Abstract
The conversion of testosterone into dihydrotestosterone by steroid 5α-reductase is a key reaction in androgen action, and is essential both for the formation of the male phenotype during embryogenesis and for androgen-mediated growth of tissues such as the prostate1,2. Single gene defects that impair this conversion lead to pseudohermaphroditism in which 46 X, Y males have male internal urogenital tracts, but female external genitalia3. We have described the isolation of a human 5α-reductase complementary DNA from prostate4. Subsequent cloning and genetic studies showed that this gene (designated 5α-reductase 1) was normal in patients with 5α-reductase deficiency26. We report here the isolation of a second 5α-reductase cDNA by expression cloning and the polymerase chain reaction. The biochemical and pharmacological properties of this cDNA-encoded enzyme (designated 5α-reductase 2) are consistent with it being the major isozyme in genital tissue. A deletion in this gene is present in two related individuals with male pseudohermaphroditism caused by 5α-reductase deficiency. These results verify the existence of at least two 5α-reductases in man and provide insight into a fundamental hormone-mediated event in male sexual differentiation.
To isolate cDNAs encoding additional 5α-reductases, a size-fractionated and oriented cDNA library was constructed from human prostate poly(A)+ messenger RNA in a pCMV expression vector.5 The size of cDNA pools that could be screened for expression of 5α-reductase in cultured 293 cells was determined in serial dilution transfection experiments using an expression vector containing the 5α-reductase 1 cDNA and an irrelevant cDNA library. Enzyme activity in transfected cells could be detected over background (threefold) when the 5α-reductase 1 cDNA was diluted 104-fold. On the basis of this information, 200 pools of this size from the prostate cDNA library were transfected into 293 cells and assayed for expression of 5α-reductase activity as described in the legend to Fig. 1. Two pools were identified which expressed a 5α-reductase activity.
FIG. 1.
cDNA and amino-acid sequence of human 5α-reductase 2. Nucleotides are numbered at right. Amino acids are numbered above the protein sequence.
METHODS. An oriented and size-fractionated cDNA library was constructed in a pCMV vector5 from human prostate poly(A)+ mRNA using a kit purchased from GIBCO-BRL. The cDNA was electroporated into Escherichia coli HB101 cells, and pools of about 104 independent cDNAs were grown overnight in 10 ml cultures of superbroth media20. Plasmid DNA was prepared using Quiagentip 100 columns and 5 μg aliquots were transfected through a calcium phosphate procedure21 into 60 mm dishes of human embryonic kidney 293 cells (ATCC #CRL 1573). To enhance expression, 0.5 μg of a plasmid (pVA1) containing the adenovirus VAI gene was cotransfected with the pooled cDNAs21. On day 2 of the transfection experiments, 14C-testosterone (120 d.p.m. nmol−1) was added to the medium at a final concentration of 1 μM, and conversion into dihydrotestosterone was determined 18 h later as described previously4,6. A pool expressing 5α-reductase enzyme activity was subsequently screened with a probe generated by a polymerase chain reaction in which two oligonucleotides, GA(A/G)TGGTG(T/C)T(T/A)(T/C)GCN(C/T)TNGC and TTIGG(A/G)TAITC(T/C)TC(A/G)AA(T/C)TT, encoding amino acids 205 to 211 and 243 to 249, respectively, of the human and rat 5α-reductase proteins4,6, were used to amplify random-primed cDNA synthesized from 0.4 μg total human prostate RNA. The reaction conditions were those of Strathmann et al.22, except that 30-s incubations at 94 °C, 40 °C, and 72 °C were used in place of those described. About one hybridization-positive was found per 104 colonies screened from the expressing pool. The DNA sequence of one such cDNA was determined on an Applied Biosystems Model 373 fluorescence sequencer after subcloning fragments into bacteriophage M13 vectors.
Simultaneously with the expression cloning studies, pairs of oligonucleotides derived from conserved regions of the human 5α-reductase 1 and rat cDNAs6, were used in polymerase chain reactions7 that used cDNA template reverse transcribed from human prostate RNA. One pair, derived from the region of the cDNAs encoding the carboxy terminus of the protein, generated inter alia a 91-base pair product whose DNA sequence was 57% identical to the corresponding region of the human 5α-reductase 1 cDNA. When this product was used to screen an expressing pool of prostate cDNAs, hybridization positives were obtained at a frequency of about 1 in 104. This result, combined with DNA sequence analysis of a hybridization-positive clone (see below), indicated that both approaches had identified the same cDNA.
The DNA sequence of the 2.437-kilobase cDNA insert in the expression plasmid encoded a hydrophobic protein of 254 amino acids and contained a long 3′-untranslated region (Fig. 1). The sequence of the predicted protein (5α-reductase 2) was 50% identical to that of human 5α-reductase 1 and 46% identical to the rat 5α-reductase enzyme (Fig. 2). All three proteins shared almost identical hydropathy plots despite their relatively low sequence identity (not shown). A search of the data bases indicated that residues 10–85 of human 5α-reductase 2 shared a 38% sequence identity with residues 231–305 of the tobacco chloroplast NADP-ubiquinone oxidoreductase chain 5 protein8, and that residues 9–72 shared a 39% identity with residues 222–281 of the pol polyprotein of the Cas-Br-E murine leukemia virus9. Surprisingly, the entire 5α-reductase 2 protein is 28% identical to residues 264–462 of the Epstein–Barr virus terminal proteins10, suggesting the latter proteins may bind steroids or NADPH.
FIG. 2.
Alignment of 5α-reductase proteins. The amino-acid sequences in single-letter code of the human 5α-reductase 2, 5α-reductase 1 (ref. 4), and rat 5α-reductase6 proteins are aligned. Identities between two or more enzymes are boxed in black. Numbers above the sequences refer to the human 5α-reductase 2 protein.
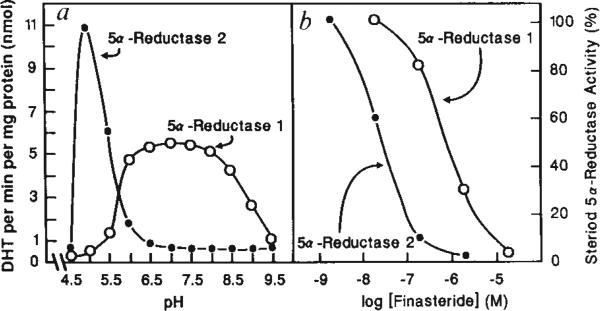
Cell extracts from 293 cells transfected with the expression vector containing the 5α-reductase 2 cDNA actively reduced testosterone to dihydrotestosterone, indicating that the cDNA does encode a 5α-reductase enzyme. The data in Fig. 3a show that 5α-reductase 2, like the major enzyme expressed in human genital skin fibroblasts11 and prostate12, has an acidic pH optimum whereas 5α-reductase 1 has a more basic pH optimum.
FIG. 3.
Characterization of expressed 5α-reductase isozymes, a, pH optima. Enzyme activity was assayed at the indicated pH in cell extracts prepared from 293 cells transfected with the 5α-reductase 1 or 2 cDNA. Nontransfected cells express negligible amounts of enzyme activity, b, Inhibition by finasteride. Reductase enzyme activity obtained in extracts of transfected 293 cells in the absence of inhibitor is defined as 100%. In the presence of Increasing concentrations of finasteride, progressively less 5α-reductase enzyme activity was detected.
METHODS. An expression plasmid containing the 5α-reductase 2 cDNA (Fig. 1) or the 5α-reductase 1 cDNA (ref. 4) was transfected into 293 cells as described in the legend to Fig. 1. In a, 48 h after transfection, cell extracts were prepared as described previously4 and 10 μg cell protein were assayed for 5α-reductase enzyme activity in 0.1 M Tris-citrate buffers at the indicated pH with 10 μM 14C-testosterone (120 d.p.m. pmol−1) as substrate and 10 mM NADPH as cofactor. In b, 5 μg of transfected cell protein were assayed in duplicate for 5α-reductase activity in the presence of the indicated concentration of finasteride (MK-906)12 (17β-(N-t-butyl)carbamoyl-4-aza-5α-androst-l-ene-3-one, a kind gift of G. Rasmusson, Merck, Sharp and Dohme), 4 μM 14C-testosterone (120 d.p.m. pmol−1) and 10 mM NADPH. In both panels, conversion into dihydrotesterone was determined after 10-min incubations by thin layer chromatography as described previously6. Protein concentrations in cell extracts were measured by Lowry assay23.
The central role of 5α-reductase in prostate development and maintenance13 has led to the discovery and development of powerful inhibitors of this enzyme as potential therapeutics for the treatment of prostate disorders such as benign prostatic hyperplasia14,15. The 4-azasteroid finasteride (MK-906), competitively inhibits the major human prostate 5α-reductase enzyme12,26, but is a poor inhibitor of the 5α-reductase 1 enzyme3. Expression of the 5α-reductase 2 cDNA in 293 cells results in the synthesis of an enzyme that is markedly inhibited by finasteride (50% inhibitory concentration IC50 = 30nM), whereas expression of the 5α-reductase 1 cDNA yields a poorly inhibited enzyme (IC50 = 900nM) (Fig. 3b). More detailed kinetic experiments indicated finasteride inhibition constant (Ki) values of about 5 nM and 230 nM for the 5α-reductase 2 and 1 enzymes, respectively (not shown).
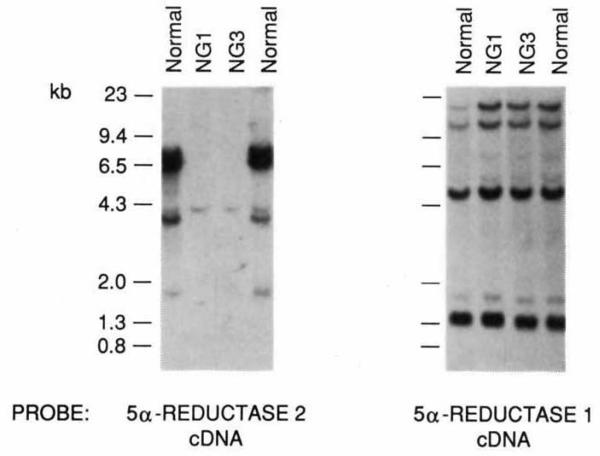
Fibroblasts cultured from genital skin biopsies of 5α-reductase-deficient subjects have reduced or abnormal acidic pH optimum enzyme activity3,16. To determine if mutations in the gene encoding the 5α-reductase 2 cDNA could be detected in these subjects, DNAs from multiple unrelated affected individuals were screened for gene rearrangements by Southern blotting. As a control, the same DNAs were screened with a probe from the 5α-reductase 1 cDNA. The data of Fig. 4 indicate that a deletion in the 5α-reductase 2 gene was found in two related pseudohermaphrodites from the Simbari Anga linguistic group in the Highlands of Papua New Guinea17 but was not present in the DNA of a normal individual from this tribe. The deletion seems to have removed a majority of the 5α-reductase 2 gene from the affected individuals, as only a single weakly hybridizing fragment is visible on the autoradiogram (Fig. 4). No gross rearrangements in the 5α-reductase 2 gene were detected in affected individuals derived from 19 different pedigrees from throughout the world, indicating that, as with many other genetic diseases18, most of the mutations cannot be detected by Southern blotting.
FIG. 4.
Deletion of 5α-reductase 2 gene in subjects with 5α-reductase deficiency. DNA isolated from two normal individuals and two related 5α-reductase deficiency subjects from a geographically isolated tribe in the Highlands of Papua New Guinea17 was screened by Southern blotting using the indicated 5α-reductase cDNA probes. The normal DNA analysed in the most left-hand lane was isolated from an individual from the same New Guinea tribe as the NG1 and NG3 subjects, whereas the normal DNA analysed in the right-hand lane was isolated from a caucasian American. The filter was screened with the 5α-reductase 2 cDNA probe first, and then stripped and reprobed with the 5α-reductase 1 cDNA probe. A deletion of all but a weakly hybridizing fragment of about 4.5 kilobases in the DNA of the affected NG1 and NG3 individuals is apparent from the autoradiogram obtained with the 5α-reductase 2 probe. All individuals have a normal 5α-reductase 1 gene.
METHODS. Genomic DNA was isolated from peripheral blood samples from the indicated individuals and prepared and analysed by high-stringency Southern blotting as described previously24. Aliquots of DNA (20 μg) were digested with Hindlll before electrophoresis on agarose gels. Three single-stranded 32P-labelled probes spanning the coding region of the 5α-reductase 2 cDNA shown in Fig. 1 were prepared as described by Church and Gilbert25 and used in hybridization. After autoradiography for 5 days at −70 °C, the filter was stripped20 and reprobed with a random hexanucleotide 32P-labelled probe20 corresponding to the full-length 5α-reductase 1 cDNA (ref. 4).
Our results confirm the existence of two functional 5α-reductase genes in man. The 5α-reductase 2 cDNA and expressed enzyme described here seems to represent the major isozyme of genital tissue based on its acidic pH optima, exquisite sensitivity to finasteride, and deletion in two related individuals with 5α-reductase deficiency. By contrast, the 5α-reductase 1 cDNA and enzyme is expressed at much lower amounts in the prostate4, has a basic pH optimum, is insensitive to finasteride, and is normal in multiple 5α-reductase deficient patients26. The isolation of two 5α-reductases raises interesting questions for future study concerning the tissue specificity and regulation of these isozymes and their roles in androgen physiology.
Male pseudohermaphroditism reflects the incomplete establishment of phenotypic sex in 46 X, Y individuals and in a majority of cases is due to mutations in the androgen receptor or 5α-reductase3. These mutations are formally analogous to those described in nematodes and yeast that disrupt intercellular signalling systems controlling vulval development19 or mating type27, respectively. Thus, the androgen receptor and 5α-reductase are components of an intercellular signalling pathway that ultimately determines cell fate in the bipotential analage of the mammalian external genitalia. How the androgen receptor receives and transmits the inductive signal provided by dihydrotestosterone, as well as further parallels between mammalian and invertebrate sexual differentiation are topics for future study.
ACKNOWLEDGEMENTS
We thank J. Cormier and D. Davis for technical assistance. J. McConnell for surgical samples, J. Imperato-McGinley for blood samples, J. L. Goldstein, M. S. Brown and J. D. Wilson for critical reading of the manuscript. This work was supported by the NIH. the Welch Foundation, and the Perot Family Foundation. The sequence data presented in Fig. 1 will appear in the EMBL, GenBank and DDBJ Nucleotide Sequence Databases under the accession number M74047.
References
- 1.Wilson. J. D. Harvey Led. 1985;79:145–172. [PubMed] [Google Scholar]
- 2.Wilson JD. Endocrinology. 1975;25:491–508. [Google Scholar]
- 3.Griffin JE, Wilson JD. In: The Metabolic Basis of Inherited Disease. Scriver CR, Beaudet AL, Sly WS, Valle D, editors. McGraw-Hill; New York: 1989. pp. 1919–1944. [Google Scholar]
- 4.Andersson S, Russell DW. Proc. natn. Acad. Sci. U.S.A. 1990;87:3640–3644. doi: 10.1073/pnas.87.10.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersson S, Davis D, Dahlback, Jornvail H, Russell DW. J. biol. Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 6.Andersson S, Bishop RW, Russell DW. J. biol. Chem. 1989;264:16249–16255. [PMC free article] [PubMed] [Google Scholar]
- 7.Saiki RK, et al. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 8.Shinozaki K, et al. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rassart E, Nelbach L, Jolicoeur P. J. Virol. 1986;60:910–919. doi: 10.1128/jvi.60.3.910-919.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laux G, Perricaudet M, Farrell PJ. EMBO J. 1988;7:769–774. doi: 10.1002/j.1460-2075.1988.tb02874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore RJ, Griffin JE, Wilson JD. J. biol. Chem. 1975;250:7168–7172. [PubMed] [Google Scholar]
- 12.Liang T, Cascieri MA, Cheung AH, Reynolds GF, Rasmusson GH. Endocrinology. 1985;117:571–579. doi: 10.1210/endo-117-2-571. [DOI] [PubMed] [Google Scholar]
- 13.George FW, Russell DW, Wilson JD. Proc. natn. Acad. Sci. U.S.A. 1991;88:8044–8047. doi: 10.1073/pnas.88.18.8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brooks JR, et al. Endocrinology. 1981;109:830–836. doi: 10.1210/endo-109-3-830. [DOI] [PubMed] [Google Scholar]
- 15.Vermeulen A, Giagulli VA, DeSchepper P, Buntinx A, Stoner E. Prostate. 1989;14:45–53. doi: 10.1002/pros.2990140106. [DOI] [PubMed] [Google Scholar]
- 16.Moore RJ, Wilson JD. J. biol. Chem. 1976;251:5895–5900. [PubMed] [Google Scholar]
- 17.Imperato-McGinley J, et al. Clin. Endocrin. 1991;34:293–298. doi: 10.1111/j.1365-2265.1991.tb03769.x. [DOI] [PubMed] [Google Scholar]
- 18.Hobbs HH, Russell DW, Brown MS, Goldstein JL. A. Rev. Genet. 1990;24:133–170. doi: 10.1146/annurev.ge.24.120190.001025. [DOI] [PubMed] [Google Scholar]
- 19.Horovitz HR, Sternberg PW. Nature. 1991;351:535–541. doi: 10.1038/351535a0. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory. Cold Spring Harbor; New York: 1989. [Google Scholar]
- 21.Gorman CM, Gies DR, McCray G. DNA Prot. Engng. Tech. 1990;2:3–10. [Google Scholar]
- 22.Strathmann M, Wilkie TM, Simon MI. Proc. natn. Acad. Sci. U.S.A. 1989;85:7407–7409. doi: 10.1073/pnas.86.19.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowry 0H, Rosebrough NJ, Farr AL, Randall RJ. J. biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 24.Lehrman MA, et al. Science. 1985;227:140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Church GM, Gilbert W. Proc. nato. Acad. Sci. U.S.A. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins EP, Andersson S, Imperato-McGinley J, Wilson JD, Russell DW. J. clin. Invest. doi: 10.1172/JCI115574. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herskowitz I. Nature. 1989;342:749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]