Abstract
Recent analysis of a Gal4 mutant (Gap71) carrying three point mutations (S22D, K23Q and K25F) in its DNA-binding domain (DBD), has demonstrated that it cannot occupy GAL promoters efficiently in cells and that it is not mono-ubiquitylated, suggesting a functional link between this modification and stable DNA binding in cells. The mechanistic underpinning of this phenotype is that this protein is hypersensitive to a newly discovered activity of the proteasomal ATPases – their ability to actively dissociate transcription factor-DNA complexes after direct interaction with the activation domain. In this paper, we examine the roles of each of the three point mutations contained in Gap71 individually. These experiments have revealed that serine 22 is a site of phosphorylation in the Gal4 DBD and that lysine 23 is essential for S22 phosphorylation, possibly acting as part of the kinase recognition site. Mutation of either residue blocks Gal4 DBD phosphorylation, its subsequent ubiquitylation and compromises the ability of the activator to bind promoter DNA in vivo. These data represent the first report of an essential phosphorylation event critical for the activity of this paradigmatic transcription factor.
Keywords: Gal4, Phosphorylation, Proteasome, ubiquitylation
Introduction
One of the largest “protein machines”1 in the cell is the transcriptional initiation complex that forms on the promoters of actively transcribed genes. On most genes, the formation of these pre-initiation complexes (PICs) is nucleated by one or more gene-specific transactivators that bind to particular sequences in the promoter region of the target gene. In order to perform this function, activators presumably must bind stably to the promoter long enough to interact with the transcriptional machinery, coactivators and chromatin remodeling and modification complexes. This idea is supported by studies of mutant activators that alter the lifetime of their binding to promoters, which have shown that transcriptional output is linked to the kinetic stability of the complex2–5.
While most transactivator-promoter complexes are intrinsically quite stable in vitro, it has been discovered recently that in living cells there are both proteolytic and non-proteolytic activities that oppose the formation of long-lived activator-promoter complexes, possibly to limit the number of rounds of transcription that can be driven without continued responsiveness to signal transduction (for reviews, see6–9). A spate of recent studies have demonstrated a wide diversity of lifetimes of transactivator-promoter complex lifetimes in vivo10–15, arguing that there must exist mechanisms by which the effect of these destabilizing activities can be modulated.
We found recently that one such mechanism that opposes the formation of stable activator-DNA complexes in vivo involves the ATPases of the 26S proteasome16. Specifically, we demonstrated that a sub-complex of the proteasome that includes the “base” of the 19S Regulatory Particle (RP)17 (the six ATPases (Rpts1–6), Rpn1 and Rpn2), but not the proteolytic 20S core of the proteasome, binds to activators via their activation domain18, 19 and proceeds to unfold them4. If allowed to proceed unchecked, this activity strongly inhibits Gal4-mediated transcription of the GAL genes. In wild-type cells however, this inhibitory function of the proteasomal ATPases is checked by mono-ubiquitylation of the Gal4 DNA-binding domain (DBD)4. Other investigators had previously reported that certain eukaryotic transactivators require mono-ubiquitylation in order to function efficiently20–22, but the mechanistic basis of this effect is poorly understood23. This protective effect of activator mono-ubiquitylation may provide a rationalization for at least some of these observations. Recent mechanistic studies have shown that the mono-ubiquitin moiety binds directly to Rpt1 and Rpn1 in the 19S RP base and dissociates the complex form the activation domain of Gal424. In the context of a promoter-bound complex, this activity may stimulate the transfer of the ATPase complex from the activator to the PIC, where it is required to stimulate promoter escape and elongation25.
A key reagent in the discovery of the “stripping” activity of proteasomal ATPases and the protective effect of activator mono-ubiquitylation was Gap71, a derivative of Gal4 with three mutations in the DNA-binding domain26 (K23Q, S22D and K25F)27, 28. This protein fails to be ubiquitylated in a nuclear extract and is thus hypersensitive to the “stripping” activity of the 19S ATPases4. In this study, we examine the effect of mutating each of these residues in the Gal4 DNA-binding domain (DBD) individually. We show that K23 is critical for ubiquitylation but that K25 is non-essential. Surprisingly, we also find that S22 is a site of phosphorylation and that this event is essential for subsequent mono-ubiquitylation and efficient promoter binding. Whereas Gal4 phosphorylation at three C-terminal residues has been reported previously, these modifications are only correlated with, but not essential for, the activity of this paradigmatic transcription factor29–31. Thus, we have uncovered the first phosphorylation event critical for Gal4 activity.
Results
Identification of S22 and K23 as critical residues for Gal4-DNA binding in vivo
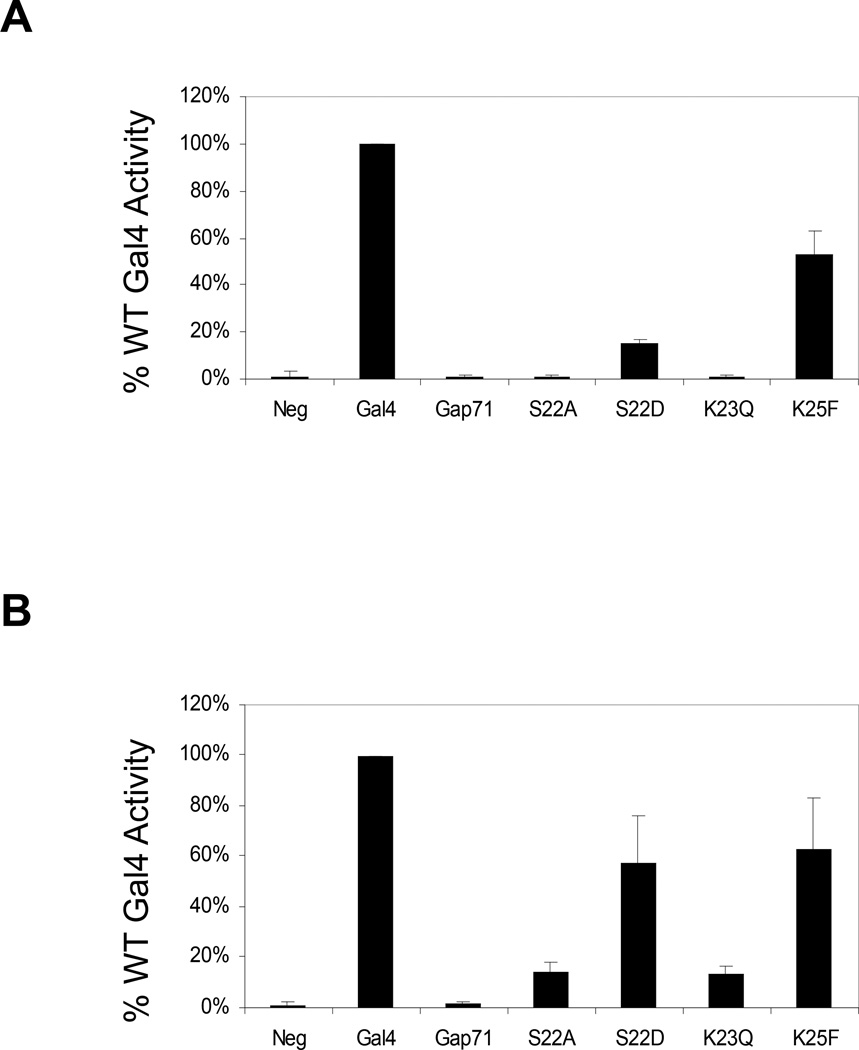
To characterize the effects of the three point mutations in Gap71 individually, site-directed mutagenesis was employed to introduce S22D, K23Q and K25F separately into Gal4. The full-length protein was expressed from a single copy plasmid and under the control of its native promoter in a Δgal4 strain. Gal4 activity was then assessed by monitoring α-galactosidase (α-gal) activity. Expression of the α-galactosidase-encoding gene (MEL1) is dependent on Gal4. As shown in Fig.1A, the K25 mutant functioned almost as well as the wild-type protein. However, the level of α-gal expression observed in the K23Q strain was similar to that observed in the absence of Gal4. Gal4(S22D) was of intermediate potency, with approximately 15% the activity of the wild-type protein. We also constructed and studied another point mutant, S22A. This protein was completely inactive (Fig. 1A).
Figure 1.
Activity of Gal4 point mutants in vivo when expressed from a single copy or multi-copy plasmid. A. Normalized values (with wild-type Gal4 set at 100%) for α-galactosidase (MEL1 gene product) activity in Δgal4 strains expressing the pSB32 S103-Gal4 derivatives. B. Same as A, but the S103-Gal4 derivatives were expressed at approximately 25-fold higher levels by using the multi-copy plasmid Yep351.
Activators must occupy their cognate promoters and recruit various transcription and chromatin-modifying proteins in order to function. To determine at which of these steps the Gal4 point mutants might be deficient, we repeated the reporter gene experiments, but expressed the Gal4 derivatives from a multi-copy plasmid, increasing the levels of the activator ≈ 25-fold relative to the cells that had been transformed with the centromeric plasmid32, 33. It is often the case that over expression can rescue the activity of activator mutants deficient in promoter occupancy33, but higher protein levels would not be expected to have an effect on activation per se, since only a fixed number of activator molecules can bind to a GAL promoter.
Fig. 1B shows the relative activity of the overexpressed proteins in driving the expression of MEL1, which encodes α-galactosidase. Gal4(K23Q) and Gal4(S22A) showed abut 18% of the activity of wild-type Gal4. However, cells harboring these proteins were able to grow on plates containing galactose as the sole carbon source, whereas cells transformed with the analogous single copy vectors did not grow at all under the same conditions (data not shown). Thus, some activity is clearly restored upon overexpression. Indeed, the promoter of the non-essential MEL1 gene contains only a single Gal4 binding site and so this may be one of the most “difficult” promoters for the mutant proteins to occupy efficiently. This model is consistent with ChIP assays examining the GAL1/10 promoter, which contains four Gal4 binding sites (see below). The S22D mutant functioned about 60% as well as the wild-type activator at these elevated protein levels. The growth of cells harboring the multi-copy Gal4(S22D) expression plasmid was essentially indistinguishable from wild-type (data not shown). These data are consistent with the idea that alterations of S22 and K23 impair the ability of Gal4 to bind to GAL promoters, though they do not rule out the possibility that these residues affect downstream steps as well.
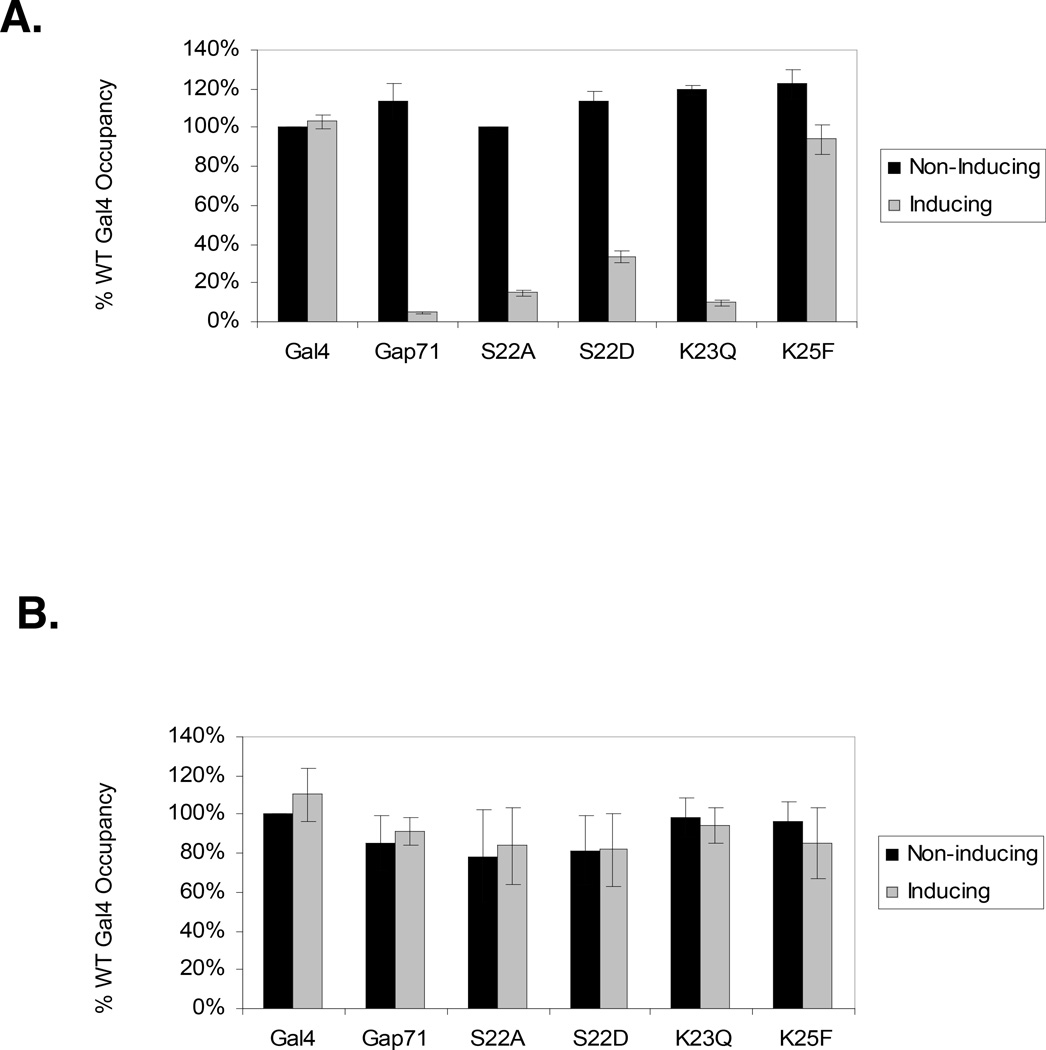
To address promoter occupancy more directly, we employed chromatin immunoprecipitation (ChIP) assays in both non-inducing (raffinose) and inducing (raffinose + galactose) media. As shown in Fig. 2A, strong ChIP signals indicated that all of the Gal4 derivatives occupy the GAL1/10 promoter in non-inducing media, conditions under which the Gal80 protein34 blocks association of the activation domain with proteasomal ATPases. However, under inducing conditions (galactose medium) a very different pattern emerged. When expressed at native levels, wtGal4, and Gal4K25F occupied the promoter efficiently, whereas little binding of Gap71 K23Q or S22A was detected. As in the in vivo activity assay, Gal4S22D exhibited intermediate binding activity. When the Gal4 proteins were expressed from the multi-copy plasmid, all of them occupied the GAL1/10 promoter, as assayed by ChIP analysis (Fig. 2B).
Figure 2.
Effect of single point mutations in the Gal4-DNA-binding domain on the occupancy of the Gal1–10 promoter, which contains three strong and one weak Gal4 binding sites. Chromatin immunoprecipitation (ChIP) assays were conducted on yeast under non-inducing (raffinose) and inducing (galactose) conditions by antibodies against the C-terminus of Gal4. A. The Gal4 derivatives were expressed at approximately native levels from a single copy plasmid with the native Gal4 promoter. Immunoprecipitated DNA was analyzed by QRT-PCR for Gal4 occupancy on the Gal1–10 promoter. B. Same as A, but the Gal4 proteins were expressed from a multi-copy plasmid that results in approximately 25-fold higher levels of the protein. These data were derived from the primary data seen in Supplementary Figure 1.
We also examined the effects of the individual mutations in the context of a Gal4–Gal1135 fusion protein, Gal4(1–147)DBD-Gal11(799–1082), that lacks a classical activation domain. Gal4 DBD-Gal11 C-terminal fusion proteins are unusually potent AD-less activators36, 37, possibly because the fusion mimics accurately a bona fide contact of the Gal4 AD with Gal1138 in the Mediator complex39, 40. Since the available data indicates that the APIS complex interacts with the Gal4 AD and a Gal4 DBD-DNA complex lacking an AD is insensitive to APIS-mediated stripping4, we wondered if this fusion would eliminate the sensitivity of some of the point mutants to this activity.
We first tested the panel in an α-galactosidase assay with the fusion protein expressed from a multi-copy plasmid (Figure 3A). The K25F mutant behaved like the wild-type protein, while the S22D mutant had partial activity. Neither Gap71, S22A nor K23Q showed significant activity levels. These results mimic the data in Figure 1. Promoter occupancy of these fusion proteins was also tested using chromatin immuno-precipitation assays (Figure 3B). The occupancy reflects the expression data. K25F behaved like wild-type Gal4. The Gap71, S22A and K23Q samples did not appear to occupy the GAL 1–10 promoter, while the S22D-containing protein displayed intermediate behavior. The close correlation observed between the effects of the point mutations in the context of wild-type Gal4 and the Gal4–Gal11 fusion protein strongly suggest that the APIS complex can associate with the latter, even though it lacks a classical AD.
Figure 3.
Analysis of the effect of various mutations on the activity of Gal4(1–147)-Gal11(799–1082). A. Normalized values (with wild-type Gal4–Gal11 set at 100%) for α-galactosidase (MEL1 gene product) activity in Δgal4 strains expressing the Yep351 Gal4–Gal11 derivatives indicated from the native GAL4 promoter. Negative control is empty vector in the same strain background. B. ChIP assays with antibodies against the N-terminus of Gal4 were done on the same Gal4–Gal11 derivatives in A. Immunoprecipitated DNA was evaluated by QRT-PCR for occupancy on the GAL1–10 promoter.
Serine 22 is a site of phosphorylation
The significantly different activities of the S22D and S22A mutants, at least when expressed from a single copy plasmid (Fig. 1A and 2A), was interesting. In some cases, an aspartic acid can serve as a phosphoserine mimic due its negative charge41, 42. We therefore considered the possibility that S22 is a site of phosphorylation in Gal4 and that this modification is essential for full activity.
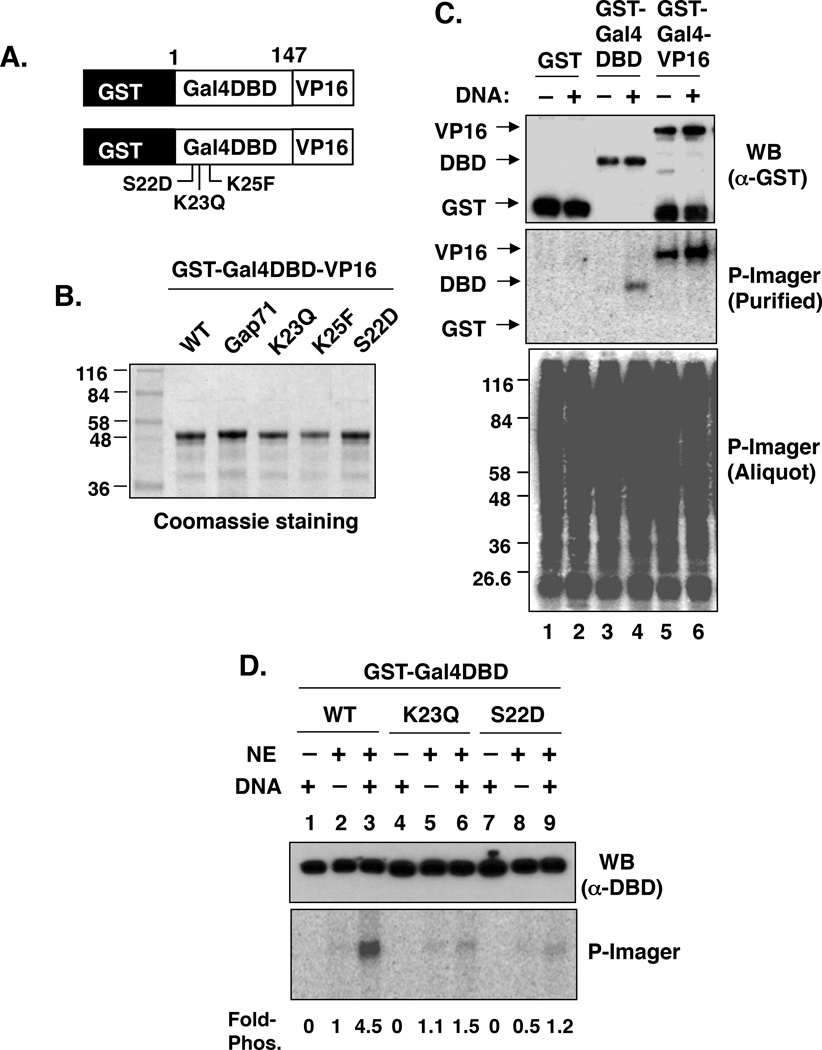
To probe for Gal4 DBD phosphorylation directly, a chimeric GST-Gal4 (1–147)-VP16 activation domain fusion protein (hereafter called GST-Gal4–VP16, was employed for in vitro phosphorylation assays (Fig. 4A), since native Gal4 is difficult to purify in quantities useful for biochemical experiments. All of the protein preparations were at least 90% pure (Fig. 4B). HeLa nuclear extract (NE) and γ-32P-ATP were mixed with GST, GST-Gal4 DBD or GST-Gal4–VP16 in the presence or absence of DNA containing five Gal4 binding sites. After a twenty-minute incubation at 30 °C, an aliquot was removed and processed for phosphorimager analysis, which showed extensive protein phosphorylation (Fig. 4C, bottom panel). The fusion proteins were then isolated on glutathione-sepharose beads, resolved by SDS-PAGE and then transferred to a membrane. Western blotting with an anti-GST antibody showed that approximately equal amounts of each protein were obtained on the glutathione-sepharose beads (Fig. 4C, top panel). Phosphorimager analysis (Fig. 4C, middle panel) revealed that GST-Gal4 DBD and GST-Gal4–VP16 had been phosphorylated, but GST had not. This experiment also showed that the phosphorylation reaction was stimulated by the presence of DNA, suggesting that the substrate is the Gal4 DBD-DNA complex and/or that the kinase is DNA-associated.
Figure 4.
Phosphorylation of the Gal4 DNA-binding domain (DBD). A. Cartoon of the general structure of the GST-Gal4–VP16 derivatives employed for these experiments. B. Coomassie-stained gel showing the preparations of each protein (bottom). Each protein was expressed in, and purified from, E. coli. Numbers on the left indicate the molecular masses (in kDa) of the protein standards. C. Phosphorylation of GST, GST-Gal4 DBD and GST-Gal4–VP16 in HeLa NE. The indicated GST fusion proteins were incubated with NE and γ-32P-ATP in the presence (+) or absence (−) of DNA containing Gal4 binding sites. An aliquot (5 µl) was removed and processed for phosphorimager analysis, which showed extensive protein phosphorylation (bottom panel). GST fusion proteins were purified on glutathione-agarose, separated on a SDS-PAGE gel and then transferred to PVDF membrane for western blotting with anti-GST (top panel) and phosphorimager analysis (middle panel). D. Effect of K23Q and S22D on DNA-stimulated Gal4 phosphorylation. Phosphorylation of wild-type GST-Gal4-DBD and the K23Q and S22D mutants was analyzed as described in panel A in the presence (+) and absence (−) of NE and DNA.
To determine if S22 is the site of phosphorylation in the Gal4 DBD, wild-type GST-Gal4 DBD and the S22D derivative were exposed to the same reaction conditions. Whereas the wild-type DBD was phosphorylated efficiently in the presence of DNA, the S22D derivative was not (Fig. 4D), consistent with S22 being the primary site of phosphorylation. Interestingly, when GST-Gal4 DBD(K23Q) was used in this assay, it was found that K23 is also required for this reaction (Fig. 4D), possibly serving as part of the kinase recognition site.
Efficient ubiquitylation of the Gal4 DBD is influenced by serine 22
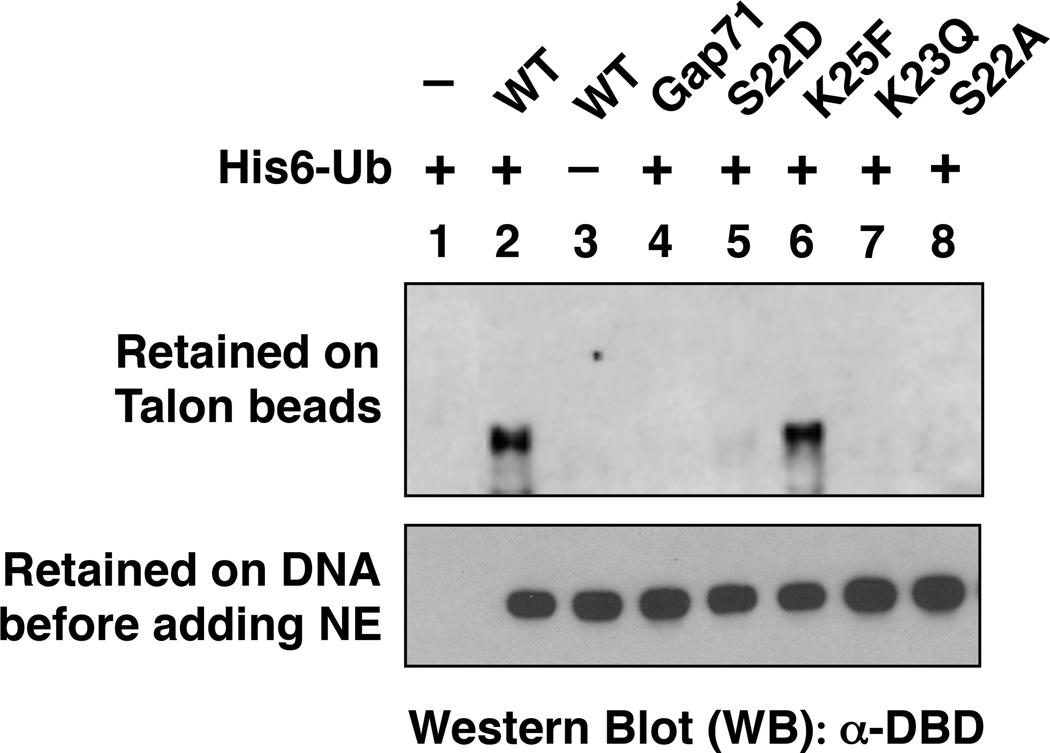
Prior phosphorylation is often essential for subsequent ubiquitylation of a nearby lysine residue by an E3 ubiquitin ligase complex43, 44. Therefore, we asked if S22 is essential for Gal4 DBD ubiquitylation. The GST-Gal4–VP16 fusion proteins (see Fig. 4A) with either the wild-type sequence or mutations at S22, K23 or K25 were bound to an immobilized DNA template containing five consensus Gal4 binding sites. Western blotting of the protein retained on the beads showed that each protein bound with approximately equal efficiency (Fig. 5A, bottom panel). This further shows that, biochemically, none of the mutations affected the intrinsic DNA binding activity of the Gal4 DBD, in agreement with the ChIP data in raffinose (Fig. 2A).
Figure 5.
Effect of point mutations in the Gal4 DNA-binding domain on ubiquitylation of GST-Gal4 DBD-VP16 in vitro. (Top) a western blot revealing the retention of GST-Gal4–VP16 derivatives on Talon beads under denaturing conditions. (Bottom) the indicated Gal4 derivative was incubated first with the immobilized DNA containing five Gal4 binding sites. The beads were pelleted and washed briefly. An aliquot was removed and Western blot analysis reveals the level of GST-Gal4–VP16 protein retained on the beads prior to NE addition in each case. In each experiment, isolated activator-DNA complexes were incubated in the presence (+) or absence (−) of His6-ubiquitin with ATP and a HeLa nuclear extract that had been pre-incubated with anti-Trp1 antibody. The proteins were then denatured and incubated with Talon beads, conditions under which only His6-ubiquitylated proteins are retained.
The immobilized protein-DNA complexes were then incubated with ATP, His6-tagged ubiquitin and HeLa nuclear extract (NE) that had been pre-incubated with anti-Trip1 (mammalian Sug1) antibody to block destabilization of the protein-DNA complex by the proteasomal ATPases4. To assay for the efficiency of ubiquitylation of these Gal4 derivates, the NE-treated complexes were exposed to immobilized metal ion affinity resin under denaturing conditions. Only proteins modified covalently with His6-Ub will be retained on the beads under these conditions. As shown in Fig. 5 (top panel), only wild type GST-Gal4–VP16 and the K25F mutant were ubiquitylated efficiently. Very low-level ubiquitylation of the S22D-containing protein could also be detected, but no ubiquitylation of the Gap71, K23Q or S22A derivates was observed. Ubiquitylation was not observed if GST-Gal4–VP16 (lane 1) or His6-Ub (lane 3) was omitted from the reaction, proving that the band observed is indeed the ubiquitylated transcription factor. This ubiquitylation must occur somewhere in the Gal4 DBD since the VP16 AD has no lysine residues and we have demonstrated that GST is not ubiquitylated4.
These data show that S22 and K23 are required for efficient mono-ubiquitylation of the Gal4 DBD. The fact that the S22D-containing protein is ubiquitylated more efficiently than the S22A-containing protein is consistent with the idea that S22 phosphorylation is required for ubiquitylation and that S22D weakly substitutes for this modification.
Is K23 the site of Gal4 DBD mono-ubiquitylation?
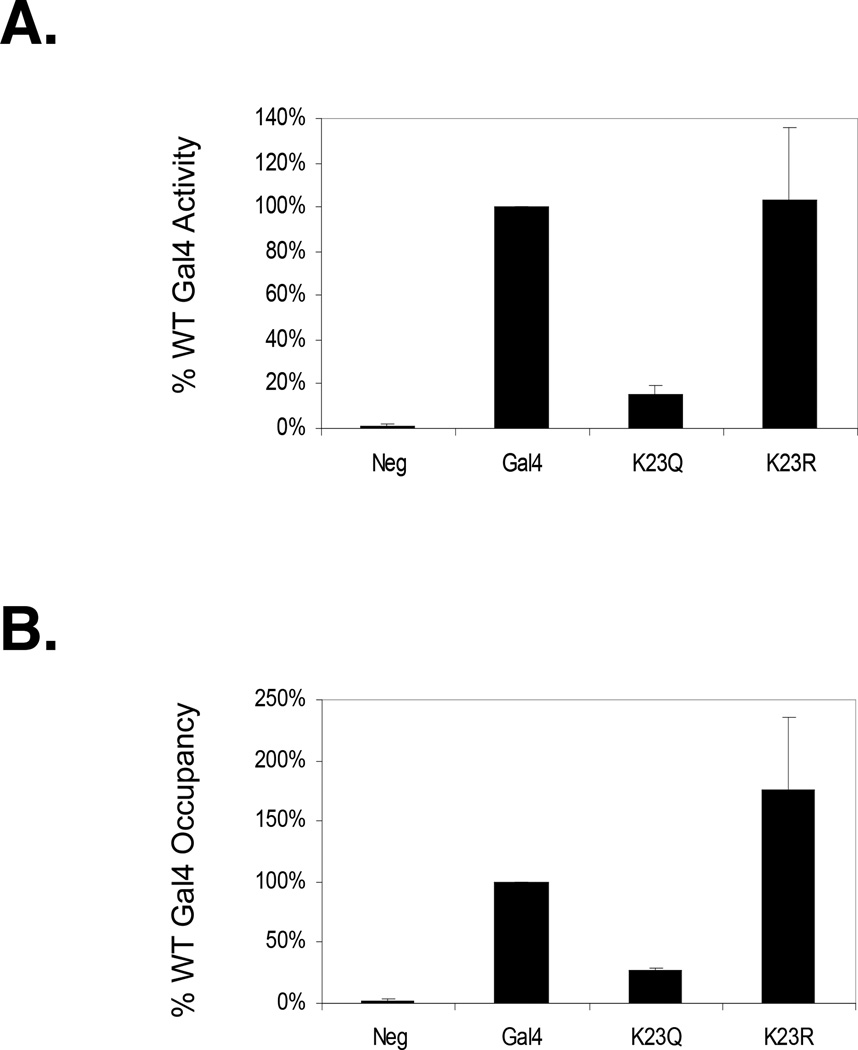
The K23Q mutation abolishes Gal4 DBD mono-ubiquitylation, possibly suggesting that K23 is the site of Gal4 DBD mono-ubiquitylation. However, S22 phosphorylation appears to be an essential pre-requisite for the ubiquitylation event and K23Q also affects this process. Thus, the K23Q mutation could affect Gal4 DBD mono-ubiquitylation indirectly. To further analyze this point, PCR-based mutagenesis was used to generate Gal4 K23R, representing a more conservative substitution for K23 than K23Q, but one which is incapable of being ubiquitylated. Gal4 K23R was expressed under the control of the native GAL4 promoter on a multi-copy plasmid and in a Δgal4 background. The results showed that Gal4 K23R was about as active as wild-type Gal4 in activation of the MEL1 gene (Fig. 6A) and occupied the GAL1–10 promoter at least as efficiently as wild-type Gal4 (Fig. 6B). As will be discussed in more detail below, these data imply that K23 is unlikely to be the site of Gal4 DBD mono-ubiquitylation. However, we cannot rule out the possibility that K23 is indeed the site of mono-ubiquitylation in the wild-type activator, but that when this residue is mutated the (as yet uncharacterized) E3 ubiquitin ligase employs a different lysine as the ubiquitin acceptor.
Figure 6.
Comparison of the activities of Gal4, Gal4 K23R and Gal4 K23Q. A. Normalized values (with wild-type Gal4 set at 100%) for α-galactosidase (MEL1 gene product) activity in Δgal4 strains expressing the indicated YEp351 S103-Gal4 derivative. Negative control is empty vector in the same strain background. B. Occupancy of the same Yep351 S103-Gal4 derivatives on the GAL1–10 promoter as analyzed by ChIP and qRT-PCR.
Ubiquitylation of Gal4–VP16 is essential for promoter occupancy in vitro and in vivo
To better connect the biochemical experiments presented in Figs. 4 and 5, which employed GST-Gal4DBD or GST-Gal4–VP16, to the effect of the mutations on native Gal4 (Figs. 1 and 2), we examined the DNA-binding properties of the various GST-Gal4 DBD-VP16 constructs in extracts and in cells. In vitro, all activators were first incubated with immobilized DNA containing five Gal4 binding sites and activator-DNA complexes were isolated. An aliquot was removed and the remaining activator-DNA complexes were further incubated with ATP, with or without HeLa NE for 30 minutes. Note that in this case, the extract had not been pre-incubated with anti-Trip1 antibody and therefore the activator-DNA complex was subject to the destabilizing activity of the proteasomal ATPases. The immobilized DNA-activator complexes were then re-isolated and analyzed by SDS-PAGE followed by Western blotting to determine the level of bound activator remaining. Whereas similar amounts of each activator were associated with the DNA in the absence of the NE (Fig. 7A, compare “NE −” lanes), the amount of protein recovered after exposure to the HeLa NE varied greatly for the different mutants (Fig. 7A, compare “NE +” lanes). The wild-type protein and K25 mutant bound efficiently, whereas Gap71 and the K23Q derivative bound poorly. The S22D mutant also was poorly retained and its behavior was reproducibly intermediate between the wild-type and the Gap71 proteins. It is interesting to note that in all cases the protein that remained bound to the DNA displayed the modest decrease in electrophoretic mobility that is indicative of mono-ubiquitylation (Fig. 7A). Thus, the evidence argues that mutations at S22 and K23 affect the efficiency of ubiquitylation and that the protein must be mono-ubiquitylated in order to bind DNA in the presence of the proteasomal ATPases.
Figure 7.
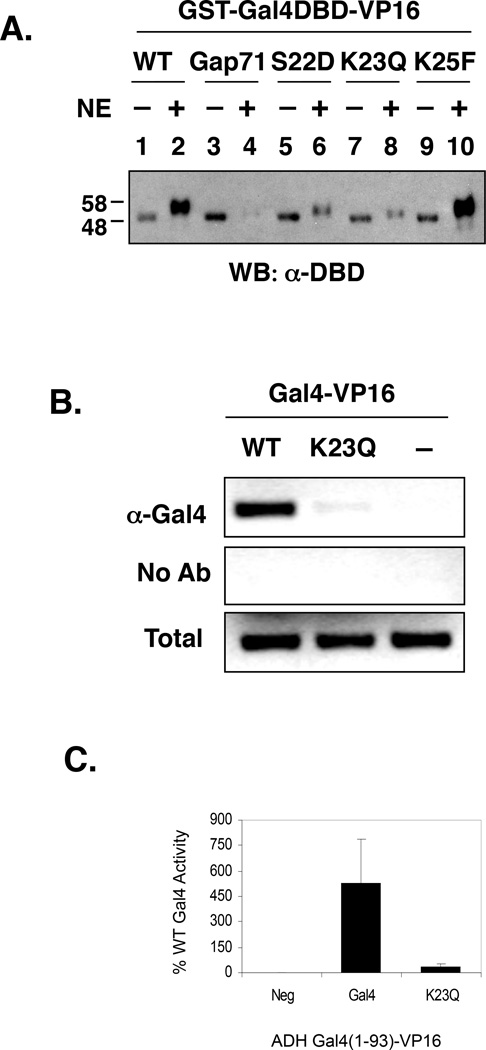
Effect of point mutations in the Gal4 DNA-binding domain on the activity of GST-Gal4 DBD-VP16 in vitro and in vivo. A. Effect of point mutations in the DNA-binding domain on GST-Gal4–VP16-DNA binding in vitro in the presence of HeLa NE. GST Gal4DBD-VP16 constructs were isolated as immobilized activator-DNA complexes (see Fig. 5 caption) and an approximately equivalent amount of each activator-DNA complex (compare lanes without NE) was incubated with NE and ATP for 30 minute at 30 °C. The material retained on the beads was re-isolated and analyzed by SDS-PAGE and Western blotting using an anti-Gal4 DBD antibody. B. Promoter binding activities of Gal4–VP16 and Gal4–VP16 K23Q in vivo as monitored by the chromatin precipitation assay. Gal4–VP16 and Gal4(K23Q)-VP16 were expressed at high levels (multi-copy plasmid with an ADH promoter) in a Δgal4 Δgal80 yeast strain. An anti-amino-terminal Gal4 antibody was employed in the immunoprecipitation step. C. Activities of WT and the K23Q mutant Gal4–VP16 in vivo as determined by an α-galactosidase reporter gene assay.
The wild-type and Gal4(K23Q)-VP16 proteins were also expressed from a multi-copy plasmid carrying an ADH promoter in a Δgal4 yeast strain and their ability to bind the GAL1 promoter was assessed by a ChIP assay. As shown in Fig. 7B, binding of the wild-type DBD-containing protein was efficient, but the signal obtained in cells expressing the K23Q mutant was barely detectable above background, even at this high level of protein. As would be expected from this result, the activity of Gal4(K23Q)-VP16 in yeast was <10% that of the corresponding wild-type protein (Fig. 7C) We conclude that this mutation, which cripples both phosphorylation and ubiquitylation of Gal4–VP16, hypersensitizes Gal4–VP16 to the proteasomal ATPase-mediated destabilization activity in HeLa NE or yeast cells. Clearly, the chimeric activator is much more sensitive than is full-length Gal4, since overexpression of the native transactivator largely rescues promoter occupancy and activity (Figs 1 and 2).
Discussion
In a recent study we reported that the proteasomal ATPases have the previously unrecognized ability to destabilize activator-DNA complexes in a non-proteolytic fashion4. Furthermore, we demonstrated that mono-ubiquitylation of the DNA-binding domain (DBD) of the activator inhibits this reaction, allowing the activator to remain stably associated with cognate DNA sites in the presence of the proteasomal proteins4. A mutant of Gal4, called Gap71 (Gal4(S22D,K23Q,K25F))27, 28, was found to be hypersensitive to this activity due to the fact that it’s DNA-binding domain fails to be ubiquitylated efficiently. Here, we constructed and analyzed the properties of the corresponding single point mutants in vivo and in vitro in the context of the chimeric activators Gal4–VP16 and Gal4–Gal11, as well as native Gal4.
Several lines of experimentation showed that mutation of S22 or K23 reproduced the phenotype of the Gap71 triple mutant, whereas mutation of K25 had little or no effect. Given that mono-ubiquitylation of the activator is essential for allowing it to resist the destabilization activity of the proteasomal ATPases4, it was not surprising that one of the lysines would prove to be a key residue. Thus, K23 is either the site of mono-ubiquitylation in the DBD or is essential for a critical ubiquitylation elsewhere (vide infra). But the finding that S22 mutation also affects Gal4DBD ubiquitylation and the ability to resist “stripping” was a surprise. The intermediate activity of the S22D mutant between that of wild-type Gal4 and the S22A mutant suggested that this residue might be the site of phosphorylation. There are several well-studied cases in which serine or threonine phosphorylation necessarily precedes ubiquitylation (for reviews, see:44–47). We note that S22 phosphorylation does not require the presence of an activation domain (see Fig. 4C), whereas efficient mono-ubiquitylation of the Gal4DBD requires both a DNA template and the presence of an activation domain4. This raised the possibility that phosphorylation of S22 is essential for subsequent ubiquitylation. Supportive of this idea was the observation that the S22A derivative was consistently less active than the S22D mutant in a variety of assays (DNA-binding, ubiquitylation, reporter gene activity, etc.). Aspartate can sometimes act as a partial mimic of phosphoserine because of its negative charge. Therefore, we asked directly if the Gal4DBD was phosphorylated. As was shown in Fig. 4, both GST-Gal4 DBD and GST-Gal4–VP16 were phosphorylated in the presence of HeLa nuclear extract, but GST was not (Fig. 4C). Mutation of S22 strongly decreased the level of phosphorylation (Fig. 4D). We conclude that S22 is a critical site of phosphorylation in the Gal4DBD that is essential for subsequent ubiquitylation (Fig. 5), efficient promoter occupancy (Fig. 2) and full transactivation function (Fig. 1).
There have been several previous reports of Gal4 phosphorylation, but these were at sites in the C-terminal portion of the protein29, 48, 49. For example, Sadowski and co-workers showed that the Srb10 kinase, a RNA polymerase II holoenzyme component, targets S699 of Gal449 and identified S837 as another site of phosphorylation29 by an unknown kinase. In both cases, phosphorylation is correlated with activation29, 31 but is not required for Gal4 activity. More recently, Tansey and co-workers published evidence suggesting that phosphorylation of a C-terminal serine is correlated with enhanced expression of GAL genes, but again abolishing this event did not compromise transcription31. Thus, our discovery of the essential role of S22 in Gal4-DNA interactions constitutes the first example of a critical phosphorylation event in this paradigmatic activator.
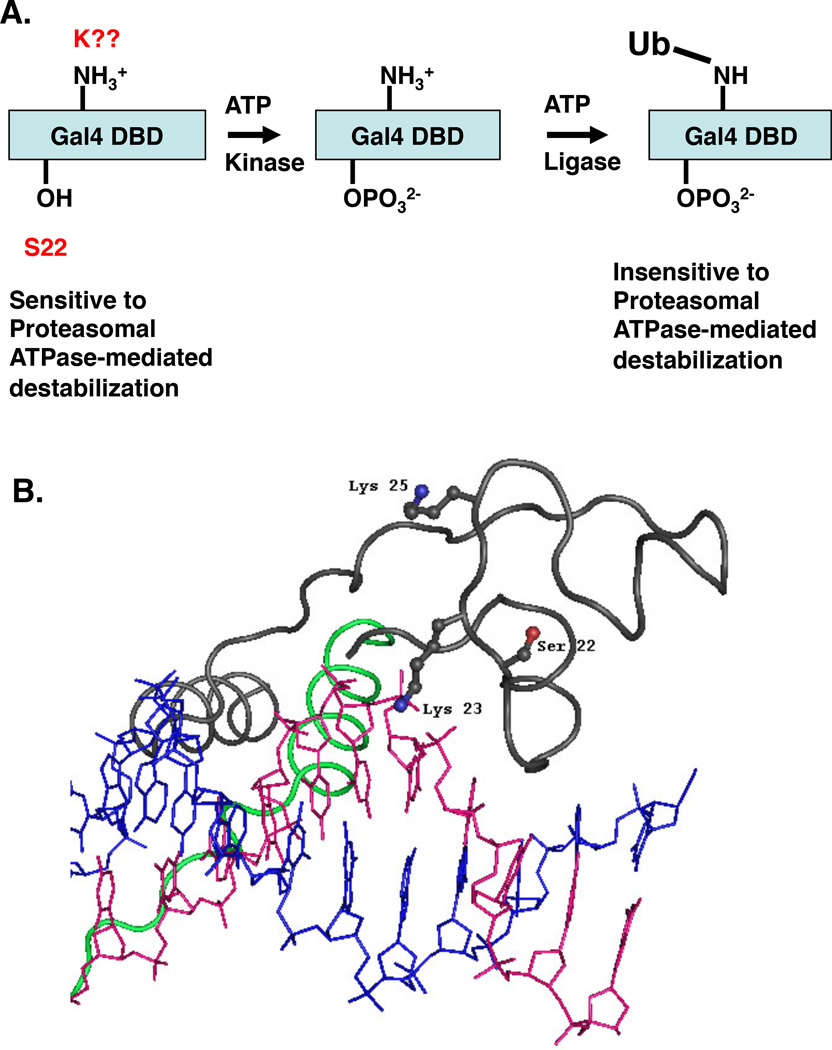
At present, the kinase that mediates phosphorylation of S22 is unknown. We have tested the ability of yeast strains deleted for all known, non-essential, nuclear kinases to grow on media containing galactose as the sole carbon source. No growth defect was observed for any of these strains (M.O. and T.K., unpublished results), indicating that the kinase is encoded by an essential gene, has not previously been documented as a nuclear protein or that more than one kinase can target the Gal4 S22 residue. The ExPASy tool NetPhosYeast predicts the phosphorylation of serine and threonine residues based on established kinase recognition sites. Based on this software program Gal4 Ser22 is most likely being modified by an uncharacterized kinase. Future studies will focus on function-based assays designed to identify this protein kinase. These efforts may be aided by some of the information reported here. For example, as shown in Fig. 4C, the presence of DNA containing Gal4 binding sites, while not essential, stimulates S22 phosphorylation, suggesting that binding to the promoter alters the conformation of Gal4 subtly, thus making it a better substrate for the kinase, or that the kinase is DNA-associated. Alternatively, it could be that the DNA serves as part of the kinase recognition site itself. Serine 22 could contribute to this interaction considering it is very close to the nucleic acid in the crystal structure of the Gal4(1–65)-DNA complex26 (Fig. 8B).
Figure 8.
A model for coupled Gal4 DNA-binding domain phosphorylation and mono-ubiquitylation. A. Schematic representation of the proposed post-translational modifications of the Gal4 DBD. Gal4 is first phosphorylated on S22, which then triggers mono-ubiquitylation of a lysine residue within the Gal4 DBD. Since K23 is required for phosphorylation of S22 and this phosphorylation is, in turn, essential for ubiquitylation, the lack of ubiquitylation of Gal4 K23Q is consistent with our model. B. Graphic showing the location of the three residues mutated in Gap71 in the context of the Gal4 DBD-DNA crystal structure26.
It was also of interest to find that the K23Q mutation, in addition to abolishing Gal4DBD ubiquitylation (Fig. 5), also strongly inhibited S22 phosphorylation (see Fig. 4D). Phosphorylation is clearly essential for subsequent ubiquitylation of the Gal4DBD, as evidenced by the failure of the S22A mutant to be ubiquitylated and the weak ubiquitylation of the S22D mutant (Fig. 5). Thus, whereas K23 is an obvious candidate for the residue that is conjugated to ubiquitin, another possibility is that K23 is instead (or additionally) required for S22 phosphorylation. For example, K23 could be an essential residue for kinase recognition.
To probe this point, the activity of a Gal4 K23R mutant was examined. We found that the transactivation and DNA-binding activity of this protein is at least as high as that of wild-type Gal4 (Fig. 6). Thus, we favor the idea that K23 is not the direct site of Gal4 DBD mono-ubiquitylation, but that S22 phosphorylation requires a positively charged residue at the adjacent position. Since Gal4 DBD mono-ubiquitylation will not occur unless S22 is phosphorylated, this model would explain the very different activities of K23Q and K23R. However, we cannot absolutely rule out the idea that a positive charge at residue 23 is required for phosphorylation and that K23 is also the site of ubiquitylation in the wild-type protein. It could be that the as yet unknown E3 ubiquitin ligase that targets the Gal4 DBD normally employs K23 as a ubiquitin acceptor, but is capable of utilizing a different lysine if this residue is mutated. Note that we have shown that genetic fusion of a single ubiquitin protein to the N-terminus of Gal4 partially reconstitutes the activity of Gap714, so it cannot be the case that the stereochemical placement of the mono-ubiquitin residue is critical. To resolve this issue, direct biochemical mapping of the ubiquitylation site on the wild-type Gal4 DBD will be required. Such efforts are in progress, but this is a technically demanding task that has so far been accomplished only rarely for gene-specific transcription factors50. This may also be complicated by the possibility that Gal4 is ubiquitylated on more than one residue, as is the case for p5350.
In summary, we have presented evidence of the first example of a critical phosphorylation event of the paradigmatic transactivator Gal4. Phosphorylation of S22 is essential for efficient subsequent mono-ubiquitylation of a lysine residue in the Gal4 DBD and thus is an important event in achieving promoter occupancy and full transactivation activity (Fig 8A).
Experimental Procedures
Plasmids and Yeast
S103-Gal4 and S103-Gap71 are under the native Gal4 promoter in the single copy vector pSB32 and also the multi-copy vector Yep351. The Gal4(1–147)-Gal11(799–1082) construct are in Yep351 under the native Gal4 promoter. pHCA Gal4(1–93)-VP16 was a generous gift from Dr. D. Picard52. Specific mutant Gal4 proteins were generated by site-directed PCR mutagenesis of pSB32 S103-Gal4, YEp351 Gal4–Gal11, or pHCA Gal4–VP16 using the following oligo pairs:
| Oligo Pair | Forward | Reverse |
|---|---|---|
| S22A | GCTCAAGTGCGCCAAAGAAAAACCGAAGTGCGCC | GGCGCACTTCGGTTTTTCTTTGGCGCACTTGAGC |
| S22D | GCTCAAGTGCGACAAAGAAAAACCGAAGTGCGCC | GGCGCACTTCGGTTTTTCTTTGTCGCACTT GAGC |
| K23Q | GCTCAAGTGCTCCCAAGAAAAACCGAAGTGCGCC | GGCGCACTTCGGTTTTTCTTGGGAGCACTTGAGC |
| K23R | GGCGCACTTCGGTTTTTCTCGGGAGCACTTGAGC | GCTCAAGTGCTCCCGAGAAAAACCGAAGTGCGCC |
| K25F | GCTCAAGTGCTCCAAAGAATTTCCGAAGTGCGCC | GGCGCACTTCGGAAATTCTTTGGAGCACTTGAGC |
Full-length gal4 mutants from the pSB32 vectors were then cloned into the YEp351 plasmid to generate the corresponding panel of multi-copy plasmids. The mutations were confirmed by sequencing (data not shown). Saccharomyces cerevisiae strains TKY54 (MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 S103-SUG1 gal4::HIS3) and TKY226 (MATa Δgal4 ura3-52 leu2 MEL1) were individually transformed with the pSB32 and Yep351 plasmid sets. The strain YJ0Z (Δgal4 Δgal80 ura3-52 leu2-3,112 his3 ade1 MEL1 GAL1::lacZ fusion)51 was transformed by the pHCA plasmids.
Construction of expression vectors and purification of recombinant proteins
Bacterial expression vectors for GST and GST fusion Gal4–VP16, Gap71–VP16 and Gal4DBD (1–147) were described {Ferdous, 2007 #3702}. Expression vectors for K23Q, K25F, S22D and S22A mutant Gal4–VP16 were generated by PCR mediated site-directed mutagenesis as described above. Plasmids pGEX-Gal4(S22D)-VP16 and pGEX-Gal4(K23Q)-VP16 were digested with EcoRI, purified from an agarose gel and then re-ligated to generate pGEX-Gal4(S22D)DBD and pGEX-Gal4(K23Q)DBD. His6-tagged ubiquitin was purified as described previously25. Expression and purification of each GST-tagged or His6-tagged protein from E. coli was done as before and purity was checked by SDS-gel electrophoresis25.
Preparation of HeLa nuclear extract
A pellet derived from a five liter culture of S3 HeLa cells was purchased from the National Cell Culture Center. HeLa Nuclear extract (NE) was prepared according to the Dignam method53. Protein concentration was determined with the Bradford method using bovine serum albumin (BSA) as a standard.
Antibodies and Western blotting
Mouse monoclonal anti-GST, anti-Gal4DBD-HRP and polyclonal anti-His5 antibodies were from Santa Cruz. Polyclonal anti-Gal4 antibody was isolated from a rabbit immunized with a C-terminal or an amino-terminal fragment of the protein. Mouse polyclonal anti-ubiquitin antibody was generated by genetic immunization as described54. Western blotting was performed as described25.
Reporter gene assay
TKY226 was transformed by the indicated plasmids for the detection of MEL activity. Cells were grown in raffinose conditions at 30 °C until they reached to an OD600 of 0.6. The cells were then induced with galactose for 30 minutes and a 50 ml aliquot was collected. Cells were disrupted by vortexing with zirconia/silica beads at 4° C in citric acid buffer (31mM citric acid, 39mM KH2PO4, pH 4.0). Protein concentration was measured by Coomassie Plus protein assay (Pierce, Rockford, IL). Equal protein amounts were added to each assay. Transcriptional activity of WT and mutant activators were analyzed by an α-galactosidase assay previously described55. The graphs represent the data from three individual experiments.
Chromatin Immunoprecipitation (ChIP) assay
Occupancy of the GAL1 promoter was analyzed in the yeast strain TKY54 by ChIP assay as described18. Samples were collected after the cells had grown to an OD600 of 0.6 on raffinose. The cultures were subsequently induced with galactose for 30 minutes and samples were collected. The exception is the yeast strains expressing the Gal4–VP16 constructs. These did not undergo galactose based induction. Poly-clonal anti-Gal4 antibody specific to the carboxy terminus was used for samples with full-length protein. Poly-clonal anti–Gal4 antibody specific to the amino terminus was used for the Gal4–VP16 and Gal4–Gall1 based panel of constructs. ‘No antibody’ controls were done and are considered in the graphed data. Control antibody experiments were previously done with non-Gal antibodies and show a pattern similar to our ‘no antibody’ controls (data not shown). Promoter occupancy of wild-type and mutant Gal4 samples were analyzed by amplifying the DNA fragment corresponding to upstream three Gal4 sites was described18.
Quantitative Real-time PCR
Amount of DNA precipitated from the UASG region was quantitated by qPCR using SYBR Green kit (Qiagen) on an i-Cycler (Bio-Rad ABI Prism 7700) and analyzed by ΔΔCT method {Livak, 2001 #3898}. Occupancy Wt Gal4 in non-inducing condition was taken as 100%. The graph shows the relative amount of DNA immunoprecipitated by the anti-Gal4 antibody after subtracting out the no antibody background. ChIPs were performed three times in triplicate. Error bars represent the standard error of the mean.
Analysis of protein phosphorylation
Reactions were performed in transcription buffer: 10% glycerol, 10mM Hepes pH 7.8, 50 mM KCl, 6.25 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, and 1 mM PMSF. The reaction mixture (30 µl) contained 375 nM GST, 75 nM GST fusion proteins, 100 µM cold ATP, 40 ug of HeLa NE, 1 µl of [γ-32P]-ATP (3000 Ci/mmol, ICN) and 0.2 µM/µl of okadaic acid in transcription buffer and was incubated for 20 minutes in a 30°C water bath. An aliquot (5 µl) was removed to analyze the overall kinase reaction and the recombinant proteins in the remaining reaction (25 µl) were purified on glutathione-sepharose beads in PBS-500. The beads were dissolved in SDS loading buffer, boiled for 5 minutes and then loaded on a 10% SDS-polyacrylamide gel. Proteins were transferred to PVDF membrane and protein phosphorylation was analyzed and quantified. The membrane was then probed with anti-GST or anti-DBD antibodies.
Isolation of the activator-DNA complex, its subsequent destabilization in HeLa NE, and the ubiquitylation status of the protein were carried out as described4.
Acknowledgments
This work was supported by the NIH (GM-71833). Kip Nalley was supported by an NIH Cardiology Training Grant Fellowship (HL-07360).
References
- 1.Alberts B. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 2.Brady KL, Ponnampalam SN, Bumbulis MJ, Setzer DR. J. Biol. Chem. 2005;280:26743–26750. doi: 10.1074/jbc.M502677200. [DOI] [PubMed] [Google Scholar]
- 3.Yang E, Henriksen MA, Sachaefer O, Zakharova N, Darnell JE. J. Biol. Chem. 2002;277:13455–13462. doi: 10.1074/jbc.M112038200. [DOI] [PubMed] [Google Scholar]
- 4.Ferdous A, Sikder D, Gillette TG, Nalley K, Kodadek T, Johnston SA. Genes & Dev. 2007;20:112–123. doi: 10.1101/gad.1493207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Archer CT, Delahodde A, Gonzalez F, Johnston SA, Kodadek T. J. Biol. Chem. 2008;283:12614–12623. doi: 10.1074/jbc.M801050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tansey W. Genes & Dev. 2001;15:1045–1050. doi: 10.1101/gad.896501. [DOI] [PubMed] [Google Scholar]
- 7.Collins GA, Tansey WP. Curr. Op. In Gentics & Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Hager GL, Nagaich AK, Johnson TA, Walker DA, John S. Biochim. Biophys. Acta. 2004;1677:46–51. doi: 10.1016/j.bbaexp.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Kodadek T, Sikder D, Nalley K. Cell. 2006;127:261–264. doi: 10.1016/j.cell.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Yu P, Kodadek T. J. Biol. Chem. 2007;282 doi: 10.1074/jbc.M707557200. In press. [DOI] [PubMed] [Google Scholar]
- 11.Métivier R, Penot G, Hübner MR, Reid G, Brand H, Ko M, Gannon F. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 12.Reid G, Hübner MR, Metivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F. Mol. Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 13.McNally JG, Müller WG, Walker D, Wolford R, Hager GL. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 14.Nalley K, Johnston SA, Kodadek T. Nature. 2006;442:1054–1057. doi: 10.1038/nature05067. [DOI] [PubMed] [Google Scholar]
- 15.Yao J, Munson KM, Webb WW, Lis JT. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- 16.Baumeister W, Walz J, Zuhl F, Seemuller E. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 17.Saeki Y, Toh-e A, Yokosawa H. Biochem. Biophys. Res. Comm. 2000;273:509–515. doi: 10.1006/bbrc.2000.2980. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez F, Delahodde A, Kodadek T, Johnston SA. Science. 2002;296:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- 19.Archer C, Burdine L, Kodadek T. Molecular BioSystems. 2005;1:366–372. doi: 10.1039/b510019d. [DOI] [PubMed] [Google Scholar]
- 20.Salghetti SE, Caudy AA, Chenoweth JG, Tansey WP. Science. 2001;293:1651–1653. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- 21.Bres V, Kiernan RE, Linares LK, Chable-Bessia C, Plechakova O, Treand C, Emiliani S, Peloponese J-M, Jeang K-T, Coux O, Scheffner M, Benkirane M. Nature Cell Biol. 2003;5:754–761. doi: 10.1038/ncb1023. [DOI] [PubMed] [Google Scholar]
- 22.Greer SF, Zika E, Conti B, Zhu XS, Ting JP. Nature Immunol. 2003;4:1074–1082. doi: 10.1038/ni985. [DOI] [PubMed] [Google Scholar]
- 23.Kurosu T, Peterlin BM. Curr. Biol. 2004;14:1112–1116. doi: 10.1016/j.cub.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 24.Archer CT, BUrdine L, Liu B, Ferdous A, Johnston SA, Kodadek T. J. Biol. Chem. 2008;283 doi: 10.1074/jbc.M803075200. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferdous A, Gonzalez F, Sun L, Kodadek T, Johnston SA. Mol. Cell. 2001;7:981–991. doi: 10.1016/s1097-2765(01)00250-7. [DOI] [PubMed] [Google Scholar]
- 26.Marmorstein R, Carey M, Ptashne M, Harrison SC. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 27.Corton JC, Johnston SA. Nature. 1989;340:724–727. doi: 10.1038/340724a0. [DOI] [PubMed] [Google Scholar]
- 28.Corton JC, Moreno E, Johnston SA. J. Biol. Chem. 1998;273:13776–13780. doi: 10.1074/jbc.273.22.13776. [DOI] [PubMed] [Google Scholar]
- 29.Sadowski I, Neidbala D, Wood K, Ptashne M. Proc Natl Acad Sci U S A. 1991;88:10510–10514. doi: 10.1073/pnas.88.23.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parthun MR, Jaehning JA. Mol Cell Biol. 1992;12:4981–4987. doi: 10.1128/mcb.12.11.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muratani M, Kung C, Shokat KM, Tansey WP. Cell. 2005;120:887–899. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 32.Xu H, Kodadek T, Johnston SA. Proc. Natl. Acad. Sci. USA. 1995;92:7677–7680. doi: 10.1073/pnas.92.17.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vashee S, Xu H, Johnston SA, Kodadek T. J. Biol. Chem. 1993;268:24699–24706. [PubMed] [Google Scholar]
- 34.Johnston SA, Salmeron JM, Jr, Dincher SS. Cell. 1987;50:143–146. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- 35.Sakurai H, Hiraoka Y, Fukasawa T. Proc. Natl. Acad. Sci. USA. 1993;90:8382–8386. doi: 10.1073/pnas.90.18.8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaudreau L, Adam M, Ptashne M. Mol. Cell. 1998;1:913–916. doi: 10.1016/s1097-2765(00)80090-8. [DOI] [PubMed] [Google Scholar]
- 37.Gaudreau L, Keaveney M, Nevado J, Zaman Z, Bryant GO, Struhl K, Ptashne M. Proc. Natl. Acad. Sci. USA. 1999;96:2688–2693. doi: 10.1073/pnas.96.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeong C-J, Yang S-H, Xie Y, Zhang L, Johnston SA, Kodadek T. Biochemistry. 2001;40:9421–9427. doi: 10.1021/bi010011k. [DOI] [PubMed] [Google Scholar]
- 39.Lee YC, Park JM, Min S, Han SJ, Kim Y-J. Mol. Cell. Biol. 1999;19:2967–2976. doi: 10.1128/mcb.19.4.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kornberg RD. Trends Biochem. Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 41.Sarcevic B, Mawson A, Baker RT, Sutherland RL. Embo J. 2002;21:2009–2018. doi: 10.1093/emboj/21.8.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gburcik V, Bot N, Maggiolini M, Picard D. Mol. Cell. Biol. 2005;25:3421–3430. doi: 10.1128/MCB.25.9.3421-3430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ. Genes & Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Mol. Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 45.Ravid T, Hochstrasser M. Curr Biol. 2004;14:R898–R900. doi: 10.1016/j.cub.2004.09.074. [DOI] [PubMed] [Google Scholar]
- 46.Brooks CL, Gu W. Curr. Op. Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 47.Joazeiro CA, Weissman AM. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 48.Mylin LM, Bhat JP, Hopper JE. Genes & Dev. 1989;3:1157–1165. doi: 10.1101/gad.3.8.1157. [DOI] [PubMed] [Google Scholar]
- 49.Hirst M, Kobor MS, Kuriakose N, Greenblatt J, Sadowski I. Mol. Cell. 1999;3:673–678. doi: 10.1016/s1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez MS, Desteroo JMP, Lain S, Lane DP, Hay RT. Mol. Cell. Biol. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leuther KK, Johnston SA. Science. 1992;256:1333–1335. doi: 10.1126/science.1598579. [DOI] [PubMed] [Google Scholar]
- 52.Louvion JF, Havaux-Copf B, Picard D. Gene. 1993;131:129–134. doi: 10.1016/0378-1119(93)90681-r. [DOI] [PubMed] [Google Scholar]
- 53.Dignam JD, Lebovitz RM, Roeder RG. Nuc. Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chambers RS, Johnston SA. Nature Biotech. 2003;21:1088–1092. doi: 10.1038/nbt858. [DOI] [PubMed] [Google Scholar]
- 55.Johnston SA, Hopper JE. Proc. Natl. Acad. Sci. USA. 1982;79:6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]