Abstract
AlkB proteins are evolutionary conserved Fe(II)/2-oxoglutarate-dependent dioxygenases, which remove alkyl and highly promutagenic etheno (ε)-DNA adducts, but their substrate specificity has not been fully determined. We developed a novel assay for the repair of ε-adducts by AlkB enzymes using oligodeoxynucleotides with a single lesion and specific DNA glycosylases and AP-endonuclease for identification of the repair products. We compared the repair of three ε-adducts, 1,N6-ethenoadenine (εA), 3,N4-ethenocytosine (εC) and 1,N2-ethenoguanine (1,N2-εG) by nine bacterial and two human AlkBs, representing four different structural groups defined on the basis of conserved amino acids in the nucleotide recognition lid, engaged in the enzyme binding to the substrate.
Two bacterial AlkB proteins, MT-2B (from Mycobacterium tuberculosis) and SC-2B (Streptomyces coelicolor) did not repair these lesions in either double-stranded (ds) or single-stranded (ss) DNA. Three proteins, RE-2A (Rhizobium etli), SA-2B (Streptomyces avermitilis), and XC-2B (Xanthomonas campestris) efficiently removed all three lesions from the DNA substrates. Interestingly, XC-2B and RE-2A are the first AlkB proteins shown to be specialized for ε-adducts, since they do not repair methylated bases. Three other proteins, EcAlkB (Escherichia coli), SA-1A, and XC-1B removed εA and εC from ds and ssDNA but were inactive toward 1,N2-εG. SC-1A repaired only εA with the preference for dsDNA. The human enzyme ALKBH2 repaired all three ε-adducts in dsDNA, while only εA and εC in ssDNA and repair was less efficient in ssDNA. ALKBH3 repaired only εC in ssDNA Altogether, we have shown for the first time that some AlkB proteins, namely ALKBH2, RE-2A, SA-2B and XC-2B can repair 1,N2-εG and that ALKBH3 removes only εC from ssDNA. Our results also suggest that the nucleotide recognition lid is not the sole determinant of the substrate specificity of AlkB proteins.
Keywords: AlkB; Etheno adducts; 1,N6-Ethenoadenine; 3,N4-Ethenocytosine; 1,N2-Ethenoguanine; DNA repair
1. Introduction
AlkB proteins belong to the superfamily of Fe(II)/2-oxoglutarate-dependent dioxygenases, and the first AlkB member was discovered in Escherichia coli (EcAlkB) [1–5]. EcAlkB is a nucleic acid repair enzyme that hydroxylates alkyl groups bound to the nitrogen atoms at position 1 of purines and 3 of pyrimidines. The resulting unstable hydroxylalkyl group is then spontaneously released as an aldehyde, which leads to the regeneration of the unmodified base (Fig. 1) [2,3]. The main substrates repaired by EcAlkB are 1-methyladenine (m1A) and 3-methylcytosine (m3C) in single-stranded (ssDNA) and double-stranded DNA (dsDNA) [2,3,6–8]. Intriguingly, EcAlkB is also able to remove methyl lesions from RNA, suggesting a role for AlkB proteins in RNA repair [9–12]. In addition to the methyl lesions, EcAlkB repairs larger groups, e.g. ethyl, hydroxyethyl, propyl, hydroxypropyl, and some exocyclic DNA adducts (ε-adducts) [13–15].
Fig. 1.
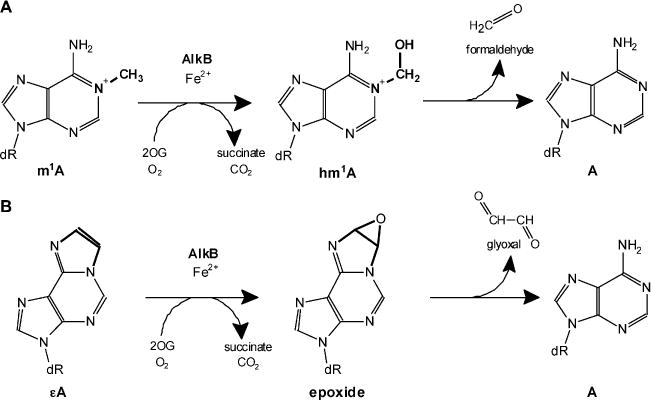
The mechanism of AlkB-mediated DNA repair through oxidative dealkylation of 1-methyladenine (m1 A) (A) and 1,N6-ethenoadenine (εA) (B).
Homologues of EcAlkB are present in numerous bacteria, eukaryotes, and certain plant RNA viruses. Nine AlkB homologues have been identified in humans: ALKBH1-ALKBH8 (AlkB homologue) and fat mass and obesity-associated protein FTO [1,16–18]. ALKBH2 and ALKBH3 are the sole human homologues that complement AlkB deficiency in E. coli regarding reactivation of ssDNA phages in response to methylating agents (e.g., methyl methane sulfonate (MMS) [9,13]. ALKBH3 demethylates m1A and m3C in ssDNA and RNA but is inactive toward these lesions in dsDNA [8–10, 19–21]. ALKBH2 repairs alkylated bases only in DNA, preferentially in dsDNA [9,19,20]. Among vertebrate AlkB proteins, only ALKBH2 is considered a true repair enzyme, since ALKBH2-null mice accumulate m1A in their genomes, but ALKBH3-null mice do not [22]. Recently, the function of ALKBH proteins has been expanded beyond nucleic acid repair. FTO and ALKBH5 were found to demethylate N6-methyladenine in mRNA [23–25], whereas ALKBH8 catalyzes the final two step modifications of wobble uridine in tRNA anticodons [26–29]. Finally, ALKBH1 and ALKBH4 have been proposed to demethylate lysine in histone H2A and actin, respectively [30,31].
AlkB proteins also repair the highly mutagenic exocyclic ε-adducts [15,32–36], e.g. 1,N6-ethenoadenine (sA) and 3,N4-ethenocytosine (εC). During the AlkB-mediated repair by oxidative dealkylation, the etheno bridge is converted to an epoxide and the product undergoes non-enzymatic hydration to a glycol which is then released as glyoxal (Fig. 1B) [32,34]. These lesions, as well as N2,3-ethenoguanine (N2,3-εG), and 1,N2-ethenoguanine (1,N2-εG) are formed in the reaction of a variety of reactive bifunctional electrophilic agents with a nitrogen atom of the DNA base, followed by dehydration and ring closure [37,38]. These ubiquitous lesions are generated in cellular DNA either by endogenous metabolic processes, e.g., oxidative stress-induced lipid peroxidation, or by the reaction of the DNA with diverse chemical carcinogens, e.g., bioactivated derivatives of vinyl chloride or urethane [39–42]. Since the additional exocyclic ring disrupts the Watson-Crick hydrogen bonding, the ε-lesions generate a broad spectrum of base substitutions (transitions and transversions) or frameshift mutations [41,43]. It is estimated that in mammalian DNA from 14% to 60% of ε-adducts give rise to mutations, while only 3% of 8-oxoguanine residues are pro-mutagenic in cells with functional DNA repair [44–46]. These promutagenic properties of ε-lesions strongly underlie their contribution to carcinogenesis in mammals [47–49].
It is commonly believed that the main mechanism for removal of the ε-adducts from DNA is base excision repair (BER) initiated by specific DNA glycosylases. In mammalian cells εA and 1,N2-εG are removed by N-methylpurine-DNA glycosylase (ANPG) [50,51], and εC is repaired by mismatch-specific thymine-DNA glycosylase (TDG) [52,53]. In addition to TDG, a protein that binds methylated CpG islands (MBD4/MED1) and uracil-DNA glycosylase specific for ssDNA (SMUG1) are also capable of excising εC from the DNA [54,55]. In E. coli, N2,3-εG is excised by alkyl-N-purine-DNA glycosylase AlkA, which also excises εA, but the latter with a very low efficiency [50,56]. Mismatch-specific uracil-DNA glycosylase from E. coli, Mug excises from the DNA εC and 1,N2-εG [51,53,57]. Recently it has been shown that εA and εC can also be repaired via an alternative nucleotide incision repair (NIR) pathway, in which human AP endonuclease APE1 makes an incision 5′ to the ε-base in a DNA glycosylase-independent manner [58]. Additionally, εA and εC are removed from the DNA in vitro by EcAlkB and ALKBH2 [15,32–36].
AlkB proteins are widespread among bacteria, and some bacterial species possess two or three AlkBs, but their substrate specificities are poorly characterized. Based on sequence alignment and phylogeny analysis bacterial AlkB proteins have been divided into four groups 1A, 1B, 2A, and 2B each characterized by the presence of specific conserved amino acid residues in the nucleotide recognition lid, which has been postulated to specify substrate recognition, at least partially [59].
The main goal of this work was to gain insight into repair of ε-adducts by bacterial and human AlkB proteins. To reach this goal we developed a new, sensitive, and time and cost effective BER-mediated assay, and analyzed the ability of nine bacterial and two human AlkB proteins representing all four groups mentioned above (1A, 1B, 2A, and 2B) to remove from the DNA εA, εC and 1,N2-εG in vitro in ss and dsDNA. To the best of our knowledge repair of 1,N2-εG has not been presented before, and for several of the studied enzymes repair of εC was not studied as well. In this newly developed assay the DNA with a single lesion is subjected to AlkB protein, and subsequently digested with an appropriate DNA glycosylase excising the damaged base and AP-endonuclease, which cleaves the DNA at the site of the lesion if the damage was not repaired by AlkB protein.
The majority of the bacterial AlkBs tested were able to repair ε-lesions. Three of the proteins analyzed, SA-2B (Streptomyces avermitilis), XC-2B (Xanthomonas campestris), and RE-2A (Rhizobium etli) appeared to be specialized toward ε-adducts, since exhibited significant activity against all the ε-adducts tested without being active toward methylated bases. Altogether, our data suggest that AlkB-mediated removal of ε-lesions could be an important mechanism responsible for the repair of these lesions in some bacteria. Additionally, we analyzed the in vitro repair of εA, εC, and 1,N2-εG in ss and dsDNA by human ALKBH2 and ALKBH3. We confirmed that ALKBH2 is active toward εA and εC in dsDNA and showed a lower activity against these lesions in ss DNA Finally, we have shown for the first time that ALKBH2 repairs 1,N2-εG in dsDNA and ALKBH3 εC in ssDNA.
2. Materials and methods
2.1. Plasmids and protein expression
pET-28a(+)-derived plasmids encoding N-terminally 6× Histagged bacterial and human AlkB proteins were obtained as described previously [9,59], and were used to transform E. coli strain BL21-CodonPlus(DE3)-RIPL (Stratagene, La Jolla, CA, USA). Bacteria were cultured and when the optical density reached 1 measured at 600 nm (OD600), expression of recombinant proteins was induced by adding isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM and incubation was continued at 16 °C for 16 h. Then, the bacteria were harvested by centrifugation at 5000 ×g for 10 min at 4°C and stored at −80°C until use.
2.2. Purification of AlkB proteins
Bacterial pellets were resuspended in a buffer containing 50 mM sodium phosphate (pH 7.0), 150mM NaCl, 5mM β-mercaptoethanol, 0.5% Nonidet NP-40 detergent (v/v), 10% glycerol (v/v), 5 mM imidazole, and EDTA-free Complete-Protease Inhibitor (Roche). The cells were lysed by the addition of lysozyme to a final concentration of 1 mg/mL, incubation on ice for 30 min, and subsequent sonication with three 11 W pulses of 30 s (with 30 s intervals). Cell debris was removed by centrifugation at 12,000 × g for 10 min at 4 °C. The resulting supernatant was directly mixed with HisPur™ Cobalt Resin (Thermo Scientific) and recombinant proteins were obtained by a single affinity purification step according to the manufacturer’s instructions. Protein purity and yield were assessed by 15% (w/v) SDS-PAGE followed by Coomassie Brilliant Blue (R-250) staining of the gel. Fractions containing AlkB proteins were pooled and dialysed in a buffer containing 50 mM Tris–HCl pH 7.0,100 mM NaCl, 5 mM β-mercaptoethanol. Then 1 volume of storage buffer (20 mM Tris–HCl (pH 7.5), 100mM NaCl, 50% glycerol (v/v) was added, and the concentration of purified proteins was estimated using a Bradford reagent (Sigma) assay.
2.3. Preparation of32P-labeled oligodeoxynucleotide substrates
The oligodeoxynucleotide substrates containing a single εA (40-mer, 5′-d(GCTACCTACCTAGCGACCTεACGACTGTCCCACTGCT-CGAA)-3′, Eurogentec S.A., Belgium), εC (25-mer, 5′-d(ATTCTCGTT-AGGATεCGCGTCAAGCC)-3′, Chemgenes Corporation, USA), or 1,N2-εG (18-mer, 5′-(TCACεGAATCCTTACGAGCCCCC)-3′, [60,61] were 32P-labeled at the 5′ end by T4 polynucleotide kinase (Takara) with an excess of [γ-32P]ATP (3000 Ci/mmol, Hartmann Analytic). Radiolabeled oligodeoxynucleotides were purified from unincorporated radioactivity using Micro Bio-Spin P-30 columns (Bio-Rad) according to the manufacturer’s instructions. The 32P-labeled oligodeoxynucleotides, when necessary, were annealed to their complementary oligonucleotides (present in a 2-fold molar excess) by incubation for 2 min at 90 °C and subsequent cooling to room temperature. Formation of duplexes was verified by nondenaturing PAGE.
2.4. In vitro repair assay of DNA etheno adducts
Repair reactions of the ε-adducts in DNA with bacterial AlkB proteins were performed by incubating 100 pmol of the appropriate bacterial AlkB protein with 1 pmol of ss or ds 32P-labeled DNA oligodeoxynucleotide at 37 °C for 30 min in a 50 μL reaction mixture containing 50 mM Tris-HCl (pH 7.0), 2 mM ascorbic acid, 1 mM 2-oxoglutarate, and 80 μM (NH4)2Fe(SO4)2. The repair activity of the ε-adducts by ALKBH2 and ALKBH3 was estimated by incubating 1 pmol of the appropriate 32P-labeled oligodeoxynucleotide substrate with different amounts of the tested protein (10, 25 and 50 pmol) using the reaction conditions as above. Additionally, to determine specific repair activity of ALKBH2 and ALKBH3 toward etheno lesions, reactions with increasing amount of recombinant proteins and various times of incubation were performed as indicated in Section 3. All AlkB-mediated reactions were stopped by incubation at 65 °C for 20 min. The AlkB protein was removed by addition of 1 μL of proteinase K (20 mg/mL, Sigma-Aldrich) to the reaction and further incubation for 15 min at 42 °C followed by heat-inactivation of proteinase K at 90 °C for 10 min. When ssDNA substrates were used in the reaction, prior to the BER enzyme digestion the complementary DNA oligodeoxynucleotide was added and annealed as described.
In order to distinguish the repaired oligodeoxynucleotide from the unrepaired one, the 32P-labeled DNA was incubated with the excess of the appropriate DNA glycosylase and the resulting abasic site was cleaved with the excess of human AP-endonuclease 1 (APE1). The amounts of DNA glycosylases and AP-endonuclease necessary to completely cleave oligodeoxynucleotide at the site of the lesion were verified by applying a series of enzymes dilutions in reaction buffers supplemented with BSA (0.1 mg/mL). Briefly, 5 μL of AlkB reaction mixture was incubated for 30 min at 37 °C with human alkyl-N-purine-DNA glycosylase (ANPG) for εA or with E. coli uracil-DNA glycosylase Mug for εC and 1,N2-εG, followed by cleavage with the excess of APE1 for 30 min at 37°C. Buffers for DNA glycosylases contained 70 mM Tris-HCl pH 7.8, 1 mM EDTA pH 8.0, 100 mM KCl, 5 mM β-mercaptoethanol, 100μg/mL BSA, while for APE1 endonuclease, 20 mM Tris-HCl pH 7.5, 0.1 mM EDTA pH 8.0, 50 mM KCl, 5 mM MgCl2, 1 mM β-mercaptoethanol, 100 μg/mL BSA. Two controls were performed for each reaction: a negative control containing untreated oligodeoxynucleotide, and a positive control of the activity of the BER enzymes with the oligodeoxynucleotide subjected to the action of ANPG/Mug and APE1 without previous AlkB treatment. Finally, the reaction products were separated by denaturing 20% (w/v) PAGE containing 7 M urea. The scheme of the method is shown in Fig. 2. A digital image of the separated radioactive oligodeoxynucleotides was captured by phosphorimaging using FLA-7000 (Fujifilm), and the radioactivity of the bands was quantified using Multi Gauge Analysis software (Fujifilm).
Fig. 2.
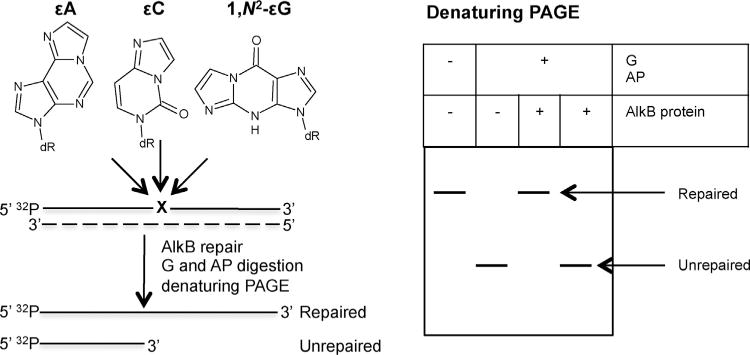
Schematic representation of the BER enzymes digestion assay for visualization of AlkB-mediated repair of ε-lesions.32 P-labeled ssDNA ordsDNA oligodeoxynucleotides containing single ε-lesion are incubated with AlkB protein. Subsequently, to visualize the AlkB-mediated repair, oligodeoxynucleotides are treated with the appropriate DNA glycosylase (G) followed by AP endonuclease 1 (AP). Reaction products are separated by denaturing PAGE and visualized by phosporimaging. The dashed line indicates the complementary, lesion-free unlabeled oligodeoxynucleotide, which is either present during the repair reaction (dsDNA repair) or is added post-repair (ssDNA repair). G/AP cleavage of the DNA substrate occurs only if the ε-lesion is not removed. A repaired substrate will appear as a slower-migrating band on PAGE.
Since we and others have previously shown that ethenoadenine decomposes slowly into derivatives, which are not recognized by ANPG glycosylase, which excises ethenoA [62,63], and other ethenoadducts are also unstable, we have taken this into consideration during calculation of the results. In each experiment the negative control reaction without AlkB protein was performed, and percent of “repair” was calculated also for this negative control reaction. Percent of oligodeoxynucleotide in apparently “repaired band” was then subtracted from the percent of repair by each AlkB protein. Such procedure enabled to avoid false results as this background was included during calculation of the percent of in vitro AlkB-mediated repair.
2.5. In vivo repair of DNA etheno adducts by the studied enzymes
pJB658-derived plasmids bearing the studied AlkB enzymes were prepared as described before [59]. Cultures of E. coli strain HK 82/F′ which were transformed with pJB658-derived plasmids bearing the studied AlkB enzymes were induced with 2 mM toluic acid (Sigma) at OD600 = 0.1, and further grown until they reached A600 = 0.5. Bacteria in 500 μL of the culture were pelleted, resus-pended in 250 μL of M9 minimum salt medium, and then mixed with an equal volume of minimum salt medium containing an appropriate concentration of chloroacetaldehyde (CAA). Bacteria were incubated with CAA for 30 min in 37 °C, subsequently diluted 10,000-fold and plated at LB plates. Colonies were counted after 24 h incubation at 37 °C.
3. Results
3.1. In vitro repair of etheno lesions in ss and dsDNA by bacterial AlkB proteins
We studied the repair activity of nine bacterial AlkB proteins (Table 1) toward εA, εC, and 1,N2-εG using a newly developed DNA glycosylase/AP-endonuclease digestion assay. The N-terminally 6× His-tagged bacterial and human AlkB proteins cloned into pET-28a(+)-derived plasmids were overexpressed in E. coli strain BL21-CodonPlus(DE3)-RIPL strains and purified by affinity chromatography on His Pur™ Cobalt Resin. The purity of received enzymes was verified by PAGE, and is shown in Supplementary data, Fig. S1. Most of the studied AlkBs were homogenous, only three of them, MT-2B, RE-2A and SC-2B contained one or few additional faint protein bands. The proteins chosen represented all four structural AlkB groups (1A, 1B, 2A, and 2B), distinguished on the basis of the amino acid sequence of their nucleotide-recognizing domains. Previously, van den Born et al. [59] showed that the majority of bacterial AlkB proteins studied in the present work remove methyl groups from m1A and m3C. They also found that AlkBs from X. campestris (XC-1B and XC-2B) and one from R. etli (RE-2A) displayed a high repair activity toward εA in both ss and dsDNA. Interestingly, XC-2B and RE-2A showed negligible repair activity toward the methyl lesions, suggesting that those enzymes might have evolved to repair bulky alkylated DNA adduct such as ε-lesions [59]. In this study we further characterized the repair potential of these AlkBs toward two additional DNA ε-adducts, εC and 1,N2-εG, using the novel assay with BER enzymes described in Section 2 (Fig. 2). Since van den Born et al. [59] characterized the εA repair activity utilizing an established digestion assay based on the methylation-sensitive restrictase Dpn II [22], we additionally reexamined the repair of this lesion but using our BER-based assay. The recombinant proteins were incubated with 5′-[32P]-labeled oligodeoxynucleotides containing a single εA, εC, or 1,N2-εG. The AlkB repair of the lesions was visualized by DNA glycosylase treatment followed by the cleavage of the resulting abasic sites with AP-endonuclease (Fig. 2), and the obtained results are shown in Fig. 3.
Table 1.
AlkB proteins tested in the study.
| Origin | AlkB | Accession No. | Gi number | Size (aa) |
|---|---|---|---|---|
| Escherichia coli | EcAlkB | NP_754639 | 26248599 | 216 |
| Mycobacterium tuberculosis | MT-2B | NP_335465 | 15840428 | 205 |
| Rhizobium etli | RE-2A | YP_472140 | 86360251 | 189 |
| Streptomyces avermitilis | SA-1A | NP_822618 | 29827984 | 222 |
| Streptomyces avermitilis | SA-2B | NP_822306 | 29827672 | 208 |
| Streptomyces coelicolor | SC-1A | NP_625335 | 21219556 | 216 |
| Streptomyces coelicolor | SC-2B | NP_631357 | 21225578 | 210 |
| Xanthomonas campestris | XC-1B | NP_636023 | 21230106 | 201 |
| Xanthomonas campestris | XC-2B | NP_636458 | 21230541 | 211 |
| Homo sapiens | ALKBH2 | NP_001001655 | 48717226 | 261 |
| Homo sapiens | ALKBH3 | NP_631917 | 21040275 | 286 |
Fig. 3.
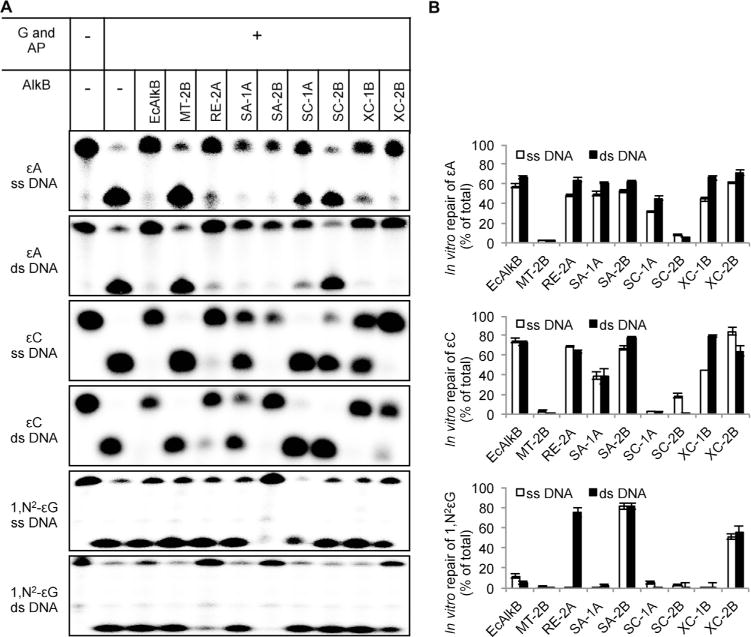
In vitro repair activity of bacterial AlkB proteins toward DNA ε-adducts. (A) 1 pmol of 32P-labeled oligodeoxynucleotide was incubated with 25 or 50pmol of tested AlkB protein for 30 min at 37 °C, digested with the excess of an appropriate DNA glycosylase (G)and APendonuclease(AP), and separated by 20%(w/v) denaturing PAGE. (B) Quantification of results from experiments exemplified in A. Values represent the means ± SD of three independent experiments.
Supplementary Fig. S1 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.dnarep.2015.02.021.
We found that AlkB from E. coli (EcAlkB), which belongs to group 1A, exhibits a substantial repair activity toward εA and εC in both ss and dsDNA, but negligible activity toward 1,N2-εG and only in ssDNA. The second AlkB protein tested belonging to group 1A, SA-1A from S. avermitilis, showed similar activity, albeit it was slightly lower toward εC in comparison with EcAlkB. Unexpectedly, the third protein from that group, SC-1A from S. coelicolor, removed only εA and was more active on the dsDNA substrate. XC-1B from X. campestris, the only AlkB protein from group 1B studied in this work, was active toward εA and εC and clearly preferred dsDNA. The AlkB proteins from group 2B showed a strongly divergent ε-base repair capacity. Those from M. tuberculosis (MT-2B) and S. coelicolor (SC-2B) were inactive toward all three ε-adducts examined, whereas SA-2B from S. avermitilis and XC-2B X campestris were very active toward all the ε-bases tested in both ss and dsDNA. Finally, the R. etli (RE-2A) AlkB protein from group 2A exhibited substantial repair activity toward εA and εC in both ss and dsDNA, and toward 1,N2-εG in dsDNA.
3.2. In vitro repair of etheno lesions in ss and dsDNA by human ALKBH2 and ALKBH3 proteins
Repair of ε-adducts by human AlkB proteins has been studied by several laboratories; however, conflicting results have been obtained. Repair of εA and εC by ALKBH2 was demonstrated by Ringvolletal. [35] and Fu and Samson [33], respectively. The repair specificity for ALKBH3 is more controversial. While Mishina et al. [34] reported that human ALKBH3 repaired εA in vitro in the specific trinucleotide substrate T(εA)T, albeit with a much lower activity than E. coli AlkB, Ringvoll et al. [35] did not confirm this finding in mammalian cells. Because of these discrepancies and the general paucity of data on those enzymes, we decided to study the repair activity of ALKBH2 and ALKBH3 toward εA, εC and 1,N2-εG in ss and dsDNA substrates in more detail using a BER enzyme-based assay. First, in order to check if ALKBH2 and ALKBH3 are able to remove ε-adducts in vitro, we incubated 5′-[32P]-labeled ss or dsDNA oligodeoxynucleotides containing εA, εC, or 1,N2-εG with 10, 25, or 50 pmol of each recombinant protein. As shown in Fig. 4, ALKBH2 repaired εA and εC in both ss and dsDNA and 1,N2-εG in dsDNA. Similar to Ringvoll et al. [35], we did not observe any repair activity of ALKBH3 with εA; 1,N2-εG was not removed by this protein either. However, we observed that ALKBH3 repaired εC in ssDNA (Fig. 4).
Fig. 4.
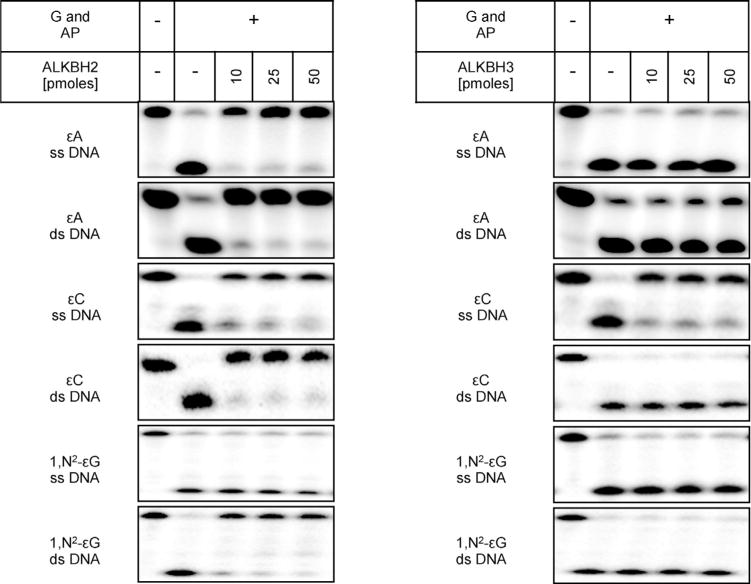
In vitro repair activity of human ALKBH2 and ALKBH3 toward DNA ε-adducts. ss or ds 32P-5′-labeled oligodeoxynucleotides (1 pmol), containing εA, εC or 1,N2-εG, were incubated with 10,25, or 50 pmol of recombinant ALKBH2 or ALKBH3 for 30 min at37 °C, digested with the appropriate DNA glycosylase (G) and AP endonuclease (AP), separated by 20% (w/v) denaturing PAGE and labeled DNA was visualized by phosphorimaging.
To characterize the repair efficiency of ALKBH2 and ALKBH3 toward ε-lesions in more detail, we measured specific activities of these enzymes toward three ε-adducts studied. The reaction duration and the amount of the recombinant protein were chosen to give a linear reaction rate. ALKBH2 exhibited a considerably higher specific repair activity against εA and εC in ds DNA than in ss DNA (Fig. 5). Thus, likewise to the methyl lesions, the ε-adducts are preferentially repaired by ALKBH2 in dsDNA. The ALKBH2 activity against these two ε-lesions in ds DNA was very similar, in both cases reaching a plateau of ~70% of the total repaired substrate after 5 min of incubation with 1 pmol of the enzyme in the reaction (Fig. 5). The measured specific activities toward εA and εC of ALKBH2 were almost identical, 69 and 66 fmol min−1 pmol enzyme−1, respectively (Table 2). In ssDNA, ALKBH2 repaired εA about 2-fold more efficiently than εC, with specific activities of 21 and 11 fmolmin−1 pmolprotein−1, respectively (Table 2). The repair of εA in ssDNA reached a plateau of 70% of the repaired substrate after 10 min of incubation with 6 pmol of the enzyme, and the repair of εC in ssDNA reached only a plateau of ~50% of the repaired substrate after 20 min with 6 pmol of the enzyme (Fig. 5). The ALKBH2-mediated repair of 1,N2-εG in dsDNA was an order of magnitude less efficient than the repair of εA and εC with the specific activity being 5 fmolmin−1 pmol ALKBH2−1 (Table 2). A plateau of ~60% of the repaired substrate was reached after 30 min of incubation at ~6pmols of the enzyme (Fig. 5). For ALKBH3, we studied only the repair of εC in ssDNA, because the earlier qualitative analysis showed that other ε-lesions were not repaired by that enzyme. The rate of εC repair was rather low, with a specific activity of 6 fmol min−1 pmol ALKBH3−1 (Table 2), and a plateau was reached at only ~35% of the repaired substrate after 20 min of incubation with 8 pmol of the enzyme (Fig. 5).
Fig. 5.
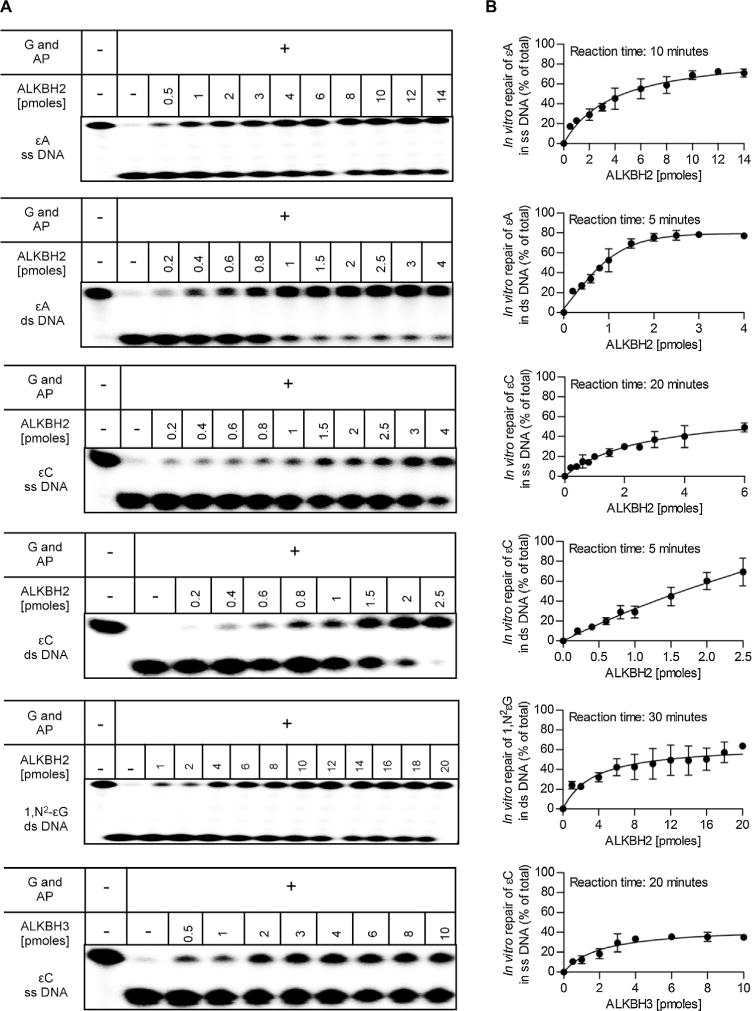
Comparison of in vitro repair efficiency of DNA ε-adducts by recombinant human ALKBH2 and ALKBH3. (A) ss or ds 32P-end-labeled oligonucleotides containing εA, εC, or 1,N2-εG (1 pmol) were incubated with increasing amounts of recombinant ALKBH2 or ALKBH3 for the indicated time at 37 °C, then digested with the appropriate DNA glycosylase (G) and AP endonuclease (AP), separated by 20% (w/v) denaturing PAGE. Radioactive DNA was visualized by phosphorimaging. (B) Quantification of results from experiments shown in A. Each data point shows the mean of three independent measurements, error bars indicate SD. Calculated specific repair activities are given in Table 2.
Table 2.
Specific repair activity of human recombinant ALKBH2 and ALKBH3 proteins against etheno lesions.
| Protein | ε-lesion | ss/ds DNA | Specific repair activity [fmol of product min−1 pmol protein−1] |
|---|---|---|---|
| ALKBH2 | εA | ss | 21.4 ± 3.3 |
| ds | 69.4 ± 3.6 | ||
| εC | ss | 11.1 ± 2.7 | |
| ds | 65.8 ± 1.2 | ||
| εG | ds | 4.6 ±1.3 | |
| ALKBH3 | εC | ss | 5.8 ± 1.1 |
3.3. In vivo repair of ε-adducts by bacterial AlkB proteins
The in vivo importance of reported in vitro repair activities of nine bacterial AlkBs was partially addressed by van den Born and coworkers [59] who tested in vivo repair of etheno lesions in ssDNA by these AlkBs, and showed that expression of SA-1A, SC-1A, RE-2A, SA-2B andXC-2B proteins in EcAlkB-deficient E. coli complemented its repair deficiency with activity comparable to that of EcAlkB. We have extended these studies on e-adducts repair in dsDNA. A set of alkB deficient E. coli strains, bearing plasmids coding for the studied AlkB proteins was constructed, and sensitivity of these strains to chloroacetaldehyde was verified (Fig. 6). AlkB deficient strain (HK 82/F′) was more sensitive to CAA than the strain bearing AlkB from E. coli (Fig. 6). Also all other studied enzymes increased viability of AlkB-deficient E. coli strain upon treatment with CAA. This was surprising, since two AlkBs, MT-2B and SC-2B did not repair any ε-adduct in vitro or repair was very inefficient. However, similar complementation was observed by van den Born and coworkers [59] when ssDNA of M13 phage damaged with CAA was transformed into E. coli expressing AlkBs studied here. This might suggest that even a remnant activity of AlkB is sufficient in vivo to protect bacteria from deleterious effects of ε-adducts.
Fig. 6.
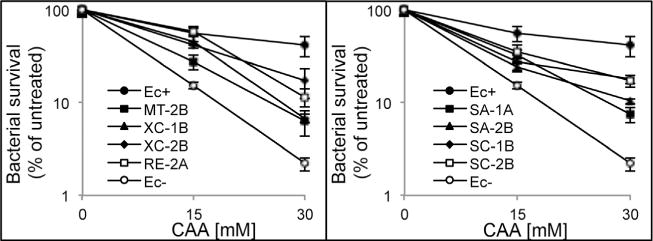
In vivo repair of chloroacetaldehyde induced damage in E. coli by bacterial AlkB proteins. AlkB-deficient E. coli carrying either an empty expression plasmid (Ec−), or corresponding plasmids for expression of the indicated bacterial AlkB proteins or EcAlkB (Ec+) were treated with the indicated concentrations of CAA, and the bacterial survival was assessed.
4. Discussion
The mutation spectra in repair mutants of E. coli exposed to chloroacetaldehyde (CAA), which efficiently creates ε-adducts of A, C, and G, suggest that their repair is executed by both base excision repair and AlkB-mediated oxidative dealkylation [36]. However, the rates of repair by AlkBs of particular lesions induced by CAA have not been established, and the removal of 1,N2-εG has not been investigated thus far. The latter, although a minor lesion in dsDNA, has strong mutagenic potential, and is introduced by chemical carcinogens to ssDNA with a similar frequency as N2,3-εG. We have established a new BER enzyme-based assay for quantifying the repair of ε-lesions in DNA by AlkB proteins. In this assay, following incubation with an AlkB protein the DNA substrates are treated with an appropriate DNA glycosylase and the resulting abasic sites are cleaved with AP-endonuclease. The DNA glycosylase/AP endonuclease cleavage occurs only if the ε-lesion is not repaired by oxidative dealkylation mediated by the AlkB protein. The method elaborated by us for measuring AlkB activity against ethenoadducts is an alternative to HPLC and restriction nuclease methods. Its simplicity and speed enables to test many AlkB enzymes at once. In comparison to HPLC method it needs much less expensive substrate (about 100–500 times less), and either less or the same amount of AlkB enzyme [15]. In comparison to restriction nucleases method, BER-mediated assay needs less enzyme (1 pmol vs 100 pmol in restriction nucleases analysis [22,59]). In addition, restriction nuclease method requires that modified base is incorporated in a specific sequence recognized by this nuclease, while in BER-mediated assay any sequence can be modified, and sequence-specificity of AlkB enzymes can be investigated. Limitation of the method is availability of BER enzymes recognizing the studied lesions. In practice, the method is currently limited to etheno-DNA adducts.
We established the in vitro repair activity toward εA, εC, and 1,N2-εG in ss and dsDNA of nine bacterial AlkBs representing each of the four structural groups defined earlier. The majority of the bacterial AlkB proteins tested, similar to EcAlkB, remove εA and εC from DNA. This in vitro repair activity toward ε-adducts suggests that AlkB proteins constitute an alternative repair mechanism to the BER pathway for removing ε-lesions from genomic DNA in vivo. The conclusion that EcAlkB plays a major role in the DNA repair in E. coli is further supported by the fact that it decreases the AT → TA mutations generated by εA [64]. Furthermore, because the bacterial AlkBs tested were active toward ε-adducts in ssDNA, they could also play an essential role in repairing ε-lesions in the replication forks and transcription bubbles during replication and transcription, respectively. It is worth mentioning that an earlier study showed that although SA-1A and SC-1A proteins efficiently reactivated CAA-treated M13 ssDNA in vivo, they displayed negligible levels of εA repair in vitro in ss and dsDNA substrates using a DpnII-based method [59]. In contrast, using our BER digestion assay we found that these proteins in fact are able to remove εA from the DNA with similar efficiency to that of EcAlkB. The in vitro repair activity of SA-1A and SC-1A toward εA (and for SA-1A also εC), demonstrated here, explains their ability to reactivate CAA-treated M13 in vivo. Our studies also show that these enzymes repair ethenoadducts in vivo in dsDNA (Fig. 6).
The present study showed for the first time that AlkB proteins can also repair the minor, but highly mutagenic 1,N2-εG adduct. The three bacterial AlkBs, i.e. SA-2B, XC-2B and RE-2A, displayed a high repair activity against all the ε-adducts tested. They are the first AlkBs for which such a substantial activity against εA, εC, and 1,N2-εG has been shown. In addition, XC-2B and RE-2A are the first AlkBs that show repair activity toward εA, εC, and 1,N2-εG without displaying any activity toward 1-me A and 3-me C.X. campestris, S. avermitilis, and R. etli are soil bacteria, and therefore they can be exposed to industrial pollutants present in the soil. It has been shown that some common industrial solvents, e.g. tetrachloroethylene, which are among the most frequent contaminants found in groundwater, can be biotransformed to vinyl chloride [65]. Given this, it seems plausible that XC-2B, SA-2B, and RE-2A AlkB proteins might have evolved to protect these soil bacteria against DNA ε-adducts introduced by vinyl chloride generated in the soil.
It has been suggested that the amino acids composition of nucleotide recognition lid determines the substrate specificity of AlkB proteins. Accordingly, AlkB proteins belonging to the same group are expected to have similar repair profiles against diverse DNA lesions. However, we observed that proteins from group 2B displayed substantial differences in their repair activities toward the ε-adducts; SA-2B and XC-2B efficiently removed all the ε-adducts tested, while MT-2B and SC-2B were not active against these lesions, and van den Born et al. [59] observed divergent repair activities of methyl lesions among this group of proteins. Also the AlkBs from group 1A displayed contrasting activities against εC, because SA-1A and EcAlkB removed εC whereas SC-1A did not. Altogether, the results obtained here (as well as the data presented by van den Born et al. [59]) indicate that AlkB proteins belonging to a given group can in fact show diverse repair activities against given DNA lesions. Thus, it is not possible to predict the substrate specificity of AlkB proteins based solely on their classification according to the amino acid composition of their nucleotide recognition lid. Indeed, recent studies have shown that substrate recognition by EcAlkB is a complex process and is also determined by aminoacids in the active site, which is located in enzyme flexible loop between aminoacids K134 to L139 [66]. The mechanism of repair of propano-type guanine adducts by AlkB from E. coli involved several stages, during which transient complexes were formed with ring-opened aldehydic form and ring-closed form, and AlkB repaired ring-opened form only [67]. Finally the efficiency of catalysis is determined by the dynamics of a specific conformational transition, which facilitates the proper sequential order of catalysis steps, that is oxidation of 2-oxoglutarate, formation of enzyme-bound oxyferryl intermediate, and hydroxylation of an alkylated nucleobase [68].
It has been demonstrated that human ALKBH2, similar to the E. coli AlkB protein, is able to repair εA and εC in DNA [33,35]. Using our BER-based digestion assay we confirmed here that ALKBH2 exhibits an in vitro activity toward εA and εC in ss and dsDNA. Similar to methylated bases, ALKBH2 repaired εA and εC more efficiently in dsDNA than in ssDNA. Based on the ability of recombinant ALKBH2 to remove εA and εC observed by us and others [33,35], it seems reasonable that this enzyme could be engaged in the repair of these lesions in vivo. However, it has been demonstrated that Alkbh2−/− mice, in contrast to Anpg−/− mice, do not accumulate εA in genomic DNA [35]. Given this, it seems that the direct reversal of εA by ALKBH2 is not sufficient to remove endogenously generated εA, and the main enzyme responsible for removing this lesion from DNA in vivo is the ANPG glycosylase [35]. However, further studies inAlkbh2−/−/Anpg−/− double mutant mice are needed to elucidate fully the importance of ALKBH2-mediated repair of εA in vivo. Likewise, the in vivo significance of εC repair by recombinant ALKBH2 observed here, as well as by Fu and Samson [33] needs further experimental verification. Some insight into this problem was recently provided by the study of Calvo et al. [69], in which a knockout of three enzymes ANPG, ALKBH2, and ALKBH3 made mice particularly susceptible to inducers of colon inflammation, and all mice died following the first instillation of a pro-inflammatory compound. It has been established that εC can be excised from the DNA in vitro by three different DNA glycosylases: TDG [52,53], MBD4/MED1 [55], and SMUG1[54]. However, the primary substrates of these glycosylases are oxidation and deamination products of 5-methylcytosine in a CpG context [70,71]. Given this, it seems likely that another repair enzyme responsible for εC repair in vivo exists, and ALKBH2 could be this one. We further found that recombinant ALKBH2 can remove 1,N2-εG from dsDNA. However, an ALKBH2-mediated repair of 1,N2-εG does not seem to play a major physiological role, because the in vitro activity of ALKBH2 against this lesion was considerably lower than the corresponding activity toward εA or εC. Finally, in agreement with the results of Ringvoll et al. [35], we showed that recombinant ALKBH3 does not remove εA from DNA. These data are in contrast with the results of Mishina et al. [34] who reported that ALKBH3 was able to repair εA in vitro. However, it should be mentioned that those authors incubated ALKBH3 with the DNA substrates for 16 h under non-physiological conditions (pH 6.0) [34]. Notably, we observed a weak repair activity of ALKBH3 against εC in ss DNA but doubt that it has a biological significance.
In conclusion, we have shown that nine of eleven AlkB proteins studied (7 bacterial and 2 human) are able to repair ε-lesions in vitro. Additionally, we posit that DNA ε-adducts could be the primary substrates for some bacterial AlkBs in vivo since they exhibit a marked in vitro repair activity toward ε-lesions and lacking such activity toward DNA methyl lesions.
Supplementary Material
Acknowledgments
We would like to thank Prof. Jan Fronk for critical reading of the manuscript, and Prof. Elzbieta Grzesiuk for providing E. coli strain HK82. This work was financed by grant from the National Science Centre (NCN) 2011/01/M/NZ1/05301 (BT) and National Institutes of Health grants R01 ES010546 (FPG), P01 ES05355 (CJR), P01 CA160032 (CJR), P30 ES00267 (CJR. and FPG) and P30 CA068485 (C.J.R. and FPG).
Abbreviations
- ε
etheno
- εA
1,N6-ethenoadenine
- εC
3,N4-ethenocytosine
- 1,N2-εG
1,N2-ethenoguanine
- 1-meA
1-methyladenine
- 3-meC
3-methylcytosine
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Aravind L, Koonin KV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2:RESEARCH0007. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 3.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 4.Kataoka H, Sekiguchi M. Molecular cloning and characterization of the alkB gene of Escherichia coli. Mol Gen Genet. 1985;198:263–269. doi: 10.1007/BF00383004. [DOI] [PubMed] [Google Scholar]
- 5.Kataoka H, Yamamoto Y, Sekiguchi M. A new gene (alkB) of Escherichia coli that controls sensitivity to methyl methane sulfonate. J Bacteriol. 1983;153:1301–1307. doi: 10.1128/jb.153.3.1301-1307.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli, Proc. Natl Acad Sci U S A. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falnes PO. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:6260–6267. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koivisto P, Robins P, Lindahl T, Sedgwick B. Demethylation of 3-methylthymine in DNA by bacterial and human DNA dioxygenases. J Biol Chem. 2004;279:40470–40474. doi: 10.1074/jbc.M407960200. [DOI] [PubMed] [Google Scholar]
- 9.Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 10.Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, et al. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol Cell. 2004;16:107–116. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Vagbo CB, Svaasand EK, Aas PA, Krokan HE. Methylation damage to RNA induced in vivo in Escherichia coli is repaired by endogenous AlkB as part of the adaptive response. DNA Repair(Amst) 2013;12:188–195. doi: 10.1016/j.dnarep.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 12.van den Born E, Omelchenko MV, Bekkelund A, Leihne V, Koonin EV, et al. Viral AlkB proteins repair RNA damage by oxidative demethylation. Nucleic Acids Res. 2008;36:5451–5461. doi: 10.1093/nar/gkn519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, et al. Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci U S A. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koivisto P, Duncan T, Lindahl T, Sedgwick B. Minimal methylated substrate and extended substrate range of Escherichia coli AlkB protein, a 1-methyladenine-DNAdioxygenase. J Biol Chem. 2003;278:44348–44354. doi: 10.1074/jbc.M307361200. [DOI] [PubMed] [Google Scholar]
- 15.Maciejewska AM, Poznanski J, Kaczmarska Z, Krowisz B, Nieminuszczy J, et al. AlkB dioxygenase preferentially repairs protonated substrates: specificity against exocyclic adducts and molecular mechanism of action. J Biol Chem. 2013;288:432–441. doi: 10.1074/jbc.M112.353342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, et al. The obesity-associated FTOgene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics. 2003;4:48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Pulido L, Andrade-Navarro MA. The FTO (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC Biochem. 2007;8:23. doi: 10.1186/1471-2091-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falnes PO, Bjoras M, Aas PA, Sundheim O, Seeberg E. Substrate specificities of bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:3456–3461. doi: 10.1093/nar/gkh655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee DH, Jin SG, Cai S, Chen Y, Pfeifer GP, et al. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J Biol Chem. 2005;280:39448–39459. doi: 10.1074/jbc.M509881200. [DOI] [PubMed] [Google Scholar]
- 21.Mishina Y, Lee CH, He C. Interaction of human and bacterial AlkB proteins with DNA as probed through chemical cross-linking studies. Nucleic Acids Res. 2004;32:1548–1554. doi: 10.1093/nar/gkh319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ringvoll J, Nordstrand LM, Vagbo CB, Talstad V, Reite K, et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1 me A and 3 me C lesions in DNA. EMBO J. 2006;25:2189–2198. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Y, Jia G, Pang X, Wang RN, Wang X, et al. FTO-mediated formation of N6-hydroxy methyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun. 2013;4:1798. doi: 10.1038/ncomms2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, et al. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol Cell Biol. 2010;30:2449–2459. doi: 10.1128/MCB.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Y, Dai Q, Zhang W, Ren J, Pan T, et al. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew Chem Int Ed Engl. 2010;49:8885–8888. doi: 10.1002/anie.201001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, et al. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated intranslational decoding. Mol Cell Biol. 2010;30:1814–1827. doi: 10.1128/MCB.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Born E, Vagbo CB, Songe-Moller L, Leihne V, Lien GF, et al. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat Commun. 2011;2:172. doi: 10.1038/ncomms1173. [DOI] [PubMed] [Google Scholar]
- 30.Li MM, Nilsen A, Shi Y, Fusser M, Ding YH, et al. ALKBH4-dependent demethylation of actin regulates actomyosin dynamics. Nat Commun. 2013;4:1832. doi: 10.1038/ncomms2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ougland R, Lando D, Jonson I, Dahl JA, Moen MN, et al. ALKBH1 is a histone H2A dioxygenase involved in neural differentiation. Stem Cells. 2012;30:2672–2682. doi: 10.1002/stem.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delaney JC, Smeester L, Wong C, Frick LE, Taghizadeh K, et al. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat Struct Mol Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 33.Fu D, Samson LD. Direct repair of 3,N(4)-ethenocytosine by the human ALKBH2 dioxygenase is blocked by the AAG/MPG glycosylase. DNA Repair (Amst) 2012;11:46–52. doi: 10.1016/j.dnarep.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishina Y, Yang CG, He C. Direct repair of the exocyclic DNA adduct 1,N6-ethenoadenine by the DNA repair AlkB proteins. J Am Chem Soc. 2005;127:14594–14595. doi: 10.1021/ja055957m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ringvoll J, Moen MN, Nordstrand LM, Meira LB, Pang B, et al. AlkB homologue 2-mediated repair of ethenoadenine lesions in mammalian DNA. Cancer Res. 2008;68:4142–4149. doi: 10.1158/0008-5472.CAN-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maciejewska AM, Ruszel KP, Nieminuszczy J, Lewicka J, Sokolowska B, et al. Chloroacetaldehyde-induced mutagenesis in Escherichia coli: the role of AlkB protein in repair of 3,N(4)-ethenocytosine and 3,N(4)-alpha-hydroxyethanocytosine. Mutat Res. 2010;684:24–34. doi: 10.1016/j.mrfmmm.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Bolt HM. Roles of etheno-DNA adducts in tumorigenicity of olefins. Crit Rev Toxicol. 1988;18:299–309. doi: 10.3109/10408448809037469. [DOI] [PubMed] [Google Scholar]
- 38.Guengerich FP. Cytochrome P450 oxidations in the generation of reactive electrophiles: epoxidation and related reactions. Arch Biochem Biophys. 2003;409:59–71. doi: 10.1016/s0003-9861(02)00415-0. [DOI] [PubMed] [Google Scholar]
- 39.Chung FL, Pan J, Choudhury S, Roy R, Hu W, et al. Formation of trans-4-hydroxy-2-nonenal- and otherenal-derived cyclic DNA adducts from omega-3 and omega-6 polyunsaturated fatty acids and their roles in DNA repair and human p53 gene mutation. Mutat Res. 2003;531:25–36. doi: 10.1016/j.mrfmmm.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Marnett LJ. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 41.Singer BBH. Exocyclic DNA Adducts in Mutagenesis and Carcinogenesis. IARC Press; Lyon: 1999. [PubMed] [Google Scholar]
- 42.Winczura A, Zdzalik D, Tudek B. Damage of DNA and proteins by major lipid peroxidation products in genome stability. Free Radic Res. 2012;46:442–459. doi: 10.3109/10715762.2012.658516. [DOI] [PubMed] [Google Scholar]
- 43.Barbin A. Etheno-adduct-forming chemicals: from mutagenicity testing to tumor mutation spectra. Mutat Res. 2000;462:55–69. doi: 10.1016/s1383-5742(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 44.Basu AK, Wood ML, Niedernhofer LJ, Ramos LA, Essigmann JM. Mutagenic and genotoxic effects of three vinyl chloride-induced DNA lesions: 1,N6-ethenoadenine, 3, N4-ethenocytosine, and 4-amino-5-(imidazol-2-yl) imidazole. Biochemistry. 1993;32:12793–12801. doi: 10.1021/bi00210a031. [DOI] [PubMed] [Google Scholar]
- 45.Pandya GA, Moriya M. 1,N6-ethenodeoxyadenosine, a DNA adduct highly mutagenic in mammalian cells. Biochemistry. 1996;35:11487–11492. doi: 10.1021/bi960170h. [DOI] [PubMed] [Google Scholar]
- 46.Moriya M, Zhang W, Johnson F, Grollman AP. Mutagenic potency of exocyclic DNA adducts: marked differences between Escherichia coli and simian kidney cells. Proc Natl Acad Sci U S A. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbin A, Bartsch H, Leconte P, Radman M. Studies on the miscoding properties of 1,N6-ethenoadenine and 3, N4-ethenocytosine, DNA reaction products of vinyl chloride metabolites, during in vitro DNA synthesis. Nucleic Acids Res. 1981;9:375–387. doi: 10.1093/nar/9.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller JA, Miller EC. The metabolic activation and nucleic acid adducts of naturally-occurring carcinogens: recent results with ethyl carbamate and the spice flavors safrole and estragole. Br J Cancer. 1983;48:1–15. doi: 10.1038/bjc.1983.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nair J, Sone H, Nagao M, Barbin A, Bartsch H. Copper-dependent formation of miscoding etheno-DNA adducts in the liver of Long Evans cinnamon (LEC) rats developing hereditary hepatitis and hepatocellular carcinoma. Cancer Res. 1996;56:1267–1271. [PubMed] [Google Scholar]
- 50.Saparbaev M, Kleibl K, Laval J. Escherichia coli, Saccharomyces cerevisiae, rat and human 3-methyladenine DNA glycosylases repair 1,N6-ethenoadenine when present in DNA. Nucleic Acids Res. 1995;23:3750–3755. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saparbaev M, Langouet S, Privezentzev CV, Guengerich FP, Cai H, et al. 1,N(2)-ethenoguanine, a mutagenic DNA adduct, is a primary substrate of Escherichia coli mismatch-specific uracil-DNA glycosylase and human alkylpurine-DNA-N-glycosylase. J Biol Chem. 2002;277:26987–26993. doi: 10.1074/jbc.M111100200. [DOI] [PubMed] [Google Scholar]
- 52.Hang B, Medina M, Fraenkel-Conrat H, Singer B. A 55-kDa protein isolated from human cells shows DNA glycosylase activity toward 3,N4-ethenocytosine and the G/T mismatch. Proc Natl Acad Sci U S A. 1998;95:13561–13566. doi: 10.1073/pnas.95.23.13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saparbaev M, Laval J. 3, N4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase. Proc Natl Acad Sci U S A. 1998;95:8508–8513. doi: 10.1073/pnas.95.15.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, et al. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J Biol Chem. 2002;277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 55.Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Genuardi M, et al. Investigation of the substrate spectrum of the human mismatch-specific DNA N-glycosylase MED1 (MBD4): fundamental role of the catalytic domain. J Cell Physiol. 2000;185:473–480. doi: 10.1002/1097-4652(200012)185:3<473::AID-JCP19>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 56.Matijasevic Z, Sekiguchi M, Ludlum DB. Release of N2,3-ethenoguanine from chloroacetaldehyde-treated DNA by Escherichia coli 3-methyladenine DNA glycosylase II. Proc Natl Acad Sci U S A. 1992;89:9331–9334. doi: 10.1073/pnas.89.19.9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gallinari P, Jiricny J. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature. 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 58.Prorok P, Saint-Pierre C, Gasparutto D, Fedorova OS, Ishchenko AA, et al. Highly mutagenic exocyclic DNA adducts are substrates for the human nucleotide incision repair pathway. PLoS ONE. 2012;7:e51776. doi: 10.1371/journal.pone.0051776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van den Born E, Bekkelund A, Moen MN, Omelchenko MV, Klungland A, et al. Bioinformatics and functional analysis define four distinct groups of AlkB DNA-dioxygenases in bacteria. Nucleic Acids Res. 2009;37:7124–7136. doi: 10.1093/nar/gkp774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodenough AK, Kozekov ID, Zang H, Choi JY, Guengerich FP, et al. Site specific synthesis and polymerase bypass of oligonucleotides containing a 6-hydroxy-3,5,6,7-tetrahydro-9H-imidazo[1,2-a]purin-9-one base, an intermediate in the formation of 1,N2-etheno-2′-deoxyguanosine. Chem Res Toxicol. 2005;18:1701–1714. doi: 10.1021/tx050141k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langouet S, Muller M, Guengerich FP. Misincorporation of dNTPs opposite 1,N2-ethenoguanine and 5,6,7,9-tetrahydro-7-hydroxy-9-oxoimidazo[1,2-a] purine in oligonucleotides by Escherichia coli polymerases I exo- and II exo-, T7 polymerase exo-, human immunodeficiency virus-1 reverse transcriptase, and rat polymerase beta. Biochemistry. 1997;36:6069–6079. doi: 10.1021/bi962526v. [DOI] [PubMed] [Google Scholar]
- 62.Basu AK, McNulty JM, McGregor WG. Solution conformation and mutagenic specificity of 1,N6-ethenoadenine. IARC Sci Publ. 1999:325–333. [PubMed] [Google Scholar]
- 63.Speina E, Ciesla JM, Wojcik J, Bajek M, Kusmierek JT, et al. The pyrimidine ring-opened derivative of 1,N6-ethenoadenine is excised from DNA by the Escherichia coli Fpg and Nth proteins. J Biol Chem. 2001;276:21821–21827. doi: 10.1074/jbc.M100998200. [DOI] [PubMed] [Google Scholar]
- 64.Maciejewska AM, Sokolowska B, Nowicki A, Kusmierek JT. The role of AlkB protein in repair of 1,N(6)-ethenoadenine in Escherichia coli cells. Mutagenesis. 2011;26:401–406. doi: 10.1093/mutage/geq107. [DOI] [PubMed] [Google Scholar]
- 65.Vogel TM, McCarty PL. Biotransformation of tetrachloroethylene to trichloroethylene, dichloroethylene, vinyl chloride, and carbon dioxide under methanogenic conditions. Appl Environ Microbiol. 1985;49:1080–1083. doi: 10.1128/aem.49.5.1080-1083.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu C, Yi C. Switching demethylation activities between AlkB family RNA/DNA demethylases through exchange of active-site residues. Angew Chem Int Ed Engl. 2014;53:3659–3662. doi: 10.1002/anie.201310050. [DOI] [PubMed] [Google Scholar]
- 67.Singh V, Fedeles BI, Li D, Delaney JC, Kozekov ID, et al. Mechanism of repair of acrolein- and malondialdehyde-derived exocyclic guanine adducts by the alpha-ketoglutarate/Fe(II)dioxygenase AlkB. Chem Res Toxicol. 2014;27:1619–1631. doi: 10.1021/tx5002817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ergel B, Gill ML, Brown L, Yu B, Palmer AG, 3rd, et al. Protein dynamics control the progression and efficiency of the catalytic reaction cycle of the Escherichia coli DNA-repair enzyme AlkB. J Biol Chem. 2014;289:29584–29601. doi: 10.1074/jbc.M114.575647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Calvo JA, Meira LB, Lee CY, Moroski-Erkul CA, Abolhassani N, et al. DNA repair is indispensable for survival after acute inflammation. J Clin Invest. 2012;122:2680–2689. doi: 10.1172/JCI63338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morera S, Grin I, Vigouroux A, Couve S, Henriot V, et al. Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA. Nucleic Acids Res. 2012;40:9917–9926. doi: 10.1093/nar/gks714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sjolund AB, Senejani AG, Sweasy JB. MBD 4 and TDG: multifaceted DNA glycosylases with ever expanding biological roles. Mutat Res. 2013;743–744:12–25. doi: 10.1016/j.mrfmmm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


