluid; Ef = Enterococcus faecium; Efs = Enterococcus faecalis; Kp = Klebsiella pneumoniae; Pa = Pseudomonas aeruginosa; PCR = polymerase chain reaction; Sa = Staphylococcus aureus.
Present at < 200 copies by PCR in BALF but not EBCF.
Present in clinical culture at ≤ 103 CFU/mL, not detected in EBCF or BALF by PCR.
The binary outcome data (target amplified or not EBCF vs BALF) are shown in Table 3. The point estimate of the overall proportion of agreement between EBCF and BALF is 49 of 51, in excess of 96%. A 99% CI of the proportion of agreement is 0.81 to 1.0. Given the nominal nature of the data, the modest sample size, and the possibility that the seven distinct PCR measurements are correlated, an appropriately conservative omnibus measure of the degree of concordance between the EBCF and BALF results is provided by the statistic κ of 0.95 (99% CI, 0.87-1.0) (Table 4). If each organism and sample is treated as independent and the data pooled, then 7 × 51 = 357 observations are available. Under these assumptions, an overall correlation (Cohen’s W or φ) in excess of 0.98 (99% CI, 0.97-0.98) is calculated. The 99% CI for overall combined sensitivity is 0.92 to 0.98, and the overall combined specificity is 0.98 to 1.0 (Table 4).
TABLE 3 ] .
Individual Organism Detection by PCR in EBCF and BALF
| Target | Sensitivity | 99% CI | Specificity | 99% CI |
| Acinetobacter baumannii | 1 | 0.87-1.0 | 1 | 0.87-1.0 |
| Escherichia coli | 1 | 0.87-1.0 | 1 | 0.87-1.0 |
| Enterococcus faecalis | 0.75 | 0.56-0.88 | 1 | 0.87-1.0 |
| Enterococcus faecium | 1 | 0.87-1.0 | 1 | 0.87-1.0 |
| Klebsiella pneumoniae | 1 | 0.87-1.0 | 1 | 0.87-1.0 |
| Pseudomonas aeruginosa | 1 | 0.87-1.0 | 1 | 0.87-1.0 |
| Staphylococcus aureus | 0.95 | 0.79-0.99 | 1 | 0.87-1.0 |
See Table 2 legend for expansion of abbreviations.
TABLE 4 ] .
Omnibus Statistical Measures of Agreement
| Statistic | Point Estimate | 99% CI |
| Targets, No. | 357 | N/A |
| Sensitivity | 0.96 | 0.92-0.98 |
| Specificity | 1.0 | 0.98-1.0 |
| Correlation, ϕ | 0.98 | 0.97-0.98 |
| Agreement | 0.96 | 0.81-1.0 |
| Concordance, κ | 0.95 | 0.87-1.0 |
N/A = not applicable.
Quantitative Agreement of RT-PCR Between EBCF and BALF
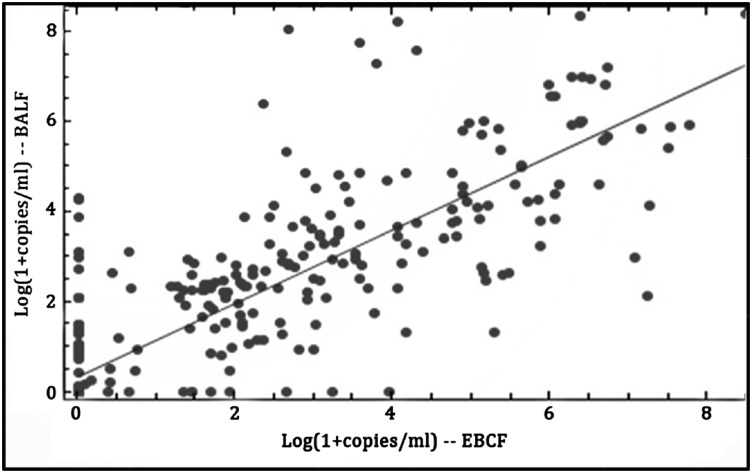
The quantitative correlation between each of the 51 matched EBCF and BALF samples was compared. Linear regression of the matched EEBCF/BALF pairs for each of the seven species-specific DNA targets and a universal 16S rDNA target is shown in Figure 3. A point estimate of the Pearson product moment correlation coefficient is 0.85 (99% CI, 0.81-0.88). While overall correlation is good, some samples demonstrated poor agreement, with low number of copies in one sample and high number in the other.
Figure 3 –
Linear regression of the bacterial target DNA/mL of patient sample for the 51 time-matched sample pairs EBCF vs BALF. The real-time polymerase chain reaction data from the seven specific targets and a universal 16s rDNA target, listed in Table 1, are shown. The Pearson product moment correlation is 0.85 (99% CI, 0.81-0.88), indicating a significant positive correlation. BALF = BAL fluid; EBCF = exhaled breath condensate fluid.
Analyses of EBCF and BALF for SP-B and DPPC were positive in all cases. Six consecutive EBCF samples from one patient provided an average of 310 ng/mL of SP-B and 261 ng/mL DPPC, with SDs of 428 ng/mL and 326 ng/mL, respectively. A matched BALF sample from the same patient contained comparable amounts (SP-B, 256 ng/mL; DPPC, 180 ng/mL).
Discussion
Tools to diagnosis VAP in a rapid, specific fashion would improve patient care quality. Studies in the literature support reduced morbidity and mortality when appropriate antibiotic therapy is initiated early in the course of VAP,20‐23 yet existing diagnostic approaches rely on symptoms that correspond with an advanced infectious process. In addition, traditional culture-based techniques require at least 48 h to establish or rule out infection, identify specific pathogens, and determine antibiotic sensitivity. These limitations contribute to poor antibiotic stewardship and substantially increased hospital costs.
Exhaled breath contains fluid particles carrying pathogens from the lower respiratory system32‐35 and prior study demonstrates that HCH/HME filters are reservoirs for pathogenic bacterial DNA.36 Our current study assesses the qualitative and quantitative correlation of DNA in EBCF with BALF and microbiologic cultures for suspected pneumonia. Results indicate that RT-PCR of EBCF correlates highly, both qualitatively and quantitatively, with RT-PCR and standard microbiologic cultures of BALF, with the point estimate of their concordance (κ) > 95%. However, Figure 3 demonstrates that some samples demonstrated a low number of DNA copies in one specimen type but high number in the other. The origin of this disagreement is unclear at present and further study and refinement is required to advance this application.
The qualitative data demonstrate substantial agreement of RT-PCR between EBCF and BALF and that PCR is more sensitive than standard clinical quantitative culture of BALF. In two cases, K pneumoniae was identified on clinical culture at levels lower than the threshold for VAP. Whether these are contaminates or represent a failure of the PCR analysis cannot be determined. However, in all cases, RT-PCR detected the pathogens involved in VAP established by quantitative BAL and could have been used to eliminate empirical coverage for either gram-positive or gram-negative pathogens in 28 patients, and eliminate empirical coverage in 16 patients without detectable pathogens. The quantitative agreement of RT-PCR of EBCF and BALF is also strong. However, translating the number of copies of DNA to CFU detected by clinical culture requires further research. In addition, a better understanding of those mismatches is paramount to advance this line of research.
Another potential application of this technique could be to demonstrate changes in the lung microbial community that occur over the period of mechanical ventilation and precede the development of VAP. Results from serial sample analyses of HMEs from a small number of patients who develop VAP suggest that derangements in lung microbiota may precede the clinical suspicion of pneumonia, and these changes are detectable by EBCF analysis (data not shown). However, this study was not designed to examine this application and further research is being conducted to examine this concept.
Although significant literature examines exhaled breath in a variety of settings,49‐52 no studies have examined serial EBCF collected from reservoirs such as HCH/HME filters, or used quantitative bacterial DNA analysis. While the knowledge that pathogenic microorganisms are carried in the exhaled breath of humans and animals is not novel, quantification by genetic analysis over time is.32,33,53,54 Most studies of exhaled breath examine volatile and nonvolatile compounds expressed over brief periods (seconds to minutes). Studies interrogating bacterial genetic material with PCR assays have either failed to test their system against the rigors of the ICU environment or continue to rely on invasive BAL techniques to acquire samples.55,56 Other exhaled breath studies use short, minute-scale durations, increasing variability, and false-negative results.57,58
Our study has several limitations. This study only examines HMEs from patients with clinically suspected VAP and thus does not provide any information regarding EBCF in patients without suspicion. Additionally, in two cases, clinical laboratory detected K pneumoniae at low levels (< 103 CFU/mL) not present in either the EBCF or BALF by PCR. Whether this indicates a lack of PCR sensitivity or contamination of the clinical specimen cannot be determined. Whether all pathogens are carried equally in exhaled breath condensate under all patient conditions cannot be determined by this study design. RT-PCR of EBCF and BALF detected low number of copies of pathogenic DNA in several instances of negative cultures, suggesting greater sensitivity. While RT-PCR of EBCF and BALF correlate well, translating this to quantitative microbiologic results requires additional studies.
In conclusion, RT-PCR of EBCF appears adequately sensitive and specific to study the lung microbiota in critically ill patients who are ventilated. These results introduce the potential opportunity to diagnose VAP noninvasively in hours, rather than days, and may allow for early, targeted antibiotic therapy and the elimination of empirical therapy when PCR is negative. While these findings are preliminary and require additional investigation, they do provide substantial rationale for continued research in this area. Future application of this novel noninvasive approach to define normal and aberrant lung microbes in patients who are ventilated may ultimately improve the expediency and specificity of VAP diagnosis.
Acknowledgments
Author contributions: A. K. M. had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the analysis. A. K. M., P. R. N., J. M. J., R. J. I., and E. M. B. contributed substantially to study design; E. M. B. contributed to sample acquisition and analysis; A. K. M., P. R. N., J. M. J., R. J. I., and E. M. B. contributed to data analysis and interpretation; J. S. B., J. R.-K., and W. P. D. contributed to data interpretation; and A. K. M., J. S. B., J. R.-K., W. P. D., P. R. N., J. M. J., R. J. I., and E. M. B contributed to writing the manuscript.
Financial/nonfinancial disclosures: The authors have reported to CHEST that no potential conflicts of interest exist with any companies/organizations whose products or services may be discussed in this article.
Role of sponsors: Funding support was used for supplies and associated cost of PCR analysis.
ABBREVIATIONS
- BALF
BAL fluid
- CFU
colony-forming unit
- DPPC
dipalmitoylphosphatidylcholine
- EBCF
exhaled breath condensate fluid
- HCH/HME
hygroscopic condenser humidifier/heat and moisture exchanger
- PCR
polymerase chain reaction
- RT-PCR
real-time polymerase chain reaction
- SICU
surgical ICU
- SP-B
surfactant-associated protein B
- VAP
ventilator-associated pneumonia
- VUMC
Vanderbilt University Medical Center
Footnotes
FOR EDITORIAL COMMENT SEE PAGE 1448
FUNDING/SUPPORT: This study was partially supported by the Vanderbilt Institute for Clinical and Translational Research [Grant CTSA 1 UL1 RR024975].
Reproduction of this article is prohibited without written permission from the American College of Chest Physicians. See online for more details.
References
- 1.Minei JP, Nathens AB, West M, et al. ; Inflammation and the Host Response to Injury Large Scale Collaborative Research Program Investigators. Inflammation and the host response to injury, a large-scale collaborative project: patient-oriented research core—standard operating procedures for clinical care. II. Guidelines for prevention, diagnosis and treatment of ventilator-associated pneumonia (VAP) in the trauma patient. J Trauma. 2006;60(5):1106-1113. [DOI] [PubMed] [Google Scholar]
- 2.Rello J, Ollendorf DA, Oster G, et al. ; VAP Outcomes Scientific Advisory Group. Epidemiology and outcomes of ventilator-associated pneumonia in a large US database. Chest. 2002;122(6):2115-2121. [DOI] [PubMed] [Google Scholar]
- 3.Safdar N, Dezfulian C, Collard HR, Saint S. Clinical and economic consequences of ventilator-associated pneumonia: a systematic review. Crit Care Med. 2005;33(10):2184-2193. [DOI] [PubMed] [Google Scholar]
- 4.Cavalcanti M, Ferrer M, Ferrer R, Morforte R, Garnacho A, Torres A. Risk and prognostic factors of ventilator-associated pneumonia in trauma patients. Crit Care Med. 2006;34(4):1067-1072. [DOI] [PubMed] [Google Scholar]
- 5.Cook DJ, Kollef MH. Risk factors for ICU-acquired pneumonia. JAMA. 1998;279(20):1605-1606. [DOI] [PubMed] [Google Scholar]
- 6.Croce MA, Tolley EA, Fabian TC. A formula for prediction of posttraumatic pneumonia based on early anatomic and physiologic parameters. J Trauma. 2003;54(4):724-729. [DOI] [PubMed] [Google Scholar]
- 7.Evans HL, Warner K, Bulger EM, Sharar SR, Maier RV, Cuschieri J. Pre-hospital intubation factors and pneumonia in trauma patients. Surg Infect (Larchmt). 2011;12(5):339-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lockey DJ, Coats T, Parr MJ. Aspiration in severe trauma: a prospective study. Anaesthesia. 1999;54(11):1097-1098. [DOI] [PubMed] [Google Scholar]
- 9.Rea-Neto A, Youssef NC, Tuche F, et al. Diagnosis of ventilator-associated pneumonia: a systematic review of the literature. Crit Care. 2008;12(2):R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croce MA, Fabian TC, Schurr MJ, et al. Using bronchoalveolar lavage to distinguish nosocomial pneumonia from systemic inflammatory response syndrome: a prospective analysis. J Trauma. 1995;39(6):1134-1139. [DOI] [PubMed] [Google Scholar]
- 11.Fagon JY, Chastre J, Wolff M, et al. Invasive and noninvasive strategies for management of suspected ventilator-associated pneumonia. A randomized trial. Ann Intern Med. 2000;132(8):621-630. [DOI] [PubMed] [Google Scholar]
- 12.Meduri GU, Mauldin GL, Wunderink RG, et al. Causes of fever and pulmonary densities in patients with clinical manifestations of ventilator-associated pneumonia. Chest. 1994;106(1):221-235. [DOI] [PubMed] [Google Scholar]
- 13.Michel F, Franceschini B, Berger P, et al. Early antibiotic treatment for BAL-confirmed ventilator-associated pneumonia: a role for routine endotracheal aspirate cultures. Chest. 2005;127(2):589-597. [DOI] [PubMed] [Google Scholar]
- 14.Croce MA, Swanson JM, Magnotti LJ, et al. The futility of the clinical pulmonary infection score in trauma patients. J Trauma. 2006;60(3):523-527. [DOI] [PubMed] [Google Scholar]
- 15.Fartoukh M, Maitre B, Honoré S, Cerf C, Zahar JR, Brun-Buisson C. Diagnosing pneumonia during mechanical ventilation: the clinical pulmonary infection score revisited. Am J Respir Crit Care Med. 2003;168(2):173-179. [DOI] [PubMed] [Google Scholar]
- 16.Lauzier F, Ruest A, Cook D, et al. ; Canadian Critical Care Trials Group. The value of pretest probability and modified clinical pulmonary infection score to diagnose ventilator-associated pneumonia. J Crit Care. 2008;23(1):50-57. [DOI] [PubMed] [Google Scholar]
- 17.Luyt CE, Chastre J, Fagon JY. Value of the clinical pulmonary infection score for the identification and management of ventilator-associated pneumonia. Intensive Care Med. 2004;30(5):844-852. [DOI] [PubMed] [Google Scholar]
- 18.Schurink CA, Van Nieuwenhoven CA, Jacobs JA, et al. Clinical pulmonary infection score for ventilator-associated pneumonia: accuracy and inter-observer variability. Intensive Care Med. 2004;30(2):217-224. [DOI] [PubMed] [Google Scholar]
- 19.Minshall CT, Eriksson EA, Hawkins KS, Wolf S, Minei JP. Early nonbronchoscopic bronchoalveolar lavage: predictor of ventilator-associated pneumonia? J Trauma Acute Care Surg. 2013;74(2):448-452. [DOI] [PubMed] [Google Scholar]
- 20.Alvarez-Lerma F; ICU-Acquired Pneumonia Study Group. Modification of empiric antibiotic treatment in patients with pneumonia acquired in the intensive care unit. Intensive Care Med. 1996;22(5):387-394. [DOI] [PubMed] [Google Scholar]
- 21.Kollef MH. Inadequate antimicrobial treatment: an important determinant of outcome for hospitalized patients. Clin Infect Dis. 2000;31(suppl 4):S131-S138. [DOI] [PubMed] [Google Scholar]
- 22.Luna CM, Vujacich P, Niederman MS, et al. Impact of BAL data on the therapy and outcome of ventilator-associated pneumonia. Chest. 1997;111(3):676-685. [DOI] [PubMed] [Google Scholar]
- 23.Rello J, Gallego M, Mariscal D, Soñora R, Valles J. The value of routine microbial investigation in ventilator-associated pneumonia. Am J Respir Crit Care Med. 1997;156(1):196-200. [DOI] [PubMed] [Google Scholar]
- 24.Barie PS, Hydo LJ, Shou J, Larone DH, Eachempati SR. Influence of antibiotic therapy on mortality of critical surgical illness caused or complicated by infection. Surg Infect (Larchmt). 2005;6(1):41-54. [DOI] [PubMed] [Google Scholar]
- 25.Fabian TC, Croce MA, Payne LW, Minard G, Pritchard FE, Kudsk KA. Duration of antibiotic therapy for penetrating abdominal trauma: a prospective trial. Surgery. 1992;112(4):788-794. [PubMed] [Google Scholar]
- 26.Harbarth S, Samore MH, Lichtenberg D, Carmeli Y. Prolonged antibiotic prophylaxis after cardiovascular surgery and its effect on surgical site infections and antimicrobial resistance. Circulation. 2000;101(25):2916-2921. [DOI] [PubMed] [Google Scholar]
- 27.Kollef MH. Ventilator-associated pneumonia. A multivariate analysis. JAMA. 1993;270(16):1965-1970. [PubMed] [Google Scholar]
- 28.May AK, Fleming SB, Carpenter RO, et al. Influence of broad-spectrum antibiotic prophylaxis on intracranial pressure monitor infections and subsequent infectious complications in head-injured patients. Surg Infect (Larchmt). 2006;7(5):409-417. [DOI] [PubMed] [Google Scholar]
- 29.Namias N, Harvill S, Ball S, McKenney MG, Salomone JP, Civetta JM. Cost and morbidity associated with antibiotic prophylaxis in the ICU. J Am Coll Surg. 1999;188(3):225-230. [DOI] [PubMed] [Google Scholar]
- 30.Trouillet JL, Chastre J, Vuagnat A, et al. Ventilator-associated pneumonia caused by potentially drug-resistant bacteria. Am J Respir Crit Care Med. 1998;157(2):531-539. [DOI] [PubMed] [Google Scholar]
- 31.Velmahos GC, Toutouzas KG, Sarkisyan G, et al. Severe trauma is not an excuse for prolonged antibiotic prophylaxis. Arch Surg. 2002;137(5):537-541. [DOI] [PubMed] [Google Scholar]
- 32.Eames I, Tang JW, Li Y, Wilson P. Airborne transmission of disease in hospitals. J R Soc Interface. 2009;6(suppl 6):S697-S702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie X, Li Y, Sun H, Liu L. Exhaled droplets due to talking and coughing. J R Soc Interface. 2009;6(suppl 6):S703-S714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edmiston CE, Jr, Sinski S, Seabrook GR, et al. Airborne particulates in the OR environment. AORN J. 1999;69(6):1169-1172. [DOI] [PubMed] [Google Scholar]
- 35.Edmiston CE, Jr, Seabrook GR, Cambria RA, et al. Molecular epidemiology of microbial contamination in the operating room environment: Is there a risk for infection? Surgery. 2005;138(4):573-579. [DOI] [PubMed] [Google Scholar]
- 36.Isaacs RJ, Debelak K, Norris PR, et al. Non-invasive detection of pulmonary pathogens in ventilator-circuit filters by PCR. Am J Transl Res. 2012;4(1):72-82. [PMC free article] [PubMed] [Google Scholar]
- 37.Carroll KC, Glanz BD, Borek AP, et al. Evaluation of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of Enterobacteriaceae. J Clin Microbiol. 2006;44(10):3506-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148(pt 1):257-266. [DOI] [PubMed] [Google Scholar]
- 39.Cherkaoui A, Emonet S, Ceroni D, et al. Development and validation of a modified broad-range 16S rDNA PCR for diagnostic purposes in clinical microbiology. J Microbiol Methods. 2009;79(2):227-231. [DOI] [PubMed] [Google Scholar]
- 40.Gu F, Li Y, Zhou C, et al. Bacterial 16S rRNA/rDNA profiling in the liquid phase of human saliva. Open Dent J. 2009;3:80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horz HP, Vianna ME, Gomes BP, Conrads G. Evaluation of universal probes and primer sets for assessing total bacterial load in clinical samples: general implications and practical use in endodontic antimicrobial therapy. J Clin Microbiol. 2005;43(10):5332-5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46(7):2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siala M, Gdoura R, Fourati H, et al. Broad-range PCR, cloning and sequencing of the full 16S rRNA gene for detection of bacterial DNA in synovial fluid samples of Tunisian patients with reactive and undifferentiated arthritis. Arthritis Res Ther. 2009;11(4):R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stowers CC, Haselton FR, Boczko EM. An analysis of quantitative PCR reliability through replicates using the C method. J Biomed Sci Eng. 2010;3(5):459-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krämer HJ, Schmidt R, Günther A, Becker G, Suzuki Y, Seeger W. ELISA technique for quantification of surfactant protein B (SP-B) in bronchoalveolar lavage fluid. Am J Respir Crit Care Med. 1995;152(5 pt 1):1540-1544. [DOI] [PubMed] [Google Scholar]
- 46.Ivanova PT, Milne SB, Byrne MO, Xiang Y, Brown HA. Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:21-57. [DOI] [PubMed] [Google Scholar]
- 47.Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998;17(8):857-872. [DOI] [PubMed] [Google Scholar]
- 48.Hanley JA. Standard error of the Kappa statistic. Psychol Bull. 1987;102(2):315-321. [Google Scholar]
- 49.Bodini A, D’Orazio C, Peroni D, et al. Biomarkers of neutrophilic inflammation in exhaled air of cystic fibrosis children with bacterial airway infections. Pediatr Pulmonol. 2005;40(6):494-499. [DOI] [PubMed] [Google Scholar]
- 50.Effros RM, Dunning MB, III, Biller J, Shaker R. The promise and perils of exhaled breath condensates. Am J Physiol Lung Cell Mol Physiol. 2004;287(6):L1073-L1080. [DOI] [PubMed] [Google Scholar]
- 51.Hunt J. Exhaled breath condensate: an overview. Immunol Allergy Clin North Am. 2007;27(4):587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson AS, Sandrini A, Campbell C, Chow S, Thomas PS, Yates DH. Comparison of biomarkers in exhaled breath condensate and bronchoalveolar lavage. Am J Respir Crit Care Med. 2007;175(3):222-227. [DOI] [PubMed] [Google Scholar]
- 53.Fabian P, McDevitt JJ, DeHaan WH, et al. Influenza virus in human exhaled breath: an observational study. PLoS One. 2008;3(7):e2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muscatello G, Gilkerson JR, Browning GF. Detection of virulent Rhodococcus equi in exhaled air samples from naturally infected foals. J Clin Microbiol. 2009;47(3):734-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bahrani-Mougeot FK, Paster BJ, Coleman S, et al. Molecular analysis of oral and respiratory bacterial species associated with ventilator-associated pneumonia. J Clin Microbiol. 2007;45(5):1588-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rice LM, Reis AH, Jr, Mistry R, et al. Design and construction of a single tube, quantitative endpoint, LATE-PCR multiplex assay for ventilator-associated pneumonia. J Appl Microbiol. 2013;115(3):818-827. [DOI] [PubMed] [Google Scholar]
- 57.Bos LD, Wang Y, Weda H, et al. A simple breath sampling method in intubated and mechanically ventilated critically ill patients. Respir Physiol Neurobiol. 2014;191:67-74. [DOI] [PubMed] [Google Scholar]
- 58.Jünger M, Vautz W, Kuhns M, et al. Ion mobility spectrometry for microbial volatile organic compounds: a new identification tool for human pathogenic bacteria. Appl Microbiol Biotechnol. 2012;93(6):2603-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]