Abstract
Strand displacement amplification is an isothermal DNA amplification reaction based on a restriction endonuclease nicking its recognition site and a polymerase extending the nick at its 3' end, displacing the downstream strand. The reaction resembles rolling-circle replication of single-stranded phages and small plasmids. The displaced sense strand serves as target for an antisense reaction and vice versa, resulting in exponential growth and the autocatalytic nature of this in vitro reaction as long as the template is the limiting agent. We describe the optimization of strand displacement amplification for in vitro evolution experiments under serial transfer conditions. The reaction was followed and controlled by use of the fluorescent dye thiazole orange binding to the amplified DNA. We were able to maintain exponential growth conditions with a doubling time of 3.0 min throughout 100 transfers or approximately 350 molecular generations by using an automatic handling device. Homology of in vitro amplification with rolling-circle replication was mirrored by the occurring evolutionary processes. Deletion events most likely caused by a slipped mispairing mechanism as postulated for in vivo replication took place. Under our conditions, the mutation rate was high and a molecular quasi-species formed with a mutant lacking internal hairpin formation ability and thus outgrowing all other species under dGTP/dCTP deficiency.
Full text
PDF
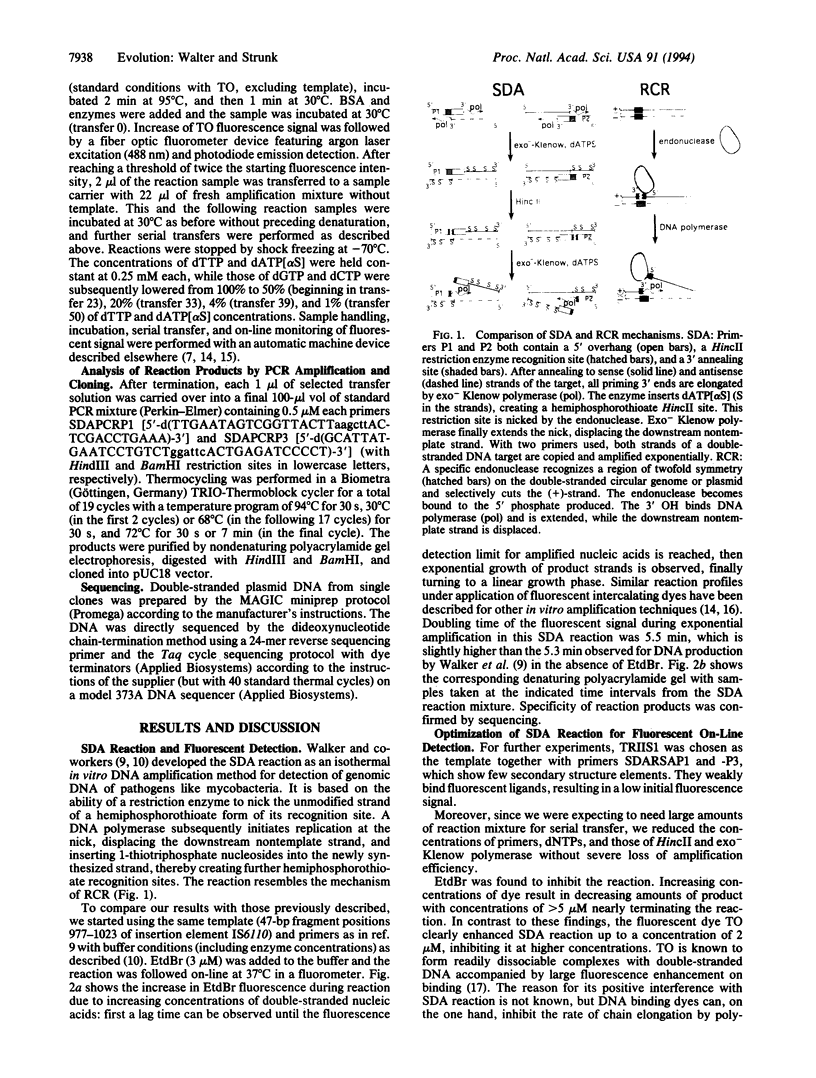
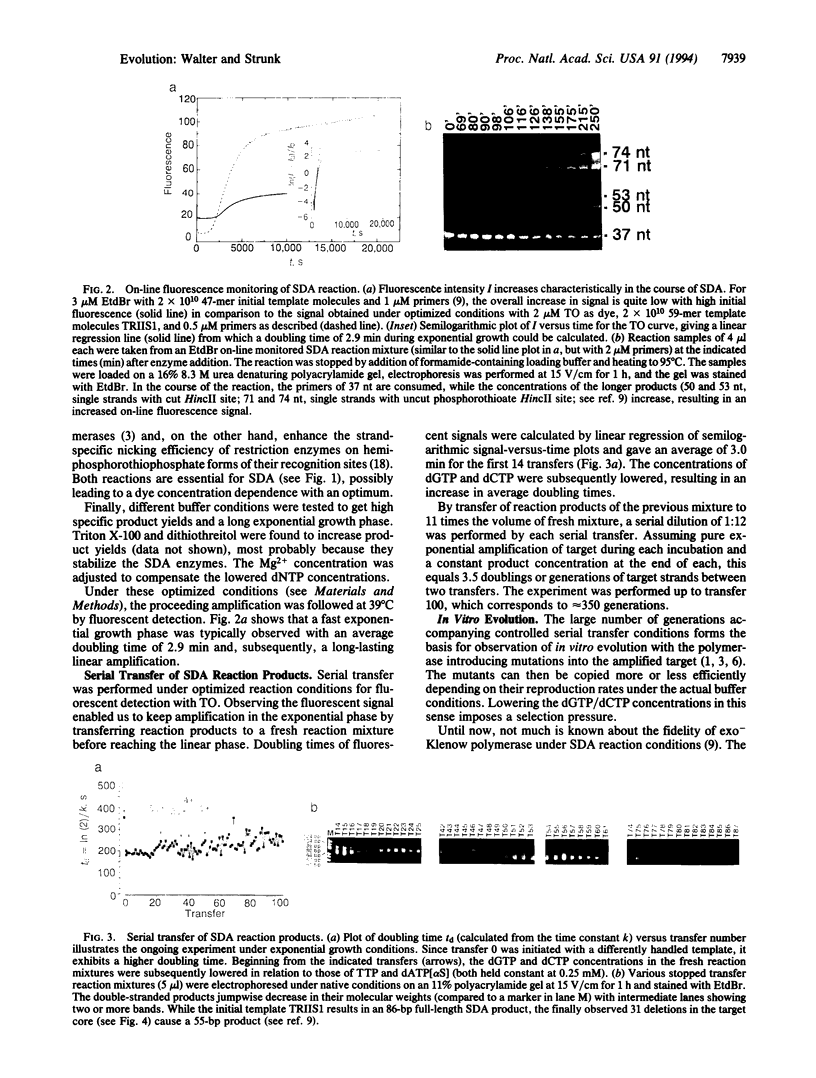


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Bebenek K., Joyce C. M., Fitzgerald M. P., Kunkel T. A. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J Biol Chem. 1990 Aug 15;265(23):13878–13887. [PubMed] [Google Scholar]
- Biebricher C. K., Eigen M., Gardiner W. C., Jr Kinetics of RNA replication: competition and selection among self-replicating RNA species. Biochemistry. 1985 Nov 5;24(23):6550–6560. doi: 10.1021/bi00344a037. [DOI] [PubMed] [Google Scholar]
- Biebricher C. K., Luce R. In vitro recombination and terminal elongation of RNA by Q beta replicase. EMBO J. 1992 Dec;11(13):5129–5135. doi: 10.1002/j.1460-2075.1992.tb05620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire V., Freemont P. S., Sanderson M. R., Beese L., Friedman J. M., Joyce C. M., Steitz T. A. Genetic and crystallographic studies of the 3',5'-exonucleolytic site of DNA polymerase I. Science. 1988 Apr 8;240(4849):199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M. The origin of genetic information: viruses as models. Gene. 1993 Dec 15;135(1-2):37–47. doi: 10.1016/0378-1119(93)90047-7. [DOI] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Fockler C., Dollinger G., Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (N Y) 1993 Sep;11(9):1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- Horiuchi K. Co-evolution of a filamentous bacteriophage and its defective interfering particles. J Mol Biol. 1983 Sep 15;169(2):389–407. doi: 10.1016/s0022-2836(83)80057-6. [DOI] [PubMed] [Google Scholar]
- Kramer F. R., Mills D. R., Cole P. E., Nishihara T., Spiegelman S. Evolution in vitro: sequence and phenotype of a mutant RNA resistant to ethidium bromide. J Mol Biol. 1974 Nov 15;89(4):719–736. doi: 10.1016/0022-2836(74)90047-3. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Eckstein F., Mildvan A. S., Koplitz R. M., Loeb L. A. Deoxynucleoside [1-thio]triphosphates prevent proofreading during in vitro DNA synthesis. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6734–6738. doi: 10.1073/pnas.78.11.6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills D. R., Kramer F. R., Dobkin C., Nishihara T., Speigelman S. Nucleotide sequence of microvariant RNA: another small replicating molecule. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4252–4256. doi: 10.1073/pnas.72.11.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills D. R., Peterson R. L., Spiegelman S. An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc Natl Acad Sci U S A. 1967 Jul;58(1):217–224. doi: 10.1073/pnas.58.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters B. P., de Boer J. H., Bron S., Venema G. Structural plasmid instability in Bacillus subtilis: effect of direct and inverted repeats. Mol Gen Genet. 1988 Jun;212(3):450–458. doi: 10.1007/BF00330849. [DOI] [PubMed] [Google Scholar]
- Rye H. S., Quesada M. A., Peck K., Mathies R. A., Glazer A. N. High-sensitivity two-color detection of double-stranded DNA with a confocal fluorescence gel scanner using ethidium homodimer and thiazole orange. Nucleic Acids Res. 1991 Jan 25;19(2):327–333. doi: 10.1093/nar/19.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers J. R., Schmidt W., Wendler A., Eckstein F. Strand specific cleavage of phosphorothioate-containing DNA by reaction with restriction endonucleases in the presence of ethidium bromide. Nucleic Acids Res. 1988 Feb 11;16(3):803–814. doi: 10.1093/nar/16.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spargo C. A., Haaland P. D., Jurgensen S. R., Shank D. D., Walker G. T. Chemiluminescent detection of strand displacement amplified DNA from species comprising the Mycobacterium tuberculosis complex. Mol Cell Probes. 1993 Oct;7(5):395–404. doi: 10.1006/mcpr.1993.1058. [DOI] [PubMed] [Google Scholar]
- Spiegelman S., Haruna I., Holland I. B., Beaudreau G., Mills D. The synthesis of a self-propagating and infectious nucleic acid with a purified enzyme. Proc Natl Acad Sci U S A. 1965 Sep;54(3):919–927. doi: 10.1073/pnas.54.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilette D., Uzest M., Ehrlich S. D., Michel B. DNA transcription and repressor binding affect deletion formation in Escherichia coli plasmids. EMBO J. 1992 Oct;11(10):3629–3634. doi: 10.1002/j.1460-2075.1992.tb05447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. T. Empirical aspects of strand displacement amplification. PCR Methods Appl. 1993 Aug;3(1):1–6. doi: 10.1101/gr.3.1.1. [DOI] [PubMed] [Google Scholar]
- Walker G. T., Fraiser M. S., Schram J. L., Little M. C., Nadeau J. G., Malinowski D. P. Strand displacement amplification--an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992 Apr 11;20(7):1691–1696. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. T., Little M. C., Nadeau J. G., Shank D. D. Isothermal in vitro amplification of DNA by a restriction enzyme/DNA polymerase system. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):392–396. doi: 10.1073/pnas.89.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston-Hafer K., Berg D. E. Palindromy and the location of deletion endpoints in Escherichia coli. Genetics. 1989 Apr;121(4):651–658. doi: 10.1093/genetics/121.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder N. D., Boeke J. D. The filamentous phage (Ff) as vectors for recombinant DNA--a review. Gene. 1982 Jul-Aug;19(1):1–10. doi: 10.1016/0378-1119(82)90183-4. [DOI] [PubMed] [Google Scholar]