Abstract
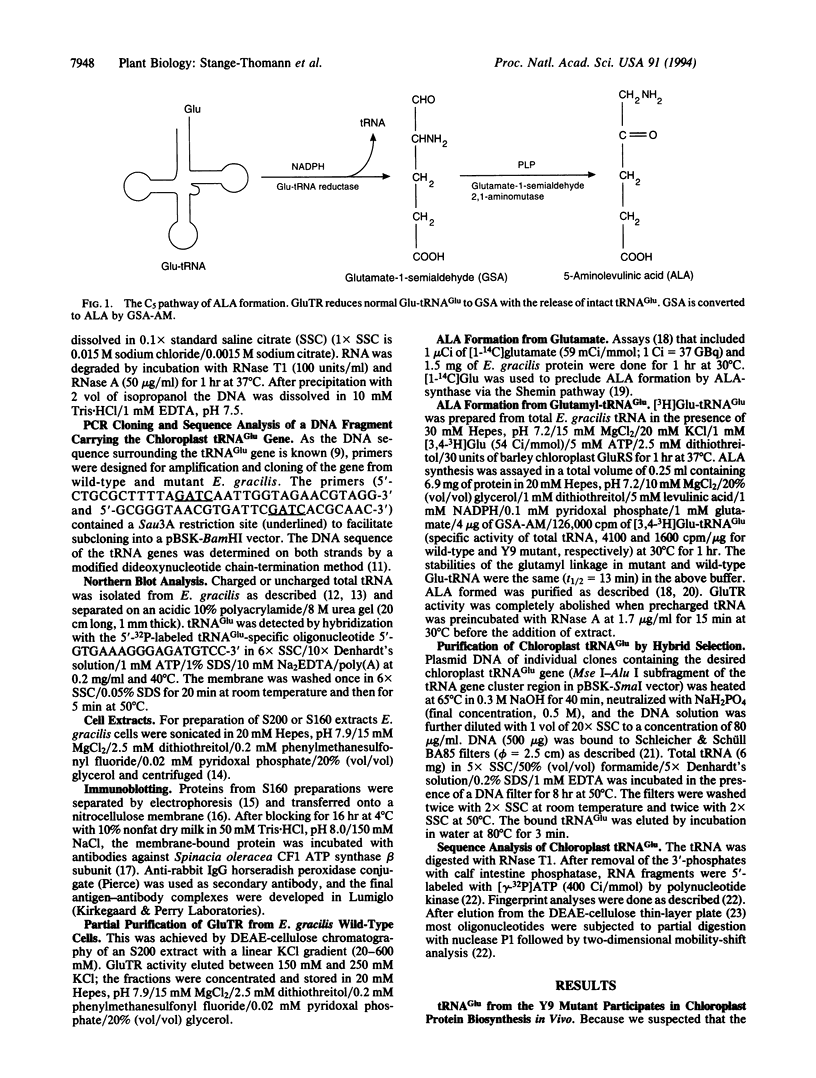
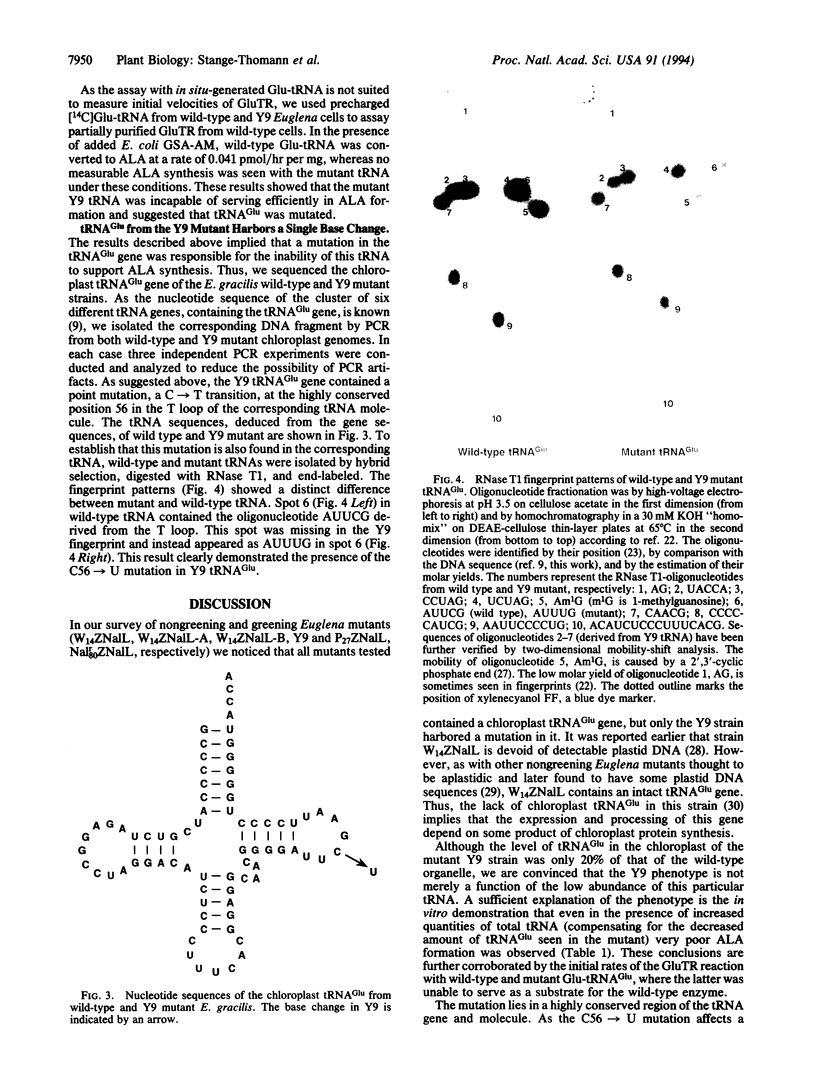
The universal precursor of tetrapyrrole pigments (e.g., chlorophylls and hemes) is 5-aminolevulinic acid (ALA), which in Euglena gracilis chloroplasts is derived via the two-step C5 pathway from glutamate charged to tRNA(Glu). The first enzyme in this pathway, Glu-tRNA reductase (GluTR) catalyzes the reduction of glutamyl-tRNA(Glu) (Glu-tRNA) to glutamate 1-semialdehyde (GSA) with the release of the uncharged tRNA(Glu). The second enzyme, GSA-2,1-aminomutase, converts GSA to ALA. tRNA(Glu) is a specific cofactor for the NADPH-dependent reduction by GluTR, an enzyme that recognizes the tRNA in a sequence-specific manner. This RNA is the normal tRNA(Glu), a dual-function molecule participating both in protein and in ALA and, hence, chlorophyll biosynthesis. A chlorophyll-deficient mutant of E. gracilis (Y9ZNalL) does not synthesize ALA from glutamate, although it contains GluTR and GSA-2,1-aminomutase activity. The tRNA(Glu) isolated from the mutant can still be acylated with glutamate in vitro and in vivo. Furthermore, it supports chloroplast protein synthesis; however, it is a poor substrate for GluTR. Sequence analysis of the tRNA and of its gene revealed a C56-->U mutation in the resulting gene product. C56 is therefore an important identity element for GluTR. Thus, a point mutation in the T loop of tRNA uncouples protein from chlorophyll biosynthesis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avissar Y. J., Ormerod J. G., Beale S. I. Distribution of delta-aminolevulinic acid biosynthetic pathways among phototrophic bacterial groups. Arch Microbiol. 1989;151(6):513–519. doi: 10.1007/BF00454867. [DOI] [PubMed] [Google Scholar]
- Beale S. I. Biosynthesis of the Tetrapyrrole Pigment Precursor, delta-Aminolevulinic Acid, from Glutamate. Plant Physiol. 1990 Aug;93(4):1273–1279. doi: 10.1104/pp.93.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton R., Sanfaçon H., Papayannopoulos I., Biemann K., Lapointe J. Glutamyl-tRNA synthetase of Escherichia coli. Isolation and primary structure of the gltX gene and homology with other aminoacyl-tRNA synthetases. J Biol Chem. 1986 Aug 15;261(23):10610–10617. [PubMed] [Google Scholar]
- Chen E. Y., Roe B. A. Sequence studies on human placenta tRNAVal : comparison with the mouse myeloma tRNAVal. Biochem Biophys Res Commun. 1977 Sep 23;78(2):631–640. doi: 10.1016/0006-291x(77)90226-1. [DOI] [PubMed] [Google Scholar]
- Domdey H., Jank P., Sänger L., Gross H. J. Studies on the primary and secondary structure of potato spindle tuber viroid: products of digestion with ribonuclease A and ribonuclease T1, and modification with bisulfite. Nucleic Acids Res. 1978 Apr;5(4):1221–1236. doi: 10.1093/nar/5.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie D., Spiegelman S. A quantitative assay for DNA-RNA hybrids with DNA immobilized on a membrane. J Mol Biol. 1965 Jul;12(3):829–842. doi: 10.1016/s0022-2836(65)80331-x. [DOI] [PubMed] [Google Scholar]
- Grimm B. Primary structure of a key enzyme in plant tetrapyrrole synthesis: glutamate 1-semialdehyde aminotransferase. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4169–4173. doi: 10.1073/pnas.87.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallick R. B., Hong L., Drager R. G., Favreau M. R., Monfort A., Orsat B., Spielmann A., Stutz E. Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res. 1993 Jul 25;21(15):3537–3544. doi: 10.1093/nar/21.15.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Hollingsworth M. J., Hallick R. B. Euglena gracilis chloroplast transfer RNA transcription units. Nucleotide sequence analysis of a tRNATyr-tRNAHis-tRNAMet-tRNATrp-tRNAGlu-tRNAGly gene cluster. J Biol Chem. 1982 Nov 10;257(21):12795–12799. [PubMed] [Google Scholar]
- Jahn D., Verkamp E., Söll D. Glutamyl-transfer RNA: a precursor of heme and chlorophyll biosynthesis. Trends Biochem Sci. 1992 Jun;17(6):215–218. doi: 10.1016/0968-0004(92)90380-r. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lapointe J., Söll D. Glutamyl transfer ribonucleic acid synthetase of Escherichia coli. I. Purification and properties. J Biol Chem. 1972 Aug 25;247(16):4966–4974. [PubMed] [Google Scholar]
- Little M. C., Hallick R. B. Chloroplast rpoA, rpoB, and rpoC genes specify at least three components of a chloroplast DNA-dependent RNA polymerase active in tRNA and mRNA transcription. J Biol Chem. 1988 Oct 5;263(28):14302–14307. [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Mayer S. M., Beale S. I. Light Regulation of delta-Aminolevulinic Acid Biosynthetic Enzymes and tRNA in Euglena gracilis. Plant Physiol. 1990 Nov;94(3):1365–1375. doi: 10.1104/pp.94.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer S. M., Beale S. I. delta-Aminolevulinic Acid Biosynthesis from Glutamatein Euglena gracilis: Photocontrol of Enzyme Levels in a Chlorophyll-Free Mutant. Plant Physiol. 1991 Nov;97(3):1094–1102. doi: 10.1104/pp.97.3.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A., Kim S. H. The three-dimensional structure of transfer RNA. Sci Am. 1978 Jan;238(1):52–62. doi: 10.1038/scientificamerican0178-52. [DOI] [PubMed] [Google Scholar]
- Rogers K. C., Söll D. Discrimination among tRNAs intermediate in glutamate and glutamine acceptor identity. Biochemistry. 1993 Dec 28;32(51):14210–14219. doi: 10.1021/bi00214a021. [DOI] [PubMed] [Google Scholar]
- Russell G. K., Draffan A. G. Light-induced Enzyme Formation in a Chlorophyll-less Mutant of Euglena gracilis. Plant Physiol. 1978 Nov;62(5):678–682. doi: 10.1104/pp.62.5.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön A., Kannangara C. G., Gough S., Söll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988 Jan 14;331(6152):187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- Schön A., Krupp G., Gough S., Berry-Lowe S., Kannangara C. G., Söll D. The RNA required in the first step of chlorophyll biosynthesis is a chloroplast glutamate tRNA. Nature. 1986 Jul 17;322(6076):281–284. doi: 10.1038/322281a0. [DOI] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- Soteropoulos P., Süss K. H., McCarty R. E. Modifications of the gamma subunit of chloroplast coupling factor 1 alter interactions with the inhibitory epsilon subunit. J Biol Chem. 1992 May 25;267(15):10348–10354. [PubMed] [Google Scholar]
- Sylvers L. A., Rogers K. C., Shimizu M., Ohtsuka E., Söll D. A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry. 1993 Apr 20;32(15):3836–3841. doi: 10.1021/bi00066a002. [DOI] [PubMed] [Google Scholar]
- Thomann H. U., Schmutzler C., Hüdepohl U., Blow M., Gross H. J. Genes, variant genes and pseudogenes of the human tRNA(Val) gene family. Expression and pre-tRNA maturation in vitro. J Mol Biol. 1989 Oct 20;209(4):505–523. doi: 10.1016/0022-2836(89)90590-1. [DOI] [PubMed] [Google Scholar]
- Ueda Y., Kumagai I., Miura K. The effects of a unique D-loop structure of a minor tRNA(UUALeu) from Streptomyces on its structural stability and amino acid accepting activity. Nucleic Acids Res. 1992 Aug 11;20(15):3911–3917. doi: 10.1093/nar/20.15.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney U., Lee C. P., RajBhandary U. L. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J Biol Chem. 1991 Dec 25;266(36):24712–24718. [PubMed] [Google Scholar]
- Zurawski G., Bottomley W., Whitfeld P. R. Structures of the genes for the beta and epsilon subunits of spinach chloroplast ATPase indicate a dicistronic mRNA and an overlapping translation stop/start signal. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6260–6264. doi: 10.1073/pnas.79.20.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]