Abstract
Reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) is a powerful technique to quantify gene expression. To facilitate gene expression study and obtain accurate results, normalization relative to stably expressed reference genes is crucial. The monarch butterfly, Danaus plexippus (L.), is one of the most recognized insect species for its spectacular annual migration across North America. Besides its great voyages, D. plexippus has drawn attention to its role as a bio-indicator, ranging from genetically modified organisms (GMOs) to natural ecosystems. In this study, nine reference genes from D. plexippus genome were selected as the candidate reference genes. The expression profiles of these candidates under various biotic and abiotic conditions were evaluated using the four readily available computational programs, BestKeeper, Normfinder, geNorm, and ΔCt method, respectively. Moreover, RefFinder, a web-based computational platform integrating the four above mentioned algorisms, provided a comprehensive ranking of the stability of these reference genes. As a result, a suite of reference genes were recommended for each experimental condition. Specifically, elongation factor 1α (EF1A) and ribosomal protein 49 (RP49) were the most stable reference genes, respectively, under biotic (development, tissue, and sex) and abiotic (photoperiod, temperature, and dietary RNAi) conditions. With the recent release of a 273-million base pair draft genome, results from this study allow us to establish a standardized RT-qPCR analysis and lay a foundation for the subsequent genomic and functional genomic research in D. plexippus, a major bio-indicator and an emerging model for migratory animals.
Introduction
The advent of next-generation sequencing technologies has led to a significant increase in transcriptomic and genomic output for various organisms [1–3]. Validation of gene expression has become a standard for reporting and assessing the quality of these transcriptomic and genomic resources [4, 5]. Reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) is a powerful technique to target and quantify gene expression [6], however, there remain limitations can significantly influence the normalization of gene expression, such as variations in RNA extraction, RNA quality and integrity, cDNA quality, and PCR efficiency [7–10]. In RT-qPCR, a commonly used technique to normalize the gene expression data is to introduce one or multiple reference genes, which are stably expressed across various experimental conditions and serve as the internal control [6, 9, 10]. Although these reference genes have been defined functionally as ‘constitutively expressed to maintain cellular function’, they do not necessarily meet prerequisites for a good reference gene that can be ‘expressed at constant levels across various biotic and abiotic conditions [4, 8, 10]. Many studies have proved that some commonly used reference genes express differentially across different experimental conditions; researchers have documented that multiple reference genes should be used for accurate normalization [4, 8, 9, 11, 12].
RNA interference (RNAi) is a biological process in which RNA molecules inhibit gene expression, typically by causing the destruction of specific mRNA molecules. In view of its high sequence specific reality, RNAi has drawn great attention from the crop protectors who recognized it as a novel and environmentally friendly ways for insect pest control [13, 14]. Recently, RNAi-based transgenic crops targeting insects have been developed [15–17]. One of the major ecological concerns with respect to the bio-safety of transgenic crops on the environment is their potential effects on non-target organisms [14]. The monarch butterfly is recognized as an important symbol of biodiversity and had been selected as a surrogate species for the risk assessment of Bacillus thuringiensis (Bt) and RNAi-based transgenic crops. Given the nature of RNAi mechanisms, non-target effects could occur via the modulation of gene expressions in these organisms [13]. Therefore, RT-qPCR will be a major research tool to evaluate potential non-target effects of this new biotechnology.
The monarch butterfly, Danaus plexippus (L.) (Lepidoptera: Danainae), is famous for its late summer/autumn southward migration from the United States and southern Canada to Mexico, and northward return in spring, which occurs over the lifespan of three to four generations of the butterfly [18]. In addition to this migration, D. plexippus has drawn attention as a surrogate species for the ecological risk assessment of genetically modified organisms (GMOs) [19–22] and as an eco-indicator for global climate change [23, 24]. The recent release of a 273-million base pair draft genome, including 16,866 protein-coding genes [25], provides an unprecedented opportunity to investigate the genetic basis governing monarch migration, including but not limited to genes involved in their migratory behavior, circadian rhythm, juvenile hormone modulation, and warning coloration [25–27]. Recent studies have identified the specific areas in the genome of the monarch that regulate migration. No genetic difference was observed between migrating and non-migrating monarchs but certain genes are expressed solely in migrating monarchs [25, 27].
The objective of this study was to determine suitable reference genes with stable expression in D. plexippus across various biotic and abiotic conditions. Here, nine reference genes from D. plexippus genome were selected as candidate reference genes [25], including elongation factor 1α (EF1A), glyceralde hyde-3-phosphate dehydro-genase (GAPDH), nicotinamide adenine dinucleotide (NADH), cyclophilins A (CypA), ribosomal protein S5 (RPS5), ribosomal protein 49 (RP49), vacuolar-type H + -ATPase (v-ATPase), 28S ribosomal RNA (28S), and 18S ribosomal RNA (18S). All of these candidates have been used previously as endogenous references for gene expression analyses [4, 8–12]. Stability of these genes was investigated under three biotic (developmental stage, tissue type, and sex) and three abiotic (temperature, photoperiod, and dietary RNAi) conditions. As a result, different sets of reference genes were recommended for each experimental condition. To validate the recommendations, effectiveness of these candidates were further examined by RT-qPCR analysis of a circadian clock gene timeless (tim) [28].
Materials and Methods
Ethics Statement
Eggs of Danaus plexippus were collected from common milkweed, Asclepias syriaca, near Ames, Iowa spring and summer 2013. No specific permit was required for the field collection. Danaus plexippus is a common American butterfly species in the United States. The permit to move live plant pests, noxious weeds, and soil were authorized by the United States Department of Agriculture Animal and Plant Health Inspection Service (Permit number: P526P-13-03521). Four to five generations of monarch adults were screened for presence of the protozoan parasite, Ophryocystis elektroscirrha, in order to eliminate it from the population. D. plexippus larvae from the maintenance colony were reared on either common milkweed or tropical milkweed, Asclepias curassavica. Larvae during the experiments were fed honeyvine milkweed, Cynanchum laeve (syn. Ampelamus albidus) at 25 ± 1°C (16L: 8D), and adults were fed 15% sugar solution.
Experimental conditions
Biotic factors
The different developmental stages including eggs, 1st, 2nd, 3rd, 4th instar larvae (collected at the first day of each instar), pupae, and adults. Tissues, including head, midgut, and carcass, were dissected from the 4th instar larvae. Male and female adults were collected separately to examine expression profiles of candidate genes between sexes.
Abiotic factors
To examine temperature influence, 2nd instar D. plexippus were exposed to 10°C, 22°C, and 37°C for 2 h. For photoperiod, 2nd instars were treated with exposure to 16:8 h, 12:12 h, and 8:16 h light: dark regime for 3 d. For the double-stranded RNA (dsRNA) feeding bioassay, there were three treatments including dsRNAs synthesized from D. plexippus vATPase subunit A (dsDP), β-glucuronidase (GUS) (dsGUS), and water control. Neonates (< 12 h) were given 1.6 μl of water solutions containing 5.0 μg/μl dsRNA each day for two days. Larvae were collected on the third day of the experiment. Altogether, 16μg of dsRNA was ingested by each D. plexippus larva.
For egg samples, six individuals were collected for each replicate; for pupa samples, one pupa was collected for each replicate. For the other biotic and abiotic conditions, approximately five individuals were collected for each treatment with three replicates. All samples were snap frozen with liquid nitrogen in 1.5 ml centrifuge tubes and then stored at -80°C.
Total RNA extraction and cDNA synthesis
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) according to previously described methods [29, 30]. First-strand cDNA was synthesized from 700 ng of total RNA using the M-MLV reverse transcription kit (Invitrogen, Carlsbad, CA) according to manufacturer’s recommendations. The cDNA was diluted 10-fold for the subsequent RT-qPCR studies.
Reference gene primer design and RT-qPCR
Nine reference genes were selected (Table 1). The primers for the RT-qPCR were designed online (https://www.idtdna.com/Primerquest/Home/Index). The information of RT-qPCR amplification reaction and program were described in detail in our previous study [29, 30]. The standard curve and PCR efficiency of each candidate reference gene were constructed and calculated according to previously described methods [29, 30].
Table 1. Primers used for RT-qPCR.
| Gene | Accessionnumber | Primer sequences (5’-3’) | Length(bp) | Efficiency(%) | R2 |
|---|---|---|---|---|---|
| EF1A | DQ157894 | F: TGTCGCTTTCGTACCCATTT | 114 | 97.1 | 0.989 |
| R: CCTTCAGCCTTACCCTCTTTAC | |||||
| NADH | U32457 | F: GGTGTTTTAATTGGGGTTGC | 122 | 102.3 | 0.9992 |
| R:GCATCAGAAAAAGTTTGTAATAATCC | |||||
| GAPDH | EU141486 | F: CGTTCCCGTAGCTAATGTATCC | 93 | 106.1 | 0.9985 |
| R: GCTTCCTTGACCTTCTGTTTAATG | |||||
| CypA | EHJ70135 | F: CAACGGCTCTCAATTCTTCATC | 89 | 99.9 | 0.9898 |
| R: CCATGCCTTCAACCACATTAC | |||||
| RPS5 | EU141382 | F: GTCGTGTGAACCAGGCTATT | 98 | 100.5 | 0.9991 |
| R: GAGCTCATCAGCAACACATTC | |||||
| RP49 | AY971345 | F: CCGGAAGGTGTTAGTCCACAAC | 71 | 98.6 | 0.9997 |
| R: CGGCGCAGTACTTCCTATTCTG | |||||
| 18S | AF394668 | F: TGAGAAACGGCTACCACATC | 102 | 109.3 | 0.9997 |
| R: CGTAAGAGTCCCGTATCGTTATT | |||||
| 28S | GQ229491 | F: AACAAGTACCGTGAGGGAAAG | 94 | 102.7 | 0.9978 |
| R: CATTCGAGTTTCGCAGGTTTAC | |||||
| v-ATPase | EHJ63113 | F: AGGACGACTTCCTGCAACAGAACA | 84 | 105.9 | 0.9998 |
| R: TGTTCTTCAACATGCCCACCGTCT |
Data analysis
Stability of the nine candidate reference genes were evaluated by four computational programs, including geNorm [6], NormFinder [10], BestKeeper [31], and the ΔC t method [32]. In addition, RefFinder (http://www.leonxie.com/referencegene.php), a web-based comprehensive platform, which integrates the four above mentioned algorithms, provided the overall ranking of the stability of these reference genes.
Validation of selected reference genes
The photoperiod-relevant tim gene of D. plexippus was used to evaluate the candidate reference genes. tim expression levels were determined in 2nd instars treated for 3 d under three photoperiod conditions (16L:8D, 12L:12D, and 8L:12D). Relative quantification of tim in different samples was performed using the 2-ΔΔCt method [33].
Results
Performance and Ct values of candidate reference genes
Each of the tested genes displayed a single amplicon with the expected size on a 1.5% agarose gel (S1 Fig). Moreover, gene-specific of these candidates was affirmed by a single peak in melting curve analysis (S2 Fig). Standard curves were created for all the candidates, and the PCR efficiency and correlation coefficient for each standard curve were shown in Table 1.
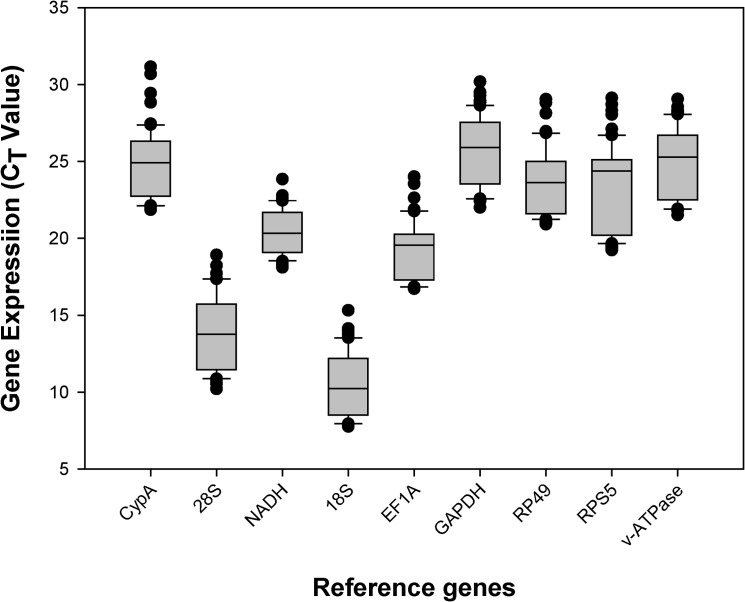
Gene expression analyses of the nine reference genes exhibited a broad C t range (Fig 1). C t values ranged from 7 to 29, while most of the values were distributed between 11 and 22. The most abundant transcripts were 18S and 28S; and the least abundant transcripts were GAPDH and CypA.
Fig 1. Expression profiles of candidate reference genes in D. plexippus.
The expression level of candidate reference genes in 60 tested samples are documented in C t-value. The dot indicates the maximum or minimum value of replicated samples, while whiskers indicate the standard error of the mean.
Stability of candidate reference genes under biotic conditions
Developmental stage
All software programs, except BestKeeper, identified EF1A as the most stable gene (Table 2).
Table 2. Stability of reference gene expression under biotic conditions.
| Biotic | Gene | geNorm | Normfinder | BestKeeper | ΔCt | ||||
|---|---|---|---|---|---|---|---|---|---|
| conditions | Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | |
| Development | EF1A | 0.53 | 1 | 0.61 | 1 | 1.35 | 6 | 1.07 | 1 |
| NADH | 1.27 | 8 | 1.66 | 9 | 0.89 | 2 | 1.81 | 9 | |
| GAPDH | 1.12 | 7 | 0.93 | 7 | 0.98 | 4 | 1.33 | 8 | |
| RP49 | 0.53 | 1 | 0.83 | 5 | 1.20 | 9 | 1.14 | 4 | |
| 18S | 0.91 | 5 | 0.81 | 4 | 0.91 | 3 | 1.24 | 5 | |
| 28S | 0.70 | 3 | 0.65 | 2 | 1.39 | 7 | 1.10 | 2 | |
| v-ATPase | 1.04 | 6 | 0.90 | 6 | 0.85 | 1 | 1.30 | 7 | |
| CypA | 0.80 | 4 | 0.99 | 8 | 1.40 | 8 | 1.30 | 6 | |
| RPS5 | 0.66 | 2 | 0.75 | 3 | 1.20 | 5 | 1.14 | 3 | |
| Tissue | EF1A | 0.45 | 1 | 0.51 | 2 | 0.80 | 7 | 0.87 | 2 |
| NADH | 0.60 | 3 | 0.71 | 5 | 0.77 | 5 | 0.98 | 3 | |
| GAPDH | 1.01 | 8 | 1.22 | 9 | 0.73 | 4 | 1.37 | 9 | |
| RP49 | 0.57 | 2 | 0.75 | 7 | 0.73 | 3 | 1.00 | 5 | |
| 18S | 0.80 | 5 | 0.82 | 8 | 1.03 | 9 | 1.07 | 8 | |
| 28S | 0.45 | 1 | 0.20 | 1 | 0.53 | 2 | 0.79 | 1 | |
| v-ATPase | 0.70 | 4 | 0.67 | 3 | 0.97 | 8 | 0.99 | 4 | |
| CypA | 0.91 | 7 | 0.68 | 4 | 0.41 | 1 | 1.00 | 6 | |
| RPS5 | 0.87 | 6 | 0.75 | 6 | 0.77 | 6 | 1.05 | 7 | |
| Sex | EF1A | 0.41 | 3 | 0.64 | 5 | 0.90 | 6 | 0.98 | 4 |
| NADH | 1.23 | 8 | 2.21 | 9 | 1.45 | 9 | 2.27 | 9 | |
| GAPDH | 0.71 | 6 | 0.53 | 4 | 0.72 | 3 | 1.15 | 6 | |
| RP49 | 0.35 | 2 | 0.10 | 1 | 0.91 | 4 | 0.89 | 1 | |
| 18S | 0.29 | 1 | 0.15 | 2 | 0.68 | 2 | 0.89 | 2 | |
| 28S | 0.29 | 1 | 0.35 | 3 | 0.82 | 5 | 0.93 | 3 | |
| v-ATPase | 0.93 | 7 | 1.24 | 7 | 0.64 | 1 | 1.55 | 8 | |
| CypA | 0.56 | 5 | 1.31 | 8 | 1.27 | 8 | 1.39 | 7 | |
| RPS5 | 0.43 | 4 | 0.70 | 6 | 0.91 | 7 | 1.01 | 5 |
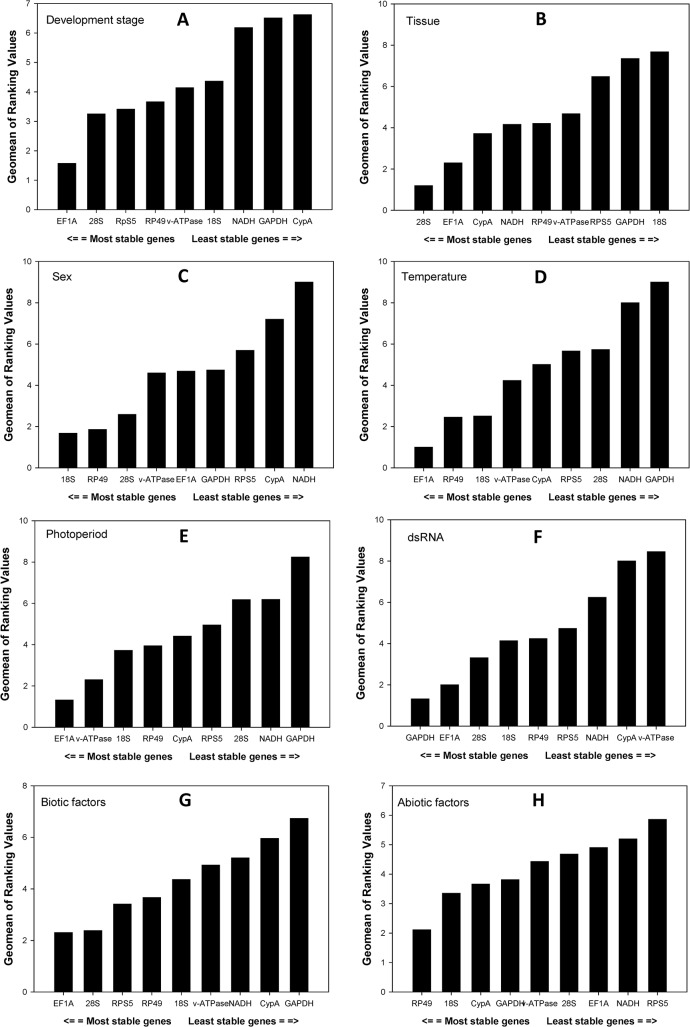
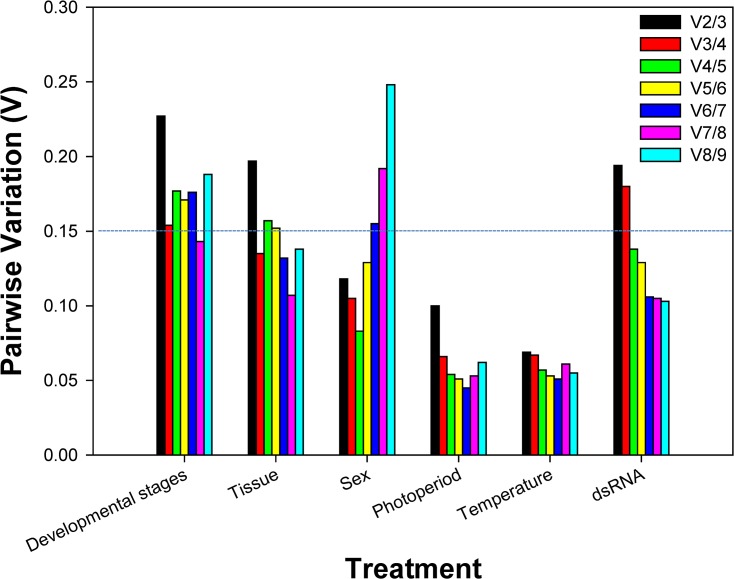
According to RefFinder, the overall order from the most stable to the least stable reference genes across different developmental stages was: EF1A, 28S, RPS5, RP49, v-ATPase, 18S, NADH, GAPDH, CypA (Fig 2A). geNorm analysis revealed that the pair-wise variation value V3/4 was 0.154, close to the proposed 0.15 cut-off (Fig 3), suggesting that three reference genes were required for reliable normalization throughout developmental stages.
Fig 2. Stability of candidate reference genes expression under different treatment by RefFinder.
A lower Geomean value indicates more stable expression.
Fig 3. Determination of the optimal number of reference genes.
Pairwise variation (V) is an index for determining the optimal number of reference genes for accurate RT-qPCR normalization. A cut-off value for pairwise variation of 0.15 was recommended [6].
Tissue
Majority of the software programs ranked 28S as the most stable gene (Table 2). Based on RefFinder, the overall order from the most to the least stable reference genes in different tissue was: 28S, EF1A, CypA, NADH, RP49, v-ATPase, RPS5, GAPDH, 18S (Fig 2B). geNorm analysis revealed that the first V-value < 0.15 showed at V3/4 (Fig 3), suggesting that three reference genes were sufficient for reliable normalization in different tissue types.
Sex
According to the results generated by geNorm, 18S was ranked as the most stable gene whereas the other three programs identified it as the second stable gene (Table 2). According to RefFinder, the overall order from the most stable to the least stable reference genes in both female and male adults was: 18S, RP49, 28S, v-ATPase, EF1A, GAPDH, RPS5, CypA, NADH (Fig 2C). geNorm analysis revealed that the first V-value < 0.15 showed at V2/3 (Fig 3), suggesting that two reference genes were enough for accurate normalization in female and male adults.
Stability of candidate reference genes under abiotic conditions
Temperature
All software programs identified EF1A as the most stable gene (Table 3). According to RefFinder, the overall order from the most stable to the least stable reference genes under the temperature stress was: EF1A, RP49, 18S, v-ATPase, CypA, RPS5, 28S, NADH, GAPDH (Fig 2D). geNorm analysis revealed that first V-value < 0.15 showed at V2/3 (Fig 3), suggesting that two reference genes were sufficient for accurate normalization across different temperatures.
Table 3. Stability of reference gene expression under abiotic conditions.
| Abiotic | Gene | geNorm | Normfinder | BestKeeper | ΔCt | ||||
|---|---|---|---|---|---|---|---|---|---|
| conditions | Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | |
| Temperature | EF1A | 0.18 | 1 | 0.05 | 1 | 0.15 | 1 | 0.31 | 1 |
| NADH | 0.38 | 7 | 0.45 | 8 | 0.39 | 8 | 0.53 | 8 | |
| GAPDH | 0.42 | 8 | 0.49 | 9 | 0.45 | 9 | 0.55 | 9 | |
| RP49 | 0.18 | 1 | 0.25 | 4 | 0.19 | 3 | 0.37 | 3 | |
| 18S | 0.28 | 4 | 0.17 | 2 | 0.16 | 2 | 0.35 | 2 | |
| 28S | 0.30 | 5 | 0.30 | 6 | 0.28 | 5 | 0.42 | 6 | |
| v-ATPase | 0.25 | 3 | 0.26 | 5 | 0.20 | 4 | 0.39 | 4 | |
| CypA | 0.33 | 6 | 0.24 | 3 | 0.29 | 6 | 0.40 | 5 | |
| RPS5 | 0.21 | 2 | 0.37 | 7 | 0.29 | 7 | 0.44 | 7 | |
| Photoperiod | EF1A | 0.27 | 2 | 0.14 | 1 | 0.27 | 1 | 0.35 | 1 |
| NADH | 0.32 | 5 | 0.29 | 7 | 0.40 | 5 | 0.40 | 7 | |
| GAPDH | 0.37 | 7 | 0.43 | 8 | 0.46 | 8 | 0.51 | 8 | |
| RP49 | 0.28 | 3 | 0.25 | 4 | 0.31 | 3 | 0.39 | 5 | |
| 18S | 0.17 | 1 | 0.27 | 6 | 0.46 | 8 | 0.39 | 4 | |
| 28S | 0.42 | 8 | 0.55 | 9 | 0.29 | 2 | 0.60 | 9 | |
| v-ATPase | 0.17 | 1 | 0.20 | 2 | 0.46 | 7 | 0.35 | 2 | |
| CypA | 0.33 | 6 | 0.23 | 3 | 0.41 | 6 | 0.39 | 3 | |
| RPS5 | 0.30 | 4 | 0.25 | 5 | 0.38 | 4 | 0.40 | 6 | |
| dietary RNAi | EF1A | 0.37 | 1 | 0.47 | 2 | 0.54 | 4 | 0.79 | 2 |
| NADH | 0.81 | 6 | 0.65 | 6 | 0.62 | 6 | 0.91 | 6 | |
| GAPDH | 0.37 | 1 | 0.46 | 1 | 0.51 | 3 | 0.79 | 1 | |
| RP49 | 0.53 | 2 | 0.55 | 4 | 0.72 | 9 | 0.84 | 3 | |
| 18S | 0.77 | 5 | 0.66 | 7 | 0.31 | 1 | 0.92 | 7 | |
| 28S | 0.72 | 4 | 0.55 | 3 | 0.31 | 2 | 0.85 | 4 | |
| v-ATPase | 0.91 | 8 | 0.90 | 9 | 0.69 | 7 | 1.08 | 9 | |
| CypA | 0.86 | 7 | 0.87 | 8 | 0.69 | 8 | 1.05 | 8 | |
| RPS5 | 0.66 | 3 | 0.63 | 5 | 0.58 | 5 | 0.90 | 5 |
Photoperiod
All software programs identified EF1A as the most or second to most stable gene (Table 3). According to RefFinder, the overall order from the most stable to the least stable reference genes under the photoperiod stress was: EF1A, v-ATPase, 18S, RP49, CypA, RPS5, 28S, NADH, GAPDH (Fig 2E). geNorm analysis revealed that first V-value < 0.15 showed at V2/3 (Fig 3), suggesting that two reference genes were adequate for accurate normalization under the photoperiod condition.
dietary RNAi
All software programs, except BestKeeper, identified GAPDH as the most stable gene (Table 3). According to RefFinder, the overall order from the most stable to the least stable reference genes under the dietary RNAi treatment was: GAPDH, EF1A, 28S, 18S, RP49, RPS5, NADH, CypA, v-ATPase (Fig 2F). geNorm analysis revealed that the pair-wise variation value V4/5 was below the proposed 0.15 cut-off (Fig 3). This result suggests that four reference genes were required for accurate normalization under the dietary RNAi condition.
According to RefFinder, the overall order from the most stable to the least stable reference genes under biotic conditions was: EF1A, 28S, RPS5, RP49, 18S, v-ATPase, NADH, CypA, GAPDH (Fig 2G); and the overall order from the most stable to the least stable reference genes under abiotic conditions was: RP49, 18S, CypA, GAPDH, v-ATPase, 28S, EF1A, NADH, RPS5 (Fig 2H).
Validation of selected reference genes
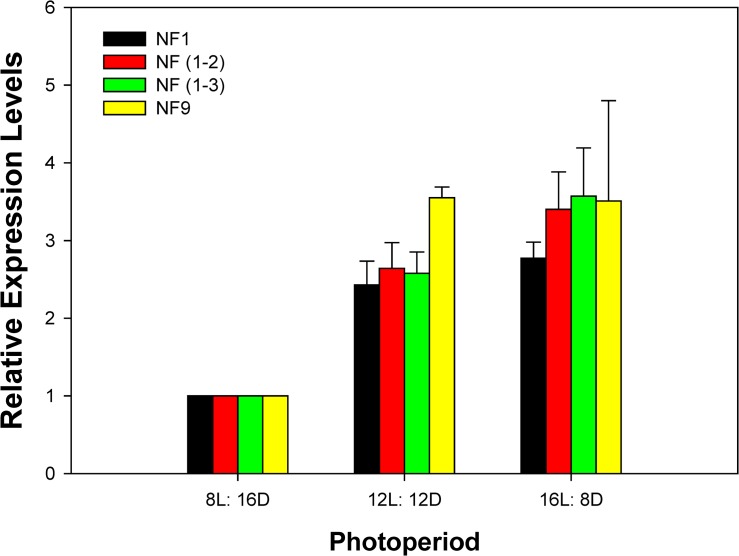
Using two best reference gene combinations for normalization, two genes [EF1A and v-ATPase; NF (1–2)] or three genes [EF1A, v-ATPase, and 18S; NF (1–3)], similar expression levels of tim occurred within each photoperiod (Fig 4). Under the 12L: 12D photoperiod condition, the expression level of tim was the highest when the reference gene with the highest Geomean value (GAPDH; NF9) was used (Fig 4).
Fig 4. Validation of recommended reference genes.
Expression profiles of tim gene under three photoperiod conditions were investigated using different combinations of reference genes. NF1, NF (1–2), NF (1–3), and NF9 indicate that the expression of tim was normalized using the best, the top two, the top three, or, the worst reference genes, respectively. Bar represents the mean and standard error of three biological replicates.
Discussion
The monarch butterfly is commonly used as a non-target insect for the ecological risk assessment of transgenic crops in the U.S.; plus interest in the migratory biology and genetics of this insect suggest future post-genome studies will occur. In this study, expression profiles of nine candidate reference genes from D. plexippus were evaluated under diverse experimental conditions. This research is the first step toward establishing a standardized RT-qPCR analysis for this insect.
Our results demonstrate that the best-suited reference genes can vary depending on biotic and abiotic factors (Tables 2 and 3). Thus, reference genes need to be selected based on the experimental conditions. Unfortunately, a universal reference gene that covers all experimental conditions is unrealistic. For example, EF1A, which plays an important role in translation by catalyzing the GTP-dependent binding of aminoacyl-tRNA to the acceptor site of the ribosome, is the most stable reference gene for biotic factors (Fig 2G). This result is consistent with previous studies in other insects [34, 35]. However, EF1A ranked poorly (7th) as a reference gene for abiotic factors (Fig 2H).
To avoid biased normalization, many researchers have started to advocate the use of multiple reference genes to analyze gene expression [4, 5, 9, 11, 12, 29, 30, 36]. Our results demonstrate that two reference genes are required for reliable normalization under photoperiod, temperature, and sex conditions; three reference genes are required for reliable normalization in different tissue types and under different developmental stages; four reference genes are sufficient to normalize the expression and provide a more conservative estimation of target gene expression under dietary RNAi condition (Fig 3). As a result, we suggest that using different combinations of reference genes are necessary for studying gene expression in D. plexippus, especially with variable experimental conditions.
This work represents an important first step toward establishing a standardized gene analysis framework for D. plexippus. Analysis of our data indicate that EF1A and RP49 are the most stable reference genes, respectively, under biotic (development, tissue, and sex) and abiotic (photoperiod, temperature, and dietary RNAi) conditions. With the recent release of D. plexippus genome, results from this study provide a critical piece for the subsequent genomics and functional genomics research in this emerging insect model, and shed light on the ecological risk assessment of RNAi-based biopesticides on this non-target bio-indicator agent.
Supporting Information
M, EZ Load 100 bp Molecular Ruler; Templates in the PCR reactions were as follows: 1) EF1A, 2) NADH, 3) GAPDH, 4) CypA, 5) RPS5, 6) RP49, 7) 18S, 8) 28S, and 9) v-ATPase.
(TIFF)
(TIFF)
Acknowledgments
The authors are grateful to anonymous reviewers and the editor for their constructive criticisms. Special thanks go to Dr. Xun Zhu for his assistance with the data analysis. The information reported in this paper (No.14-08-065) is part of a project of the Kentucky Agricultural Experiment Station and is published with the approval of the Director.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by a start-up fund from the University of Kentucky to XGZ, a grant from USDA BRAG grant (Award Agreement No.: 3048108827) to XGZ and BDS.
References
- 1. Hudson ME (2008) Sequencing breakthroughs for genomic ecology and evolutionary biology. Mol Ecol Resour 8: 3–17. 10.1111/j.1471-8286.2007.02019.x [DOI] [PubMed] [Google Scholar]
- 2. Xie W, Meng QS, Wu QJ, Wang SL, Yang X, Li RM, et al. (2012) Pyrosequencing the Bemisia tabaci transcriptome reveals a highly diverse bacterial community and a robust system for insecticide resistance. PLOS ONE 7: e35181 10.1371/journal.pone.0035181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Honeybee Genome Sequencing Consortium (2006) Insights into social insects from the genome of the honeybee Apis mellifera . Nature 443: 931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li RM, Xie W, Wang SL, Wu QJ, Yang NN, Yang X, et al. (2013) Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLOS ONE 8: e53006 10.1371/journal.pone.0053006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lourenço AP, Mackert A, dos Santos Cristino A, Simões ZLP (2008) Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie 39: 372–385. [Google Scholar]
- 6. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCR-a perspective. J Mol Endocrinol 34: 597–601. [DOI] [PubMed] [Google Scholar]
- 8. Strube C, Buschbaum S, Wolken S, Schnieder T (2008) Evaluation of reference genes for quantitative real-time PCR to investigate protein disulfide isomerase transcription pattern in the bovine lungworm Dictyocaulus viviparus . Gene 425: 36–43. 10.1016/j.gene.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 9. Guo JL, Ling H, Wu QB, Xu LP, Que YX (2014). The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Sci Rep 4: 7042 10.1038/srep07042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- 11. Zhu X, Yuan M, Shakeel M, Zhang YJ, Wang SL, Wang X, et al. (2014) Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae). PLOS ONE 9: e84730 10.1371/journal.pone.0084730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jacob F, Guertler R, Naim S, Nixdorf S, Fedier A, Hacker NF, et al. (2013). Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PLOS ONE 8: e59180 10.1371/journal.pone.0059180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huvenne H, Smagghe G (2010) Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol 56: 227–235. 10.1016/j.jinsphys.2009.10.004 [DOI] [PubMed] [Google Scholar]
- 14. Burand JP, Hunter WB (2013) RNAi: Future in insect management. J Invertebr Pathol 112: S68–S74. 10.1016/j.jip.2012.07.012 [DOI] [PubMed] [Google Scholar]
- 15. Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, et al. (2007) Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25: 1322–1326. [DOI] [PubMed] [Google Scholar]
- 16. Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, et al. (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol 25: 1307–1313. [DOI] [PubMed] [Google Scholar]
- 17. Zha W, Peng X, Chen R, Du B, Zhu L, He GC (2011) Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens . PLOS ONE 6:e20504 10.1371/journal.pone.0020504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Solensky MJ (2004) The effect of behavior and ecology on male mating success in overwintering monarch butterflies (Danaus plexippus). J Insect Behav 17: 723–743. [Google Scholar]
- 19. Jesse LCH, Obrycki JJ (2000) Field deposition of Bt transgenic corn pollen: lethal effects on the monarch butterfly. Oecologia 125: 241–248. 10.1007/s004420000502 [DOI] [PubMed] [Google Scholar]
- 20. Sears MK, Hellmich RL, Stanley-Horn DE, Oberhauser KS, Pleasants JM, Mattila HR, et al. (2001) Impact of Bt corn pollen on monarch butterfly populations: a risk assessment. Proc Natl Acad Sci U S A 98: 11937–11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wraight CL, Zangerl AR, Carroll MJ, Berenbaum MR (2000) Absence of toxicity of Bacillus thuringiensis pollen to black swallowtails under field conditions. Proc Natl Acad Sci U S A 97: 7700–7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zangerl AR, McKenna D, Wraight CL, Carroll M, Ficarello P, Warner R, et al. (2001) Effects of exposure to event 176 Bacillus thuringiensis corn pollen on monarch and black swallowtail caterpillars under field conditions. Proc Natl Acad Sci U S A 98: 11908–11912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dennis RL (1993) Butterflies and climate change (Manchester University Press; ). [Google Scholar]
- 24. Woods JN, Wilson J, Runkle JR (2008) Influence of climate on butterfly community and population dynamics in western Ohio. Environ Entomol 37: 696–706. [DOI] [PubMed] [Google Scholar]
- 25. Zhan S, Merlin C, Boore JL, Reppert SM (2011) The monarch butterfly genome yields insights into long-distance migration. Cell 147: 1171–1185. 10.1016/j.cell.2011.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stensmyr MC, Hansson BS (2011) A Genome Befitting a Monarch. Cell 147: 970–972. 10.1016/j.cell.2011.11.009 [DOI] [PubMed] [Google Scholar]
- 27. Zhan S, Zhang W, Niitepõld K, Hsu J, Haeger JF, Zalucki MP, et al. (2014) The genetics of monarch butterfly migration and warning colouration. Nature 514: 317–321. 10.1038/nature13812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu HS, Sauman I, Yuan Q, Casselman A, Emery-Le M, Emery P, et al. (2008) Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol 6: e4 10.1371/journal.pbio.1000004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang CX, Pan HP, Liu Y, Zhou XG (2014) Selection of reference genes for expression analysis using quantitative real-time PCR in the pea aphid, Acyrthosiphon pisum (Harris) (Hemiptera, Aphidiae). PLOS ONE 9: e110454 10.1371/journal.pone.0110454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang CX, Pan HP, Liu Y, Zhou XG (2015) Stably expressed housekeeping genes across developmental stages in the two-spotted spider mite, Tetranychus urticae . PLOS ONE 10: e0120833 10.1371/journal.pone.0120833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26: 509–515. [DOI] [PubMed] [Google Scholar]
- 32. Silver N, Best S, Jiang J, Thein SL (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7: 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 34. Horňáková D, Matoušková P, Kindl J, Valterová I, Pichová I (2010) Selection of reference genes for real-time polymerase chain reaction analysis in tissues from Bombus terrestris and Bombus lucorum of different ages. Anal Biochem 397: 118–120. 10.1016/j.ab.2009.09.019 [DOI] [PubMed] [Google Scholar]
- 35. Ponton F, Chapuis MP, Pernice M, Sword GA, Simpson SJ (2011) Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster . J Insect Physiol 57: 840–850. 10.1016/j.jinsphys.2011.03.014 [DOI] [PubMed] [Google Scholar]
- 36. Veazey KJ, Golding MC (2011) Golding selection of stable reference genes for quantitative RT-PCR comparisons of mouse embryonic and extra-embryonic stem cells. PLOS ONE 6: 27592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
M, EZ Load 100 bp Molecular Ruler; Templates in the PCR reactions were as follows: 1) EF1A, 2) NADH, 3) GAPDH, 4) CypA, 5) RPS5, 6) RP49, 7) 18S, 8) 28S, and 9) v-ATPase.
(TIFF)
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.