Abstract
Background
Mutations in CDH23 are responsible for Usher syndrome 1D and recessive non-syndromic hearing loss. In this study, we revealed the prevalence of CDH23 mutations among patients with specific clinical characteristics.
Methods
After excluding patients with GJB2 mutations and mitochondrial m.1555A > G and m.3243A > G mutations, subjects for CDH23 mutation analysis were selected according to the following criteria: 1) Sporadic or recessively inherited hearing loss 2) bilateral non-syndromic congenital hearing loss, 3) no cochlear malformation, 4) a poorer hearing level at high frequencies than at low frequencies, and 5) severe or profound hearing loss at higher frequencies.
Results
Seventy-two subjects were selected from 621 consecutive probands who did not have environmental causes for their hearing loss. After direct sequencing, 13 of the 72 probands (18.1%) had homozygous or compound heterozygous CDH23 mutations. In total, we identified 16 CDH23 mutations, including five novel mutations. The 16 mutations included 12 missense, two frameshift, and two splice-site mutations.
Conclusions
These results revealed that CDH23 mutations are highly prevalent in patients with congenital high-frequency sporadic or recessively inherited hearing loss and that the mutation spectrum was diverse, indicating that patients with these clinical features merit genetic analysis.
Electronic supplementary material
The online version of this article (doi:10.1186/s13023-015-0276-z) contains supplementary material, which is available to authorized users.
Keywords: CDH23, Congenital hearing loss, DFNB12, Gene mutation, Phenotype
Background
CDH23, located on chromosome 10, contains 70 exons and encodes a predicted 3354 amino acid protein. The untranslated region is followed by 27 extracellular cadherin repeat domains (exons 2–64), a single-pass transmembrane domain (exon 65), and a cytoplasmic domain (exons 66–70). Among the 113 human cadherin superfamily members, CDH23 is one of 11 that are solely responsible for hereditary diseases [1]. CDH23 is expressed in the outer and inner hair cells. The stereociliary expression of CDH23 in inner ear hair cells exhibits two patterns. One is transient expression during the development of hair bundles with procadherin 15 (PCDH15) and CDH23 forming side links and kinociliary links at their N-termini [2]. The second pattern is stable expression in tip links at the top of stereocilia from the developmental stage onwards until ear maturation. CDH23 homodimers interact in trans with PCDH15 homodimers to form filaments, and they play a key role in the mechanoelectrical transduction channel at the top of the lower stereocilia [3-5].
Mutations in CDH23 are responsible for both Usher syndrome 1D (USH1D: OMIM #601067) [6-9] and autosomal recessive non-syndromic hearing loss (DFNB12: OMIM #601386) [10-12]. Individuals with USH1D usually carry a truncated CDH23 protein because of nonsense, frameshift, or splice-site mutations, whereas those with DFNB12 usually carry missense mutations in any domain [7,10,13]. The mutation spectrums differ regionally, with the Japanese population having a unique mutation spectrum [12,14]. Recently, it was reported that 3.7% of recessive inherited hearing loss cases in the Japanese population involve homozygous or heterozygous CDH23 mutations [12].
In this study, we analyzed CDH23 in a Japanese population that was selected on the basis of DFNB12 and USH1D clinical features [12,14-16] after excluding patients with GJB2 mutations and mitochondrial m.1555A > G and m.3243A > G mutations. We identified an extremely high prevalence of CDH23 mutations in this population, and our findings expanded the CDH23 mutation spectrum, including five novel mutations.
Methods
All participating subjects, or their parents if the subjects were less than 20 years old, provided informed consent. The project was approved by the institutional review board of each institute. The patients were collected from the 11 institutes listed below: National Tokyo Medical Center, Chiba Children’s Hospital, National Mie Hospital, National Center for Child Health and Development, Hyogo Prefectural Kobe Children’s Hospital, Keio University School of Medicine, National Hospital Organization Kanazawa Medical Center, Kanagawa Children’s Medical Center, National Hospital Organization Sendai Medical Center, Kanto Rosai Hospital, and Hiroshima Prefectural Hospital. All investigations were conducted according to the principles expressed in the Declaration of Helsinki.
For the analysis of CDH23 mutations, we selected subjects who did not have hearing loss owing to environmental causes and who had participated in a genetic study at our institutes, with each subject meeting the following criteria: 1) sporadic or recessively inherited hearing loss; 2) bilateral non-syndromic congenital hearing loss; 3) a lack of cochlear malformations among those who underwent a computed tomography scan and/or magnetic resonance imaging; 4) a poorer hearing level at high frequencies (e.g., 2 kHz, 4 kHz) than at low frequencies (e.g., 250 kHz, 500 Hz); and 5) severe or profound hearing loss over 70 dB at frequencies exceeding 2 kHz. These clinical features were based on the reported phenotypes of DFNB12 and USH1D [12,14-16]. Before patient selection for CDH23 mutation analysis, genetic analyses for GJB2 and mitochondrial m.1555A > G and m.3243A > G mutations were conducted in all patients, according to published methods [17,18]. Among 621 probands who participated in a genetic study at our institutes from November 2008 to June 2012 (the first subject group), 72 subjects were selected for CDH23 mutation analysis based on these criteria.
For CDH23 analysis, DNA was extracted from peripheral blood using standard procedures. We PCR-amplified all CDH23 exons using the primers listed in Additional file 1: Table S1. The PCR products were purified and subjected to sequencing using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, CA, USA). SeqScape 2.6 software (Applied Biosystems) was used to analyze data against an NCBI human primary reference sequence (GRCh37.p13). Before SeqScape 2.6 analysis, all known CDH23 mutations within 69 coding regions and flanking 10-bp sequences of transcript variant 1 archived in the BIOBASE HGMD database (last visited on October 2014; BIOBASE, MA, USA) were entered into the SeqScape project files to enable detection of novel variants. Descriptions of DNA and amino acid positions in the literature [6-10,12,14,19] and the HGMD database were reassigned through original test programs to present the information in a consistent format. Prediction of the effect of missense variants on CDH23 protein function was performed using Polymorphism Phenotyping [PolyPhen2 [20] and Protein Variation Effect Analyzer (PROVEAN v1.1.3.) [21].
Frameshift (splice site within ± 2) and nonsense mutations were categorized as pathogenic. The pathogenicity of splice site alterations larger than ± 2 was predicted by NNSPLICE 0.9 version [22]. Novel missense variants were defined as pathogenic if they were 1) non-synonymous; 2) exhibited a low carrier rate (<1%) in 96 normal in-house control Japanese subjects and in public databases [(dbSNP135 [23], 1000GENOME [24], and NHLBI Exome Variant Server [25]]; 3) exhibited high amino acid conservation among 12 primate, 45 mammal, and 43 vertebrate species using UCSC Conservation [26]; 4) demonstrated consistency with phenotypes in family members; and 5) were detected in patients with hearing loss identified as heterozygous in association with another previously reported or determined heterozygous mutation. Variants who failed to meet criteria 1–4 were defined as non-pathogenic. Those who met criteria 1–4 but failed to fulfill criterion 5 were considered to have uncertain pathogenicity. For the purpose of this study, frameshift, splice site, and nonsense mutations were categorized as “truncating” mutations, and missense and in-frame insertion or deletion mutations were considered “non-truncating” mutations.
In addition, we further examined CDH23 mutations retrospectively in subjects who participated in a genetic study at our institutes from April 2001 to October 2008 (the second subject group). For this analysis, we selected subjects who had obvious progressive hearing loss as determined by repeated audiometry, profound hearing loss over 80 dB at frequencies above 2 kHz, as well as fulfilling the five aforementioned criteria for the first subject group. For these subjects, we additionally performed Sanger sequencing for CDH23 mutations.
Results
In the first subject group, we analyzed a total of 621 probands, and 492 underwent clinical selection for CDH23 gene testing after excluding individuals with GJB2 mutations and mitochondrial deafness. Figure 1 presents the study procedure together with the selection process for the primary screening. Using this procedure, 72 subjects were selected for CDH23 mutation analysis (14.6% of the selected individuals). Homozygous or compound heterozygous CDH23 mutations were identified in 13 subjects (18.1% of tested subjects), and nine subjects had heterozygous mutations (12.5% of tested subjects).
Figure 1.
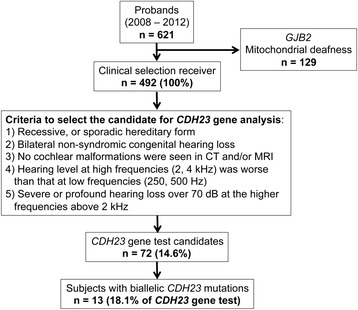
Flow chart of the selection and exclusion processes and the number of subjects at each point.
In the second subject group, 546 probands participated in the genetic study. After retrospective selection using the more stringent criteria, 15 subjects were selected. After direct sequencing for CDH23 mutations, three subjects were identified with homozygous or compound heterozygous pathogenic CDH23 mutations (20.0% of tested subjects), and three subjects were shown to have heterozygous mutations (20.0% of tested subjects).
A total of 16 pathogenic CDH23 mutations were found in 56 subjects from 28 families. Homozygous or compound heterozygous pathogenic mutations were found in 21 subjects, which included 16 probands. Heterozygous mutations were found in 12 probands. CDH23 mutations included 11 previously reported mutations [p.P240L, p.P346S, p.E956K, p.D1626A, p.Q1716P, p.R2029W, p.R1417W, p.E2438K, p.L2473P, p.R2489H, and c.6712 + 1G > A [7,12,14,27], as well as four novel mutations (Table 1). Twelve of the mutations were missense, one was a frameshift and three were splice-site mutations. Fifteen variants were considered to have uncertain pathogenicity (Table 1). These include five variants previously reported as uncertain [12,14,28,29], as well as 10 novel variants. We also found 49 non-pathogenic CDH23 variants, including 12 new variants (Additional file 2: Table S2). All the variants reported in this study were registered in the LOVD-CDH23 database [30].
Table 1.
Possible pathologic and uncertain variants found in this study
| Number in probands (in 346 allele) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Types of variants | Location | Amino acid change | Nucleotide change | Genomic position(Chr10) | Domain | Evolutionary conservation* | Homozygote | Compound heterozygote | Heterozygote | Allele frequency in normal controls | Allele frequency in ESP6500 | dbSNP135 | PolyPhen2 score | PROVEAN score | Novel or Known | Reference |
| Missense | Exon 7 | p.P240L | c.719C > T | 73330641 | EC3 | 11/11, 44/44, 40/40 | 4 | 7 | 6 | 0/192 | 0 | rs121908354 | 0.999 | −3.051 | Known | [12] |
| Exon 11 | p.P346S | c.1036C > T | 73377052 | - | 12/12, 42/42, 41/41 | 0 | 1 | 0 | 0/192 | 0 | None | 0.989 | −5.768 | Known | [27] | |
| Exon 16 | p.G539D | c.1616G > A | 73437314 | EC5 | 12/12, 43/43, 42/42 | 0 | 1 | 0 | 0/190 | 0 | None | 0.092 | −4.272 | Novel | ||
| Exon 18 | p.D645G | c.1934A > G | 73442277 | EC6 | 12/12, 45/45, 42/42 | 0 | 1 | 0 | 0/192 | 0 | None | 1.000 | −5.596 | Novel | ||
| Exon 25 | p.E956K | c.2866G > A | 73464800 | EC9 | 12/12, 45/45, 42/42 | 0 | 4 | 0 | 0/192 | 0 | None | 0.999 | −2.939 | Known | [14] | |
| Exon 35 | p.R1417W | c.4249C > T | 73498294 | EC13 | 12/12, 32/44, 0/40 | 0 | 0 | 1 | 0/192 | 0 | None | 0.453 | −1.346 | Known | [12] | |
| Exon 39 | p.D1626A | c.4877A > C | 73537468 | EC15 | 12/12, 45/45, 41/42 | 0 | 1 | 0 | 0/192 | 0 | None | 0.952 | −5.3 | Known | [14] | |
| Exon 39 | p.Q1716P | c.5147A > C | 73538025 | EC16 | 12/12, 45/45, 40/41 | 0 | 2 | 0 | 0/192 | 0 | None | 0.972 | −2.68 | Known | [12] | |
| Exon 46 | p.R2029W | c.6085C > T | 73550924 | EC19 | 12/12, 45/45, 39/40 | 0 | 2 | 2 | 0/192 | 0 | None | 0.999 | −4.659 | Known | [12] | |
| Exon 52 | p.E2438K | c.7312G > A | 73559336 | EC23 | 12/12, 43/44, 15/41 | 0 | 2 | 0 | 0/192 | 0 | None | 0.314 | −0.383 | Known | [14] | |
| Exon 53 | p.L2473P | c.7418 T > C | 73560448 | EC23 | 11/11, 45/45, 36/36 | 0 | 0 | 1 | 0/192 | 0 | None | 0.998 | −5.603 | Known | [12] | |
| Exon 53 | p.R2489H | c.7466G > A | 73560496 | EC23 | 12/12, 45/45, 36/36 | 0 | 0 | 1 | 0/192 | 0 | rs141986620 | 0.459 | −1.219 | Known | [12] | |
| Frameshift | Exon 48 | p.L2223Wfs | c.6667del C | 73553352 | - | - | 0 | 1 | 0 | 0/192 | 0 | None | - | - | Novel | |
| Exon 63 | p.N3044Tfs | c.9129 del G | 73571123 | - | - | 0 | 1 | 0 | 0/190 | 0 | None | - | - | Novel | ||
| Putative spice site | IVS11 | - | c.1135-1G > T | 73403617 | - | 12/12, 41/41, 4/4 | 0 | 0 | 1 | 0/192 | 0 | None | - | - | Novel | |
| IVS48 | - | c.6712 + 1G > A | 73553398 | - | 712/12, 44/44, 41/41 | 0 | 1 | 0 | 0/192 | 0 | None | - | - | Known | [11] | |
| Uncertain pathogenicity variants | ||||||||||||||||
| Missense | Exon 13 | p.V424M | c.1270G > A | 73405717 | EC4 | 12/12, 44/45, 0/36 | 0 | 0 | 1 | 0/190 | 0 | rs2305207 | 0.557 | −0.572 | Novel | |
| Exon 13 | p.D428N | c.1282G > A | 73405729 | EC4 | 12/12, 39/45, 0/36 | 0 | 0 | 4** | 0/190 | 0 | rs188376296 | 0.103 | −0.045 | Novel | ||
| Exon 35 | p.I1406V | c.4216A > G | 73498261 | EC13 | 12/12, 43/44, 40/40. | 0 | 0 | 1 | 1/189 | 0 | rs192459984 | 0.029 | −0.304 | Novel | ||
| Exon 38 | p.G1583S | c.4747G > A | 73501580 | EC15 | 12/12, 44/44, 41/43 | 0 | 0 | 1 | 0/192 | 0 | None | 1.000 | −4.967 | Novel | ||
| Exon 40 | p.V1711I | c.5131G > A | 73538009 | EC16 | 12/12, 29/45, 11/41 | 0 | 0 | 1 | 1/192 | 1/12645 | rs181611778 | 0.998 | −0.599 | Known | [12] | |
| Exon 42 | p.G1799W | c.5395G > T | 73544070 | EC17 | 11/11, 45/45, 42/43 | 0 | 0 | 1** | 0/192 | 0 | None | 0.998 | −6.237 | Novel | ||
| Exon 44 | p.V1908I | c.5722G > A | 73545397 | EC9 | 11/12, 28/45, 33/41 | 0 | 0 | 1 | 0/192 | 0 | None | 0.005 | 0.125 | Known | [14] | |
| Exon 48 | p.D2202E | c.6606C > A | 73553291 | EC21 | 12/12, 45/45, 41/42 | 0 | 0 | 1** | 0/190 | 0 | None | 1.000 | −3.068 | Known | [28] | |
| Exon 48 | p.Q2227P | c.6680A > C | 73553365 | EC21 | 12/12, 44/44, 23/41 | 0 | 0 | 1 | 0/192 | 0 | None | 0.729 | −0.775 | Known | [29] | |
| Exon 57 | p.D2717N | c.8149G > A | 73566009 | EC25 | 12/12, 45/45, 42/42 | 0 | 0 | 1** | 0/190 | 0 | None | 0.999 | −1.909 | Novel | ||
| Exon 61 | p.G2912S | c.8734G > A | 73569588 | EC27 | 11/11, 45/45, 42/42 | 0 | 0 | 1 | 0/192 | 0 | None | 0.774 | −2.221 | Known | [14] | |
| Exon 70 | p.V3343M | c.10027G > A | 73574997 | - | 12/12, 45/45, 16/37 | 0 | 0 | 1** | 0/188 | 0 | None | 0.670 | −0.427 | Novel | ||
| Putative splice site | IVS2 | - | c.68-3C > T | 73206072 | - | 12/12, 40/44, 18/19 | 0 | 0 | 1 | 0/188 | 0 | rs142456469 | - | - | Novel | |
| IVS7 | - | c.625-5C > T | 73330542 | - | 11/12, 41/44, 0/40 | 0 | 0 | 1 | 0/192 | 0 | None | - | - | Novel | ||
| IVS52 | - | c.7362 + 10G > A | 73559396 | - | 12/12, 11/42, 5/20 | 0 | 0 | 1 | 0/192 | 0 | None | - | - | Novel | ||
*Evolutionary conservation showed up to 12 primates, 45 mammals, and 43 vertebrates, respectively.
**Combination of uncertain pathogenicity variants were counted as heterozygotes, not as compound heterozygotes.
Overlapping audiograms of all 21 subjects with biallelic CDH23 mutations are shown in Figure 2. The ages of these patients ranged from 0 to 25 years (median: 5 years). The average hearing level was 90.1 ± 12.3 (SD) dB. The average hearing level at 250 and 500 Hz was 70.5 ± 22.7 dB, whereas that at 2000 and 4000 Hz was 101.9 ± 7.8 dB. In addition, 10 of 21 subjects displayed obvious progressive hearing loss, as assessed by comparing audiograms obtained at younger ages. At the time of this study, no subjects were diagnosed with Usher syndrome, which, in addition to hearing loss, is associated with vestibular dysfunction and visual impairment due to retinitis pigmentosa. Information for vision, fundoscopy, and motor milestones in 21 subjects with biallelic CDH23 mutations is shown in Table 2.
Figure 2.
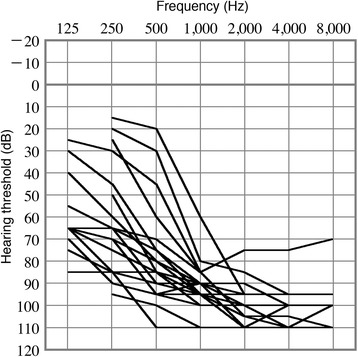
Overlapping audiograms of subjects with hearing loss caused by CDH23 mutations. Overlapping audiograms of the better-hearing ear of 20 subjects with homozygous or compound heterozygous CDH23 mutations.
Table 2.
Clinical and genetic data in the patients with biallelic CDH23 mutations
| Case ID* 1 | Age at the latestexamination | Genotypes* 2 | Vision | Fundoscopy | Motor milestones* 3 |
|---|---|---|---|---|---|
| 1 | 25y11m | p.P240L / p.P240L | Normal | Not done | Normal |
| 2 | 25y7m | p.P240L / p.P240L | Normal | Not done | Normal |
| 3 | 25y2m | p.P240L / p.P240L | Normal | Not done | Normal |
| 4 | 24y2m | p.P240L / p.P240L | Normal | Not done | Normal |
| 5 | 17y3m | p.P240L / p.R2029W | Normal | Normal | Normal |
| 6 | 14y1m | p.E2438K / p.N3044Tfs | Normal | Not done | Normal |
| 7 | 13y3m | p.P240L / p.R2029W | Myopia | Normal | Normal |
| 8 | 13y2m | p.D1626A / p.L2223Wfs | Normal | Not done | Normal |
| 9 | 13y1m | p.P240L / p.P240L | Normal | Not done | Normal |
| 10 | 11y8m | p.P240L / p.P240L | Normal | Not done | Normal |
| 11 | 11y4m | p.E2438K / p.N3044Tfs | Normal | Not done | Normal |
| 12 | 11y2m | p.D645G / p.E956K | Normal | Not done | Normal |
| 13 | 9y7m | p.P240L / p.E956K | Myopia | Normal | Normal |
| 14 | 9y2m | p.Q1716P / p.E2438K | Normal | Not done | Normal |
| 15 | 8y5m | p.P240L / p.E956K | Normal | Not done | Normal |
| 16 | 7y3m | p.P240L / p.E956K | Normal | Not done | Normal |
| 17 | 7y0m | p.P240L / p.P240L | Normal | Not done | Normal |
| 18 | 6y4m | p.P346S / p.Q1716P | Normal | Not done | Normal |
| 20 | 5y9m | p.P240L / c.6712 + 1G > A | Normal | Not done | Normal |
| 21 | 3y10m | p.P240L / c.6712 + 1G > A | Hyperopia | Normal | Normal |
*1Cases were shown in the order of their ages at the latest examination.
*2Genotypes written by bold word indicate truncating mutations.
*3Motor milestones: Head control < 4 months, Walking alone < 14 months.
The most frequent mutation in this study was p.P240L, which is consistent with previous reports of the Japanese population [12,14]. The overlapping audiograms illustrated that a high proportion of subjects homozygous for the p.P240L mutation had more severe low frequency hearing loss compared with subjects with non-p.P240L homozygous mutations, indicating a genotype-phenotype correlation (Figure 3).
Figure 3.
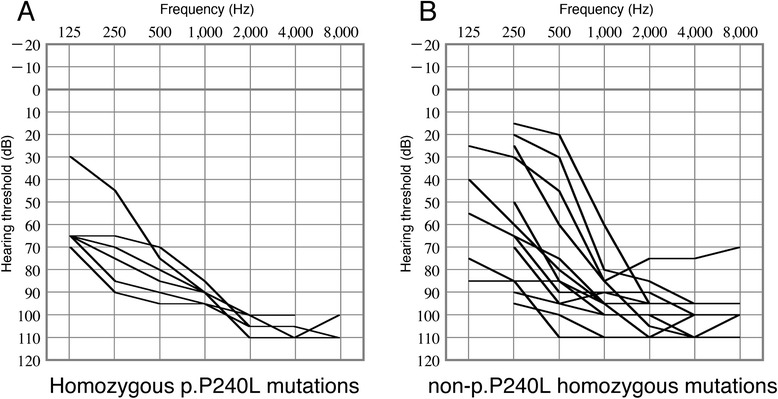
Comparison of hearing levels between individuals homozygous for p.P240L and those with other homozygous mutations. A: Overlapping audiograms of subjects with homozygous p.P240L mutations. B: Overlapping audiograms of the subjects with non-p.P240L homozygous mutations. The subjects with homozygous p.P240L mutations tended to have more severe hearing loss at lower frequencies.
Five of 21 subjects with biallelic CDH23 mutations had an allele with a truncating mutation, such as a frameshift or splice-site mutation, in association with a missense mutation on the other allele. The other 16 subjects had biallelic missense mutations. The overlapping audiograms of these subjects revealed no significant differences between the subjects with truncating mutations associated with missense mutations and those with biallelic missense mutations (Figure 4). The average hearing level of the subjects with an allele featuring truncating mutations associated with missense mutations was 90.3 ± 17.2 dB, whereas that of patients with biallelic missense mutations was 90.0 ± 10.2 dB. Because the number of subjects with truncating mutations was small, this result needs to be confirmed in a future study.
Figure 4.
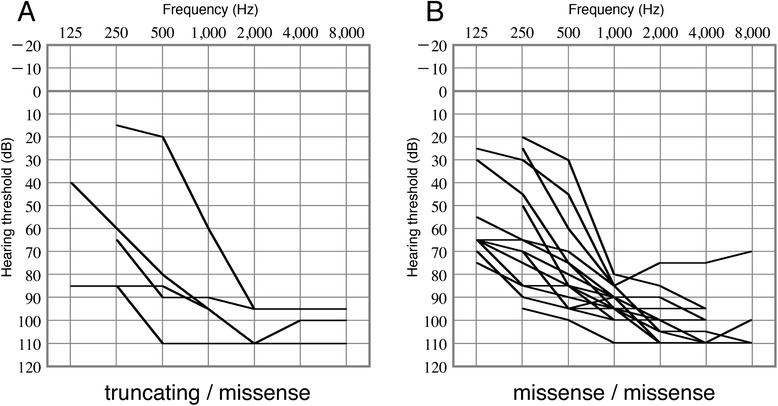
Comparison of hearing levels between patients with truncating and missense mutations and those with biallelic missense mutations. A: Overlapping audiogram of subjects with truncating and missense mutations. B: Overlapping audiograms of the subjects with biallelic missense mutations. The patterns of the audiograms were similar.
Discussion
The present results revealed an extremely high prevalence of CDH23 mutations among patients with congenital sporadic or recessively inherited high-frequency hearing loss. Among 72 probands presenting with our defined set of clinical features, 13 subjects (18.1%) exhibited homozygous or compound heterozygous CDH23 mutations. Previously, a large cohort study of a Japanese population (n = 1396) using two-step screening revealed that the frequency of biallelic CDH23 mutations was only 1.6% in patients with hearing loss [14]. The higher prevalence of CDH23 mutations in the present study can be explained by subject selection criteria; these criteria included common clinical features of patients with CDH23 mutations [12,14,16], in addition to excluding patients with GJB2 and mitochondrial m.1555A > G and m.3243A > G mutations. The present results indicate that CDH23 mutations should be considered as candidate causes of hearing loss when patients present with clinical features similar to the selection criteria used in this study, which are relatively common among individuals with early childhood hearing loss.
A strategy for clinical genetic diagnosis should consider sensitivity, specificity, and cost. The specificity of Sanger sequencing is known to be extremely high, but the cost of analyzing CDH23 by Sanger sequencing is also extremely high. The present study revealed that its sensitivity for CHD23 was 18.1% at best. Considering these data and recent advances in genetic technologies, Sanger sequencing of CDH23 is not likely to be the most practical clinical test currently available. Recently, we and others have developed genetic tests using next-generation sequencing (NGS) for non-syndromic hearing loss [31-34]. The NGS-based tests can examine almost all known deafness genes simultaneously at a cost similar to that for Sanger sequencing of CDH23 [35,36]. Currently, clinical genetic testing using NGS is available in a limited number of institutes, but these tests will become more widely available in the near future. Because the clinical features of CDH23 mutation, i.e., congenital sporadic or recessively inherited high-frequency hearing loss, are relatively common and associated with mutations in many deafness genes, genetic tests using NGS would be appropriate for patients presenting with these clinical features.
Progression is reported as an important clinical feature of hearing loss caused by CDH23 mutations [14,16]. In our study, progression of hearing level loss was only noted in 10 of 21 subjects who had biallelic mutations. In the 11 subjects who did not display obvious progressive hearing loss, most were less than 5 years old at the time of this study. In general, it is difficult to demonstrate the progression of hearing loss in early childhood; the observation times for younger patients are shorter and the evaluation is performed using different audiological tests depending on the developmental stages. The results, therefore, tend to be unstable compared with those for adults. We believe these difficulties in proving the progression of hearing loss in early childhood explain why the ratio of progressive hearing loss in the present study was lower than that reported previously [14,16], and the ratio of hearing loss progression may be higher in a follow-up study.
This study identified truncating mutations of CDH23 in combination with heterozygous missense alleles in a Japanese population of patients with non-syndromic deafness. In general, individuals with USH1D mostly carry a truncated CDH23 protein because of nonsense, frameshift, or splice-site mutations, whereas those with DFNB12 usually carry missense mutations [7,10,13]. However, USH1D caused by missense mutations has also been reported [37]. In addition, it has been reported that the DFNB12 allele is phenotypically dominant to an USH1D allele [29]. The onset of visual impairment caused by retinitis pigmentosa accompanied by USH1D generally occurs during adolescence [9]. In this study, the ages of patients with monoallelic truncating mutations were 3–14 years, and none of these patients had biallelic truncating mutations. Therefore, at present we cannot determine whether these patients have DNFB12 or USH1D, although all clinical data are compatible with DFNB12 but not with USH1D. However, the detection of a mild visual phenotype requires patients to be followed with ophthalmoscopy and at least one electroretinography. This will provide more precise genotype-phenotype correlations.
Conclusions
Our results revealed that CDH23 mutations are highly prevalent in patients with congenital high-frequency sporadic or recessively inherited hearing loss and that the mutation spectrum was diverse, indicating that patients with these clinical features would merit genetic analysis.
Acknowledgments
We thank Dr. Kaoru Ogawa at the Keio University School of Medicine, Dr. Tetsuya Takiguchi at the National Hospital Organization Kanazawa Medical Center, Dr. Noboru Ogahara at the Kanagawa Children’s Medical Center, Dr. Sho Hashimoto at the National Hospital Organization Sendai Medical Center, Dr. Tomoko Sugiuchi at the Kanto Rosai Hospital, and Dr. Shin Masuda at the Hiroshima Prefectural Hospital for assistance with sample and clinical data collection. We also thank Ms. Fumie Hiroyasu and Ms. Chie Naito for their technical support. This study was supported by a Network Research Grant for Disorders of Sensory Organs from the National Hospital Organization and Health Science Research Grants from the Ministry of Health, Labour, and Welfare of Japan (to T.M.) and MEXT KAKENHI (to K.M.; Grant number: 26462572) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Additional files
Sequencing primers used in this study.
Non-pathological variants found in this study.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KM, HM, and TM interpreted the data and drafted the manuscript. HM, KN, and YM carried out preparation and sequencing of DNA samples. KM and TM designed the study. AN, YA, SM, NM, HS, and KK contributed to the gathering and interpretation of clinical data. TM prepared the final draft of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Kunio Mizutari, Email: tari@mbf.ocn.ne.jp.
Hideki Mutai, Email: mutaihideki@kankakuki.go.jp.
Kazunori Namba, Email: nambakazunori@kankakuki.go.jp.
Yuko Miyanaga, Email: miyasama@lib.bekkoame.ne.jp.
Atsuko Nakano, Email: a.nkn@pref.chiba.lg.jp.
Yukiko Arimoto, Email: y.armt1@pref.chiba.lg.jp.
Sawako Masuda, Email: masudas@mie-m.hosp.go.jp.
Noriko Morimoto, Email: morimoto-n@ncchd.go.jp.
Hirokazu Sakamoto, Email: GED02037@nifty.com.
Kimitaka Kaga, Email: kaga@kankakuki.go.jp.
Tatsuo Matsunaga, Email: matsunagatatsuo@kankakuki.go.jp.
References
- 1.El-Amraoui A, Petit C. Usher I syndrome: unravelling the mechanisms that underlie the cohesion of the growing hair bundle in inner ear sensory cells. J Cell Sci. 2005;118:4593–603. doi: 10.1242/jcs.02636. [DOI] [PubMed] [Google Scholar]
- 2.Stover T, Diensthuber M. Molecular biology of hearing. GMS Curr Top Otorhinolaryngol Head Neck Surg. 2011;10:Doc06. doi: 10.3205/cto000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Amraoui A, Petit C. Cadherin defects in inherited human diseases. Prog Mol Biol Transl Sci. 2010;116:361–84. doi: 10.1016/B978-0-12-394311-8.00016-9. [DOI] [PubMed] [Google Scholar]
- 4.Hilgert N, Smith RJ, Van Camp G. Function and expression pattern of nonsyndromic deafness genes. Curr Mol Med. 2009;9:546–64. doi: 10.2174/156652409788488775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kazmierczak P, Sakaguchi H, Tokita J, Wilson-Kubalek EM, Milligan RA, Muller U, et al. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449:87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- 6.Bolz H, von Brederlow B, Ramirez A, Bryda EC, Kutsche K, Nothwang HG, et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet. 2001;27:108–12. doi: 10.1038/83667. [DOI] [PubMed] [Google Scholar]
- 7.Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, et al. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet. 2001;68:26–37. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oshima A, Jaijo T, Aller E, Millan JM, Carney C, Usami S, et al. Mutation profile of the CDH23 gene in 56 probands with Usher syndrome type I. Hum Mutat. 2008;29:E37–46. doi: 10.1002/humu.20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Brederlow B, Bolz H, Janecke A, Cabrera A, Rudolph G, Lorenz B, et al. Identification and in vitro expression of novel CDH23 mutations of patients with Usher syndrome type 1D. Hum Mutat. 2002;19:268–73. doi: 10.1002/humu.10049. [DOI] [PubMed] [Google Scholar]
- 10.Astuto LM, Bork JM, Weston MD, Askew JW, Fields RR, Orten DJ, et al. CDH23 mutation and phenotype heterogeneity: a profile of 107 diverse families with Usher syndrome and nonsyndromic deafness. Am J Hum Genet. 2002;71:262–75. doi: 10.1086/341558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bork JM, Morell RJ, Khan S, Riazuddin S, Wilcox ER, Friedman TB, et al. Clinical presentation of DFNB12 and Usher syndrome type 1D. Adv Otorhinolaryngol. 2002;61:145–52. doi: 10.1159/000066829. [DOI] [PubMed] [Google Scholar]
- 12.Wagatsuma M, Kitoh R, Suzuki H, Fukuoka H, Takumi Y, Usami S. Distribution and frequencies of CDH23 mutations in Japanese patients with non-syndromic hearing loss. Clin Genet. 2007;72:339–44. doi: 10.1111/j.1399-0004.2007.00833.x. [DOI] [PubMed] [Google Scholar]
- 13.Ammar-Khodja F, Faugere V, Baux D, Giannesini C, Leonard S, Makrelouf M, et al. Molecular screening of deafness in Algeria: high genetic heterogeneity involving DFNB1 and the Usher loci, DFNB2/USH1B, DFNB12/USH1D and DFNB23/USH1F. Eur J Med Genet. 2009;52:174–9. doi: 10.1016/j.ejmg.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 14.Miyagawa M, Nishio SY, Usami S. Prevalence and clinical features of hearing loss patients with CDH23 mutations: a large cohort study. PLoS One. 2012;7:e40366. doi: 10.1371/journal.pone.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McHugh RK, Friedman RA. Genetics of hearing loss: Allelism and modifier genes produce a phenotypic continuum. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:370–81. doi: 10.1002/ar.a.20297. [DOI] [PubMed] [Google Scholar]
- 16.Pennings RJ, Topsakal V, Astuto L, de Brouwer AP, Wagenaar M, Huygen PL, et al. Variable clinical features in patients with CDH23 mutations (USH1D-DFNB12) Otol Neurotol. 2004;25:699–706. doi: 10.1097/00129492-200409000-00009. [DOI] [PubMed] [Google Scholar]
- 17.Matsunaga T, Hirota E, Bito S, Niimi S, Usami S. Clinical course of hearing and language development in GJB2 and non-GJB2 deafness following habilitation with hearing aids. Audiol Neurootol. 2006;11:59–68. doi: 10.1159/000089607. [DOI] [PubMed] [Google Scholar]
- 18.Usami S, Abe S, Akita J, Namba A, Shinkawa H, Ishii M, et al. Prevalence of mitochondrial gene mutations among hearing impaired patients. J Med Genet. 2000;37:38–40. doi: 10.1136/jmg.37.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roux AF, Faugere V, Le Guedard S, Pallares-Ruiz N, Vielle A, Chambert S, et al. Survey of the frequency of USH1 gene mutations in a cohort of Usher patients shows the importance of cadherin 23 and protocadherin 15 genes and establishes a detection rate of above 90% J Med Genet. 2006;43:763–8. doi: 10.1136/jmg.2006.041954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.PolyPhen-2. http://genetics.bwh.harvard.edu/pph2/.
- 21.PROVEAN. http://provean.jcvi.org/index.php.
- 22.NNSPLICE. http://www.fruitfly.org/seq_tools/splice.html.
- 23.dbSNP. http://www.ncbi.nlm.nih.gov/snp/.
- 24.1000GENOME. http://www.1000genomes.org/.
- 25.NHLBI exome variant server. http://evs.gs.washington.edu/EVS/.
- 26.UCSC conservation. http://genome.ucsc.edu/index.html.
- 27.Shahin H, Walsh T, Rayyan AA, Lee MK, Higgins J, Dickel D, et al. Five novel loci for inherited hearing loss mapped by SNP-based homozygosity profiles in Palestinian families. Eur J Hum Genet. 2010;18:407–13. doi: 10.1038/ejhg.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang T, Wei X, Chai Y, Li L, Wu H. Genetic etiology study of the non-syndromic deafness in Chinese Hans by targeted next-generation sequencing. Orphanet J Rare Dis. 2013;8:85. doi: 10.1186/1750-1172-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultz JM, Bhatti R, Madeo AC, Turriff A, Muskett JA, Zalewski CK, et al. Allelic hierarchy of CDH23 mutations causing non-syndromic deafness DFNB12 or Usher syndrome USH1D in compound heterozygotes. J Med Genet. 2011;48:767–75. doi: 10.1136/jmedgenet-2011-100262. [DOI] [PubMed] [Google Scholar]
- 30.LOVD-CDH23 database. http://www.lovd.nl/CDH23.
- 31.Shearer AE, DeLuca AP, Hildebrand MS, Taylor KR, Gurrola J, Scherer S, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:21104–9. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shearer AE, Smith RJ. Genetics: advances in genetic testing for deafness. Curr Opin Pediatr. 2012;24:679–86. doi: 10.1097/MOP.0b013e3283588f5e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brownstein Z, Bhonker Y, Avraham KB. High-throughput sequencing to decipher the genetic heterogeneity of deafness. Genome Biol. 2012;13:245. doi: 10.1186/gb-2012-13-5-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mutai H, Suzuki N, Shimizu A, Torii C, Namba K, Morimoto N, et al. Diverse spectrum of rare deafness genes underlies early-childhood hearing loss in Japanese patients: a cross-sectional, multi-center next-generation sequencing study. Orphanet J Rare Dis. 2013;8:172. doi: 10.1186/1750-1172-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Keulenaer S, Hellemans J, Lefever S, Renard JP, De Schrijver J, Van de Voorde H, et al. Molecular diagnostics for congenital hearing loss including 15 deafness genes using a next generation sequencing platform. BMC Med Genomics. 2012;5:17. doi: 10.1186/1755-8794-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sivakumaran TA, Husami A, Kissell D, Zhang W, Keddache M, Black AP, et al. Performance evaluation of the next-generation sequencing approach for molecular diagnosis of hereditary hearing loss. Otolaryngol Head Neck Surg. 2013;148:1007–16. doi: 10.1177/0194599813482294. [DOI] [PubMed] [Google Scholar]
- 37.Becirovic E, Ebermann I, Nagy D, Zhang W, Keddache M, Black AP, et al. Usher syndrome type 1 due to missense mutations on both CDH23 alleles: investigation of mRNA splicing. Hum Mutat. 2008;29:452. doi: 10.1002/humu.9526. [DOI] [PubMed] [Google Scholar]


