Abstract
We studied the role of European rabbits (Oryctolagus cuniculus) as a reservoir for Coxiella burnetii in the Iberian region. High individual and population seroprevalences observed in wild and farmed rabbits, evidence of systemic infections, and vaginal shedding support the reservoir role of the European rabbit for C. burnetii.
Keywords: Coxiella burnetii, bacteria, lagomorph, Q fever, European rabbits, Oryctolagus cuniculus, reservoir, wildlife, zoonoses, Iberian region, Europe
Wildlife play a major role in the maintenance and transmission of multihost pathogens (1,2). Understanding the role of host species involved in multihost zoonotic pathogen maintenance and transmission is essential to prevent disease caused by these pathogens.
Coxiella burnetii, which is the cause of Q fever, is a zoonotic pathogen that infects multiple hosts (3). The implication of wildlife in the life cycle of C. burnetii has been reported worldwide (4,5), and wildlife might act as a source for humans infections (6,7).
European rabbits (Oryctolagus cuniculus) are native to the Iberian Peninsula and have been introduced into Australia, New Zealand, Chile, and Argentina (8). Domestic varieties of European rabbits are farmed worldwide. Specific ecologic traits (high density, gregarious behavior, high reproductive rate) suggest that these rabbits might become a major reservoir of zoonotic pathogens. However, whether C. burnetii can infect, replicate in, and be shed by European rabbits and contaminate the environment is not known. In this study, we investigated the role of these rabbits in a region to which Q fever is endemic.
The Study
Serum samples were collected from European wild rabbits in 13 locations in Spain, Portugal, and the Chafarinas Islands during 2003–2013 (Figure 1). Wild rabbits from 1 of the study locations (LO; Figure 1) were obtained from 2 epidemiologic scenarios (10). The first scenario involved rabbits that coexisted with farmed red deer (Cervus elaphus) (sites A and B). The second scenario involved rabbits that had not been in contact with ruminants since 2002 (site C).
Figure 1.
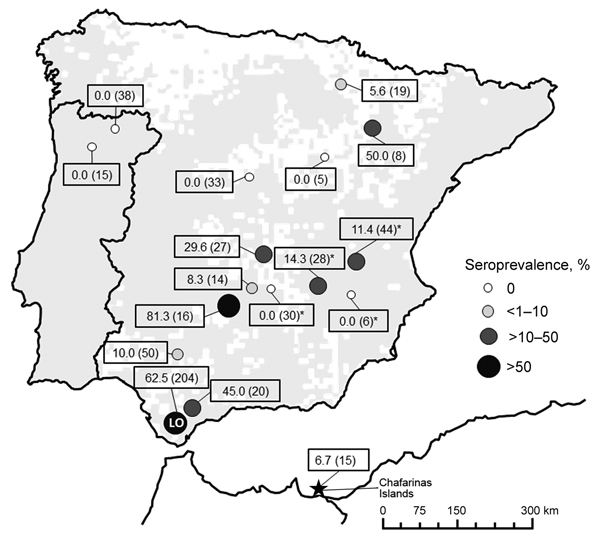
Seroprevalence of Coxiella burnetii (sample size) in wild and farmed European rabbits (Oryctolagus cuniculus), Iberian Peninsula and Chafarinas Islands. The distribution area of wild rabbits in the Iberian Peninsula (10 x 10 km Universal Transverse Mercator squares) is shown (gray shading) according to Mitchel-Jones et al. (9). LO sampling location is indicated. *Rabbit farm.
In addition to serum samples, spleen, uterus, and mammary gland samples and vaginal and uterus swab specimens were collected from rabbits surveyed at location LO. Each rabbit from this location was weighed and sexed. Serum samples were also collected from farmed rabbits on 4 farms in Spain (Figure 1). Samples were stored at −20°C until tested.
Serum samples were analyzed by using the LSIVet Ruminant Q Fever Serum/Milk ELISA Kit (Life Technologies, Carlsbad, CA, USA) and horseradish peroxidase–conjugated protein G (Sigma-Aldrich, St. Louis, MO, USA) as secondary antibody (10). Results were interpreted according to manufacturer’s recommendations.
DNA from tissues and swab specimens was extracted bu using the DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany). Swabs were incubated at 56°C for 30 min in 200 μL of AL buffer containing 20 μL of proteinase K. Swabs specimens were then vortexed for 15 s and removed. The remaining solution was incubated at 56°C for 30 min. The manufacturer’s blood extraction protocol was then used. DNA aliquots were frozen at −20°C. Negative controls (nuclease-free water; Promega, Madison, WI, USA) were included during DNA extraction.
DNA samples were analyzed by using a conventional PCR (11). PCR products were visualized by electrophoresis in 1.2% agarose gels containing 0.1 μL/mL of GelRed Nucleic Acid Gel Stain (Biotium, Hayward, CA, USA).
Logistic regression models were used to test the effect of potential factors (Table) on the individual risk of exposure to C. burnetii. Individual ELISA results were included as response variables in the models and the location origin of rabbits was used as a random factor.
Table. Variables considered as potential risk factors and outputs (coefficient/statistic) of best fitted risk factor models for Coxiella burnetii exposure in European rabbits (Oryctolagus cuniculus), Iberian Peninsula and Chafarinas Islands*.
| Variable code | Variable, units | Cbsp | CbspLO | CbsplLO | CbrtLO |
|---|---|---|---|---|---|
| Intercept | NA | 67.776/4.98† | –037270.00‡ | –2,942.687/1.15‡ | 2,925.025/0.49‡ |
| X | Longitude, decimal degrees | § | ¶ | ¶ | ¶ |
| Y | Latitude, decimal degrees | § | ¶ | § | ¶ |
| Ye | Year | −0.033/0.20‡ | § | 1.464/0.42‡ | −1.453/0.45‡ |
| Se | Season | § | § | § | § |
| Sp | Spring | 1.209/5.45‡ | § | −1.583/2.78‡ | § |
| Su | Summer | 2.257/5.45† | § | Referent | § |
| Au | Autumn | 0.043/5.45‡ | § | § | § |
| Wi | Winter | Referent | § | § | § |
| Mg | Management, wild vs. farmed | § | ¶ | ¶ | ¶ |
| Rum | Ruminants, presence vs. absence | ¶ | § | 0.059/0.0‡ | 2.004/1.08‡ |
| Sex | Sex, M vs. F | ¶ | § | −0.383/0.27‡ | 2.004/0.22‡ |
| Wg | Weight, g | ¶ | § | § | § |
*Cbsp,overall seropositivity; CbspLO, seropositivity at location LO; CbsplLO, systemic infection (C. burnetii DNA in spleen of wild rabbits); CBrtLO, shedding (C. burnetii DNA in reproductive tracts of wild rabbits); NA, not applicable. †p<0.05. ‡p>0.05. §Variable was included in each model but was not retained in the best model. ¶Variable not tested.
Logistic regression models were also used for individual exposure of rabbits from location LO to C. burnetii (ELISA), for the presence/absence of C. burnetii DNA in spleen (a proxy of systemic infection), and for the presence/absence of C. burnetii DNA in the reproductive tract (a proxy of shedding; including PCR results from uterus, and vaginal and uterus swab specimens). Location LO was included as a random factor, and sex, weight and ruminant presence/absence were also included as predictor variables (Table). Models were created by using a forward stepwise procedure. The model with the lowest Akaike information criterion (12) was selected.
Statistical analyses were performed in SPSS version 20.0 (IBM, Armonk, NY, USA). Prevalence-associated, Clopper-Pearson exact 95% CIs were estimated.
Serum samples from 572 rabbits (464 wild and 108 farmed) (Figure 1) were analyzed. Overall seroprevalence was 32.3% (95% CI 28.5%–36.4%) for wild and farmed rabbits, 37.9% (95% CI 33.5%–42.5%) for wild rabbits, and 8.3% (95% CI 3.8%–15.2) for farmed rabbits. Seroprevalence in wild rabbit populations ranged from 6.7% to 81.3%. Nine (64.3%) of 13 wild rabbit populations and 2 (50%) of 4 farms had >1 seropositive rabbit. The best model for C. burnetii exposure retained sampling year and season, and the risk for seropositivity was higher in summer (Table).
Seroprevalence at location LO was 65.2% (133/204; 95% CI 58.2%–71.7%); it was slightly lower at site C than at sites A and B (Figure 2, panel A). However, none of the considered factors were retained in the best model (Table). Six (4.4%; 95% CI 1.6%–9.4%) of 136 spleen samples analyzed at location LO were positive by PCR (4 male and 2 female rabbits). Five of the 6 spleen PCR–positive animals were seropositive. The 2 female rabbits were positive for C. burnetii DNA in vaginal swab specimens. Spleen PCR–positive rabbits were observed only at sites A and B (Figure 2, panel B).
Figure 2.
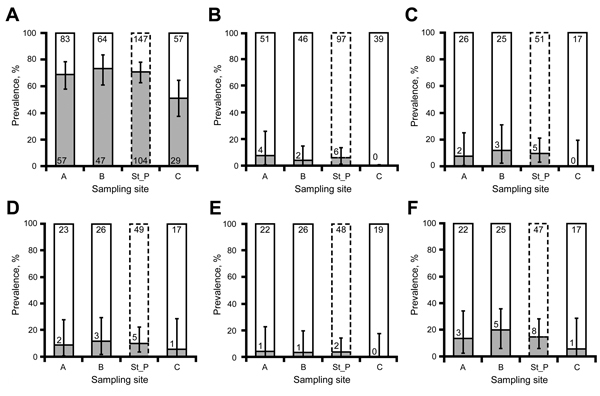
Prevalence of antibodies against Coxiella burnetii and C. burnetii DNA in European rabbits (Oryctolagus cuniculus) at sampling location LO, Iberian Peninsula. A) Antibodies; B) DNA in spleen; C) DNA in vaginal swab specimen; D) DNA in uterine swab specimen; E) DNA in uterus; F) DNA in reproductive tract (vaginal swab specimen, uterine swab specimen, uterus). Gray bars indicate seroprevalence. St_P indicates results for sites with ruminants (sites A and B); no ruminants were present at site C. Values at the top of bars indicate number of samples, and values at the bottom of bars indicate number of positive samples. Error bars indicate prevalence-associated exact 95% CIs.
The best model for the presence of C. burnetii DNA in spleen retained sampling year, season, presence of ruminants, and sex (Table). Results suggest expected higher systemic infection prevalence in rabbits coexisting with farmed red deer (Figure 2, panel B). C. burnetii DNA was detected in the reproductive tract of 9 (14.1%; 95% CI 6.6%–25.0%) of 64 female rabbits at sites A, B, and C (Figure 2, panel F). The presence of ruminants was retained in the best model for C. burnetii DNA in the reproductive tract (Table). None of the 13 mammary glands analyzed was positive for C. burnetii DNA.
Conclusions
This study provides 3 results that suggest that European rabbits might be reservoirs of C. burnetii. These 3 results are high seroprevalence of this bacteria; systemic infections; and bacterial shedding in vaginal secretions, which, in other host species, constitutes the main source for environmental contamination and transmission between species (13).
Host density is a major factor in C. burnetii prevalence in livestock (14). The highest seroprevalence values were observed at 2 locations where rabbit populations are managed for hunting purposes, which promotes high densities of rabbits. These findings suggest that rabbit density may be a major factor in the ecology of C. burnetii. In addition, the European rabbit is a gregarious species with a high reproductive rate. This rate favors transmission of C. burnetii from infected to susceptible animals, which is enhanced by replacement of C. burnetii–negative rabbits and can contribute further to spread of this bacterium in the environment.
The higher risk of exposure to C. burnetii observed during the summer might be related to increased indirect interaction with C. burnetii shed by coexisting ruminants, whose main shedding season is late spring–early summer (3). Inclusion of ruminants in the final models for systemic infection and vaginal shedding at location LO might support this hypothesis. However, further analyses, including molecular typing of circulating strains, would be needed to confirm the direction, frequency, and magnitude of interspecies interactions favoring transmission of C. burnetii.
Indirect transmission of C. burnetii between rabbits, humans, livestock, and other wild species may be enhanced in regions with high-density rabbit populations and in regions in which the European rabbit is a major game or farm species. Hunters, game keepers, rabbit farmers, veterinarians, wildlife researchers, livestock producers and livestock might be exposed to C. burnetii from rabbits (6,15). The European rabbit shows a high potential as a reservoir of C. burnetii for infection of livestock and humans in Europe.
Acknowledgments
We thank location LO farm managers, Tania Carta, María Martín, Christian Gortázar and José Antonio Ortiz for assistance during the study and Ursula Höfle for checking the English grammar of the paper.
This study was supported by European Union FP7 grant ANTIGONE (278976), European Union FP7 EMIDA ERA-NET grant APHAEA on wildlife disease surveillance in Europe, and Centro para el Desarrollo Tecnológico Industrial, Spanish Ministry for Economy and Competitiveness. F.-R-F. was supported by Juan de la Cierva and Ramón y Cajal contracts from the Spanish Ministry for Economy and Competitiveness.
Biography
Mr. González-Barrio is a doctoral student at the Spanish Wildlife Research Institute, Ciudad Real, Spain. His research interests are the epidemiology of pathogens transmitted between wildlife, livestock, and humans within a OneHealth approach; the epidemiology and diagnosis of C. burnetii infections; and development of infection control strategies for wildlife.
Footnotes
Suggested citation for this article: González-Barrio D, Maio E, Vieira-Pinto M, Ruiz-Fons F. Role of the European rabbits as reservoir for Coxiella burnetii. Emerg Infect Dis. 2015 Jun [date cited]. http://dx.doi.org/10.3201/eid2106.141537
References
- 1.Ruiz-Fons F, Segalés J, Gortázar C. A review of viral diseases of the European wild boar: effects of population dynamics and reservoir role. Vet J. 2008;176:158–69. 10.1016/j.tvjl.2007.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Viana M, Mancy R, Biek R, Cleaveland S, Cross PC, Lloyd-Smith JO, et al. Assembling evidence for identifying reservoirs of infection. Trends Ecol Evol. 2014;29:270–9. 10.1016/j.tree.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maurin M, Raoult D. Q fever. Clin Microbiol Rev. 1999;12:518–53 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ejercito CL, Cai L, Htwe KK, Taki M, Inoshima Y, Kondo T, et al. Serological evidence of Coxiella burnetii infection in wild animals in Japan. J Wildl Dis. 1993;29:481–4. 10.7589/0090-3558-29.3.481 [DOI] [PubMed] [Google Scholar]
- 5.Ruiz-Fons F, Rodríguez O, Torina A, Naranjo V, Gortázar C, de La Fuente J. Prevalence of Coxiella burnetti infection in wild and farmed ungulates. Vet Microbiol. 2008;126:282–6. 10.1016/j.vetmic.2007.06.020 [DOI] [PubMed] [Google Scholar]
- 6.Marrie TJ, Schlech WF, Williams JC, Yates L. Q fever pneumonia associated with exposure to wild rabbits. Lancet. 1986;1:427–9. 10.1016/S0140-6736(86)92380-9 [DOI] [PubMed] [Google Scholar]
- 7.González-Barrio D, Almería S, Caro MR, Salinas J, Ortiz JA, Gortazar C, et al. Coxiella burnetii shedding by farmed red deer (Cervus elaphus). Transbound Emerg Dis. 2013.http:// [DOI] [PubMed]
- 8.Monnerot M, Vigne JD, Biju-Duval C, Casane D, Callou C, Hardy C, et al. Rabbit and man: genetic and historic approach. Genet Sel Evol. 1994;26:167–82. 10.1186/1297-9686-26-S1-S167 [DOI] [Google Scholar]
- 9.Mitchell-Jones AJ, Amori G, Bogdanomicz W, Krystufek B, Beijnders PJ, Spitzenberger F, et al. , editors. Atlas of European mammals. London: Academic Press; 1999. [Google Scholar]
- 10.Maio E, Tania C, Balseiro A, Sevilla I, Romano A, Ortiz JA, et al. Paratuberculosis in European wild rabbits from Iberian Peninsula. Res Vet Sci. 2011;91:212–8. 10.1016/j.rvsc.2010.12.014 [DOI] [PubMed] [Google Scholar]
- 11.Berri M, Laroucau K, Rodolakis A. The detection of Coxiella burnetii from ovine genital swabs, milk and fecal samples by the use of a single touchdown polymerase chain reaction. Vet Microbiol. 2000;72:285–93. 10.1016/S0378-1135(99)00178-9 [DOI] [PubMed] [Google Scholar]
- 12.Akaike H. A new look at the statistical model identification. Institute of Electrical and Electronics Engineers Transactions on Automatic Control. 1974;19:716–25.
- 13.Guatteo R, Beaudeau F, Joly A, Seegers H. Coxiella burnetii shedding by dairy cows. Vet Res. 2007;38:849–60. 10.1051/vetres:2007038 [DOI] [PubMed] [Google Scholar]
- 14.Piñero A, Ruiz-Fons F, Hurtado A, Barandika JF, Atxaerandio R, García-Pérez AL. Changes in the dynamics of Coxiella burnetii infection in dairy cattle: an approach to match field data with the epidemiological cycle of C. burnetii in endemic herds. J Dairy Sci. 2014;97:2718–30. 10.3168/jds.2013-7229 [DOI] [PubMed] [Google Scholar]
- 15.Whitney EA, Massung RF, Candee AJ, Ailes EC, Myers LM, Patterson NE, et al. Seroepidemiologic and occupational risk survey for Coxiella burnetii antibodies among US veterinarians. Clin Infect Dis. 2009;48:550–7. 10.1086/596705 [DOI] [PubMed] [Google Scholar]


