Abstract
Background
Mutagenesis and labeling studies have identified amino acids from the human α1 glycine receptor (GlyR) extracellular, transmembrane (TM), and intracellular domains in mediating ethanol potentiation. However, limited high-resolution structural data for physiologically relevant receptors in this Cys-loop receptor superfamily have made pinpointing the critical amino acids difficult. Homologous ion channels from lower organisms provide conserved models for structural and functional properties of Cys-loop receptors. We previously demonstrated that a single amino acid variant of the Gloeobacter violaceus ligand-gated ion channel (GLIC) produced ethanol and anesthetic sensitivity similar to that of GlyRs and provided crystallographic evidence for ethanol binding to GLIC.
Methods
We directly compared ethanol modulation of the α1 GlyR and GLIC to a chimera containing the transmembrane domain from human α1 GlyRs and the ligand-binding domain of GLIC using two-electrode voltage clamp electrophysiology of receptors expressed in Xenopus laevis oocytes.
Results
Ethanol potentiated α1 GlyRs in a concentration-dependent manner in the presence of zinc-chelating agents, but did not potentiate GLIC at pharmacologically relevant concentrations. The GLIC/GlyR chimera recapitulated the ethanol potentiation of GlyRs, without apparent sensitivity to zinc chelation. For chimera expression in oocytes, it was essential to suppress leakage current by adding 50 μM picrotoxin to the media, a technique that may have applications in expression of other ion channels.
Conclusions
Our results are consistent with a transmembrane mechanism of ethanol modulation in Cys-loop receptors. This work highlights the relevance of bacterial homologs as valuable model systems for studying ion channel function of human receptors and demonstrates the modularity of these channels across species.
Keywords: chimera, glycine, GLIC, picrotoxin
Introduction
Alcohol effects on the nervous system are well-documented, yet underlying mechanisms remain poorly understood (Olsen et al., 2014). Pentameric ligand-gated ion channels, including glycine receptors (GlyRs), are implicated in binding pharmacologically-relevant concentrations of ethanol and mediating some of its neurodepressive effects (Perkins et al., 2010). There is also evidence for GlyRs in ethanol-mediated dopamine release in nucleus accumbens (Lido et al., 2009) and the neurocircuitry of alcohol addiction (Soderpalm and Ericson, 2013). GlyRs are promising targets for mechanistic studies, playing roles in both peripheral immobility and central reward pathways. They are amenable to efficient functional expression in heterologous cells (Perkins et al. 2010); however, identifying the critical site(s) of ethanol action in GlyRs has proven complex.
Ligand-gated ion channels in the Cys-loop receptor superfamily are structurally conserved pentamers of identical or similar subunits, each contributing to an extracellular domain that binds neurotransmitter agonists or other ligands, a helical transmembrane (TM) domain through which ions enter or exit via a central pore, and a variable intracellular loop domain involved in channel kinetics and cytoplasmic interactions (Nys et al., 2013) (Fig. 1a). Mutagenesis and labeling studies have implicated specific amino acids from the human α1 GlyR TM domain, particularly S267 and A288 (Mascia et al., 2000) and neighboring positions Q266 and M287 (Borghese et al., 2012) in ethanol potentiation. However, positions in the extracellular (Perkins et al., 2009) and intracellular domains (Talwar and Lynch, 2014, Yévenes et al., 2008) also influence ethanol potentiation. Distinguishing the relative contributions of these possible sites, and their direct or indirect involvement in ethanol binding, remains a critical challenge in the field.
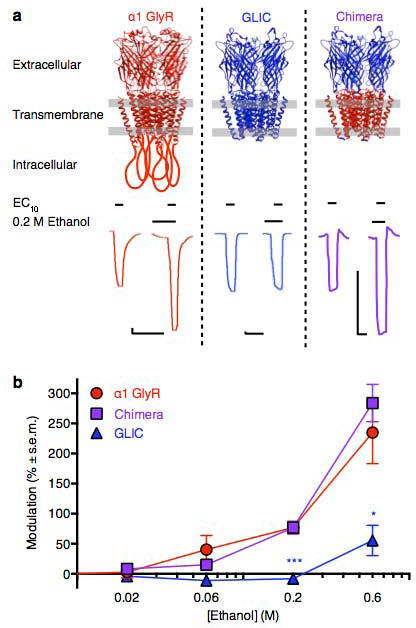
Figure 1.
Ethanol modulation of human, bacterial, and chimera receptors. (a) Top panel- structural models of receptors studied. GlyRs (red) are represented by the X-ray structure of GluCl (PDB ID 3RIF), with the intracellular domain shown as manually drawn loops. GLIC (blue) and the GLIC/GlyR chimera are represented by the X-ray structure of GLIC (PDB ID 3EAM); chimera is colored according to the source of each domain. Gray lines represent plasma membrane boundaries. Bottom panel- sample traces showing EC10 activation by glycine (GlyR) or protons (GLIC, chimera) in the absence and presence of 200 mM ethanol. Scale bars are 200 nA, 2 min. (b) Modulation of human α1 GlyRs (red), GLIC (blue), and GLIC/GlyR chimeras (purple) at EC10 activation by various concentrations of ethanol. Errors are s.e.m., n = 4–8. Significance vs. chimera, Dunnet's multiple comparison test, analysis of variance, *p < 0.05; ***p < 0.001.
A major impediment to characterizing ethanol modulation of ligand-gated ion channels, including GlyRs, at a molecular level is the limited structural data available for physiologically relevant receptors in this superfamily (Olsen et al., 2014). Cys-loop receptor homologs from lower organisms provided the first atomic-resolution structures in this channel family, and have proved valuable in modeling channel properties. In particular, crystal structures of the Gloeobacter violaceus ligand-gated ion channel (GLIC) have been determined in apparently open (Bocquet et al., 2009, Hilf and Dutzler, 2009), closed (Sauguet et al., 2014), and intermediate conformations (Prevost et al., 2012), and in complex with anesthetics and other modulators (Hilf et al., 2010, Nury et al., 2011, Pan et al., 2012). A transmembrane binding site for anesthetics identified in GLIC by X-ray crystallography (Nury et al., 2011) was subsequently validated by photoaffinity labeling not only in GLIC (Chiara et al., 2014) but also in eukaryotic receptors (Jayakar et al., 2013). Although other anesthetic-bound bacterial receptors (Spurny et al., 2012) and partial structures from nematodes (Hibbs and Gouaux, 2011), mice (Hassaine et al., 2014), and humans (Mowrey et al., 2013, Miller and Aricescu, 2014) have since been reported, the availability of structures for full-length GLIC in multiple conformational states and bound to various relevant ligands, along with its accessibility to heterologous expression and recording, continues to make it a valuable model system for structure/function studies in this family.
Wild-type GLIC was initially shown to be insensitive to pharmacologically-relevant ethanol concentrations (Weng et al., 2010); however, the single-site variant F238A (mutated at the position equivalent to ethanol-sensitive Q266 in the α1 GlyR) is potently potentiated by ethanol, n-alcohols, and anesthetics similar to GlyRs (Howard et al., 2011). Previously, we determined a co-crystal structure of this GLIC variant with ethanol bound at the TM subunit interface, including a hydrogen bond to the residue equivalent to S267 in the α1 GlyR (Sauguet et al., 2013). Structural similarities in this TM cavity between the GLIC F238A mutant and GlyR homology models (Sauguet et al., 2013) support the modular nature of functional domains in this superfamily, suggesting that GLIC is a valuable model for conserved modulatory mechanisms in human receptors. This report demonstrates that the human α1 glycine TM region of a GLIC/GlyR chimera confers ethanol modulation on the otherwise ethanol-insensitive GLIC and supports the use of prokaryotic homologs as conserved systems for modeling channel properties and deciphering sites of ethanol action.
Materials and Methods
Reagents
The synthetic GLIC/GlyR chimera in the pMT3 vector was synthesized as previously described (Duret et al., 2011), with GLIC residues 119–121 (end of Loop 7), 194–282 (TM helices M1–M3), and 286–314 (M4) replaced with equivalent residues from the α1 GlyR. Chemicals were purchased from Sigma-Aldrich. Ethanol was diluted into recording buffer immediately prior to application.
Oocyte preparation
Oocytes were obtained from anesthetized Xenopus laevis frogs via surgical ovarectomy in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals. All efforts were made to minimize suffering and the number of frogs used. Extracted ovarian tissue was placed in Modified Barth's Solution adjusted to pH 7.5 (88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 0.91 mM CaCl2, 0.82 mM MgSO4, Ca(NO3)2, 10 mM HEPES). After manual isolation with forceps, oocytes were treated with collagenase type 1A solution containing 0.5 mg/ml collagenase, 83 mM NaCl, 5 mM HEPES, 2 mM KCl, 1 mM MgCl2, adjusted to pH 7.5 for 10 min. Oocyte nuclei were injected via the animal pole with human α1 GlyR, GLIC, or GLIC/GlyR chimera cDNA (1-5 ng) using a Nanoject II microdispenser (Drummond Scientific). Injected oocytes were stored singly at 13°C in Modified Barth's Solution supplemented with 90 mg/l theophylline, 50 mg/l gentamycin, 10000 u/l penicillin, 10 mg/l streptomycin, 220 mg/l sodium pyruvate, 50 μM picrotoxin for 2–17 days.
Electrophysiology
Oocytes were fixed in wells with continuous flow of Ringer's buffer (123 mM NaCl, 2 mM KCl, 2 mM MgSO4, 2 mM CaCl2, 10 mM HEPES). For GlyR and GLIC recordings, Ringer's buffer was adjusted to pH 7.4; for chimera recordings, pH was adjusted to 8.5. For GLIC and chimera recordings, activation buffers were prepared by substituting 10 mM citrate (pH 4–6) or 10 mM MOPS (pH 6.5–7) for HEPES, then adjusting pH accordingly. For GlyR and some chimera recordings, 10 mM tricine was added prior to pH adjustment. Solutions were perfused at a rate of 2 ml/min by a peristaltic pump (Masterflex) via 18-gauge Teflon-Viton tubing. Twin microelectrodes encased in glass insulators were filled with 3 M KCl solution and inserted into oocytes. Electrodes were connected to an OC-725C voltage clamp (Warner Instruments). Membrane potentials were clamped at −70 mV and allowed to perfuse in picrotoxin-free buffer ≥5 minutes prior to recording. Electrical currents were digitized and analyzed via a PowerLab 4/30 digitizer (AD Instruments).
Activation and modulation of GlyR, GLIC, and chimera currents were measured as previously described (Howard et al., 2011, Mascia et al., 1996). Briefly, GlyR activation was initially calibrated by 15 sec application of saturating (3 mM) glycine. After 15 min washout, low-glycine solutions were tested for 30 sec periods until the EC10 concentration was identified, which was then used for subsequent measurements. GLIC and chimera receptors were activated by 1–2 min application of low-pH buffer, followed by a 5 min washout.
For all receptors, modulation was induced by pre-applying ethanol in Ringer's buffer for 1 min, followed by co-administration of EC10 buffer with an identical concentration of ethanol for the standard activation time. Due to the rapid diffusion of alcohol (Schoberth et al., 1996), pre-application times of less than 1 min have previously been taken to allow full equilibration with receptor binding sites (Hu et al., 2006, Duret et al., 2011); however, consistent use of 1-min pre-applications in this work may represent more relevant steady-state values, and may better simulate continuous physiological exposure during drinking.
Statistics and Graphics
Data are represented as mean ± s.e.m. Proton activation curves represent nonlinear regression fits to the equation R = Rmax / (1 + 10 ^ {[log(EC50 − P)] · nH}), where R is the peak channel response, Rmax is the maximal response, P is the concentration of protons, EC50 is the concentration producing 50% maximal response, and nH is the Hill coefficient; data for each receptor were normalized to its fitted maximal response. Ethanol modulation was calculated as [(RA − <R0,R1>)/<R0,R1>] × 100, where RA represents the peak EC10 channel response in the presence of ethanol, and <R0,R1> represents the mean of the pre- and post-ethanol EC10 responses. Results were analyzed with one-way analysis of variance and Dunnet's post hoc test, with significant effects set at p < 0.05. Nonlinear regression analysis and all statistical analyses were performed with Prism 5 for Mac (GraphPad Software). For clarity, sample traces were smoothed using the median filter in LabChart Pro (AD Instruments) and exported in the target color using the maximum line width. Protein structures were represented using the University of California, San Francisco Chimera package (Pettersen et al., 2004).
Results
We directly compared ethanol modulation of the GLIC/GlyR chimera to α1 GlyRs and GLIC expressed in Xenopus laevis oocytes by two-electrode voltage-clamp electrophysiology. Potentiation of α1 GlyRs by pharmacologically relevant concentrations of ethanol has been demonstrated in multiple endogenous and heterologous systems (Nys et al., 2013); however, the degree of potentiation depends on the level of channel activation (Mascia et al., 1996) and the presence of confounding modulators, including zinc at common contaminating concentrations (McCracken et al., 2010). To control these variables, we measured GlyR modulation at EC10 glycine in the presence of 10 mM tricine, a selective chelator of zinc (Paoletti et al., 2009). We considered zinc-free conditions most appropriate for comparison with GLIC and chimera receptors, which lack the zinc binding site(s) contained in the GlyR extracellular domain (Miller et al., 2005). Consistent with previous studies (McCracken et al., 2010) ethanol potentiated α1 GlyRs in a concentration-dependent manner (Fig. 1b), with in vivo immobilizing ethanol concentrations (200 mM) enhancing EC10 glycine activation >70% (Fig. 1a). In contrast, GLIC was insensitive to potentiation of EC10 proton activation (Fig. 1a), exhibiting modulation only at non-pharmacological ethanol concentrations (600 mM; Fig 1b). Although zinc has been shown to inhibit GLIC currents, it does so via a pore-blocking mechanism at concentrations higher than expected as buffer contaminants (Hilf et al., 2010); therefore, tricine was excluded from GLIC recording media.
In parallel to control experiments in GlyRs and GLIC, we were able to express GLIC/GlyR chimera receptors to higher levels than previously reported by incubating injected oocytes with 50 μM picrotoxin, a pore blocker of pentameric ligand-gated ion channels including GlyRs and GLIC (Alqazzaz et al., 2011). The presence of picrotoxin evidently reduced ion permeability of oocyte membranes in incubation medium, allowing cells to survive ≥2 weeks until substantial currents could be recorded at activation levels as low as pH 7.25 (Fig. 1a), approximately EC10 (Fig. 2a). We also increased the running buffer to pH 8.5 to minimize background currents after washout of picrotoxin. Although previous studies of picrotoxin binding to GlyRs, including at higher concentration than that used in our incubation (50 μM), have shown no irreversible or persistent channel inhibition within seconds after washout (Yang et al., 2007), we nonetheless perfused oocytes with picrotoxin-free running buffer ≥5 minutes prior to recording. Following this washout period, no increase in channel currents was observed over the course of ~2-hour recording sessions, and channel activation was comparable in magnitude and gating to previous studies in both GLIC (Howard et al., 2011) and GlyRs (Borghese et al., 2012), consistent with removal of picrotoxin.
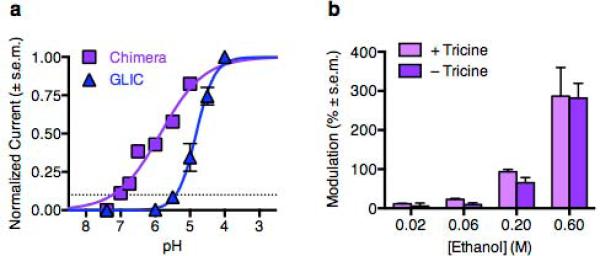
Figure 2.
Pharmacological characterization of GLIC/GlyR chimera. (a) Proton response curves for GLIC (blue) and chimera (purple) receptors. Dashes indicate EC10, ~pH 7.25 for the chimera. (b) Potentiation of chimera receptors by various concentrations of ethanol, measured as in Fig. 1, in the presence (lavender) and absence (purple) of 10 mM tricine to chelate environmental zinc. Tricine did not alter ethanol responses at any concentration tested. In a–b, errors are s.e.m., n = 2–7.
Notably, ethanol potentiation of chimera receptors was nearly superimposable with that of GlyRs (Fig. 1b), with 200 mM ethanol enhancing EC10 proton activation >70% (Fig 1a). We observed no difference in potentiation of chimera currents by any concentration of ethanol upon zinc chelation by 10 mM tricine (Fig. 2b), consistent with an absence of extracellular zinc binding sites in the GLIC extracellular domain.
Discussion
GlyRs play important roles in the central nervous system, including addiction pathways (Dutertre et al., 2012); for instance, the alcohol use disorder therapeutic acamprosate was shown to interact with GlyRs in the nucleus accumbens to reduce dopamine release and ethanol consumption (Chau et al., 2010). Because of the functional role of GlyRs in mediating the rewarding properties of ethanol, pinpointing the site of ethanol action could facilitate development of therapeutic targets for treating alcohol abuse. This long-term goal requires further knowledge of the structural/functional interaction of alcohol with human GlyRs.
Given the absence of structural data for full-length human GlyRs, homologous prokaryotic receptors such as GLIC offer a conserved model system for elucidating sites of ethanol action. Previously Duret and colleagues (Duret et al., 2011) fused the GLIC extracellular domain to the α1 GlyR TM domain (Fig. 1a); this chimeric receptor proved to be activated by protons rather than glycine, consistent with an extracellular ligand-binding domain. The structure of this GLIC/GlyR chimera (Moraga-Cid et al., 2015) subsequently provided atomic-resolution data for the GlyR TM domain in the context of a full-length functional channel. Although similar in overall topology to the NMR structure of the isolated GlyR transmembrane domain (Mowrey et al., 2013), the chimera structure is more consistent with other full-length pLGIC structures in the vicinity of the extracellular-TM domain interface, possibly reflecting the importance of structural constraints from the extracellular domain.
Initial studies indicated that n-alcohols and anesthetics potentiated the GLIC/GlyR chimera (Duret et al., 2011), similar to GlyRs rather than GLIC, which is relatively insensitive to ethanol (Weng et al., 2010). This result generally supported a TM site of action for allosteric modulators; however, only ethanol concentrations 200 mM and higher - an immobilizing or lethal dose, well above intoxicating levels - were tested. Moreover, the sensitivity of the chimera to proton activation compromised direct comparisons with previous GlyR and GLIC data. Indeed, baseline currents conducted by the chimera under standard incubation and running buffer conditions resulted in cell death shortly after injection; combined with slow expression of the chimera construct, this limitation required recording at relatively high levels of activation, ~EC30 (pH 6.8). Both GlyRs (Mascia et al., 1996) and GLIC (Howard et al., 2011) decrease in ethanol sensitivity at higher levels of activation, consistent with the general allosteric model of alcohol action involving preferential stabilization of open versus closed states of the channel (Forman, 2012). Therefore, these receptors have been predominantly characterized at EC2–10, such that initial studies of the chimera channel likely underestimated effects of channel modulation.
By standardizing activation levels and confounding modulators, our results demonstrated a remarkable conservation of ethanol potentiation in pentameric ligand-gated ion channels containing the α1 GlyR TM domain, regardless of the source of the extracellular or intracellular domains. These data are consistent with previous evidence for a TM site of action for ethanol and other allosteric modulators, including the inter-subunit binding site recently visualized in the ethanol-sensitive GLIC variant F238A (Sauguet et al., 2013). The recently published 3.5-Å structure of the GLIC/GlyR chimera confirms the presence of an equivalent cavity containing multiple amino acid residues previously shown to influence GlyR ethanol sensitivity; moreover, this pocket contained substantially fewer stabilizing interactions for alcohol in the nonconducting TM domain structure than in a model of the conducting state, consistent with a role for ethanol in preferentially stabilizing the open conformation (Moraga-Cid et al., 2015).
Given the promiscuous nature of ethanol binding (Olsen et al., 2014), it remains plausible that the mechanism of modulation of the GLIC/GlyR chimera is distinct from the predominant mechanism in native GlyRs. For example, recent electrophysiological studies in α1/3 GlyR chimeras (Sánchez et al., 2015) indicate that a complete understanding of subtype-specific alcohol modulation involves both TM and intracellular mechanisms of action. However, the dominant influence of the GlyR TM domain suggests that the intracellular loop, which is absent in both GLIC and the GLIC/GlyR chimera (Fig. 1a), may contribute to indirect modulation by ethanol via Gβɣ protein interactions (Yévenes et al., 2008) or phosphorylation (Talwar and Lynch, 2014) but is not critical in determining ethanol potentiation under the conditions studied here. Similarly, the sequence of GlyR extracellular loop 2, which has been shown to influence alcohol sensitivity (Perkins et al., 2009), differs in GLIC and therefore in the GLIC/GlyR chimera. Substitutions in this loop have facilitated the development of ultrasensitive ethanol receptors (USERs) as pharmacological tools to understand the contributions of specific receptor subtypes to alcohol responses (Naito et al., 2014); further studies may clarify whether wild-type GlyRs bind alcohol primarily via this extracellular region, or if the mechanism of modulation might be indirect, multifactorial, and/or specific to USER constructs. Furthermore, substitution of a key GlyR loop 2 position with the equivalent residue from GLIC (A52D) retained ethanol potentiation (Perkins et al., 2012), raising the possibility that ethanol-sensitive interactions in this domain are conserved between GLIC and GlyRs despite low overall sequence identity. Although further investigation is necessary, our data indicate that subtle differences in this region are unlikely to account for the differential alcohol sensitivity of GLIC versus GlyRs, and that quantitatively similar levels of modulation can be reproduced in GlyRs despite changes in the extracellular domain.
In addition to ethanol potentiation, functional properties including agonist selectivity and zinc modulation were swapped in the GLIC/GlyR chimera. Notably, the GLIC extracellular domain did not quantitatively confer proton activation, possibly reflecting additional contributions of the TM domain and/or extracellular-TM domain interface to gating. These observations may reflect recent evidence for a TM histidine residue involved in GLIC pH sensitivity (Rienzo et al., 2014, Wang et al., 2012). However, despite lacking any histidines in the pore-lining helix, the GLIC/GlyR chimera exhibited even greater sensitivity to proton activation than wild-type GLIC (Fig. 2), suggesting that other residues—likely in the GLIC extracellular domain—are also involved in proton gating. It is possible that the GlyR TM domain contains other residues that substitute for pH-sensitive sites in GLIC; however, native GlyRs have been shown to exhibit proton inhibition rather than activation (Chen et al., 2004), providing few insights into the identity of any proton-activation determinants. These challenges highlight ongoing difficulties identifying the protonation site(s) responsible for GLIC gating; a possible explanation is that pH sensing is partly or largely conserved in the GLIC/GlyR chimera, but that the interaction between the GLIC extracellular domain and the GlyR TM domain produces a lower energy barrier for the resting-open transition than the native GLIC interface. In the present work, we attempted to control for differences in proton sensitivity by measuring ethanol modulation at equivalent (EC10) levels of activation; a conserved TM site of alcohol action remains a parsimonious explanation of our modulation results. It is nonetheless plausible that the mechanisms of activation and/or modulation of the GLIC/GlyR chimera differ from native GLIC. We also cannot exclude the possibility that the modified gating properties of the GLIC/GlyR chimera indirectly increase ethanol sensitivity relative to GLIC, and only superficially mimic native GlyR modulation.
An important result of the present study is that the use of picrotoxin to block activation during expression can facilitate comparisons with wild-type receptors. Indeed, the reproducibility of GlyR ethanol potentiation in the GLIC/GlyR chimera appears all the more notable in light of imperfect transfer of proton gating from GLIC. The use of picrotoxin during expression may prove a useful tool in producing other ion channels with mild tonic activity. Simple manipulations such as this could facilitate characterization of a wider variety of native and model channels: in this case, the ability to quantify modulation of the GLIC/GlyR chimera facilitated direct comparison of a physiologically relevant target to a model protein for which direct structural data are available (Moraga-Cid et al., 2015).
The modular nature of ethanol potentiation in receptors from such disparate organisms as humans and cyanobacteria highlights the value of prokaryotic model systems for studying ion channel structure, function, and modulation of human receptors. Recent advances in structure determination of related eukaryotic receptors (Hibbs and Gouaux, 2011, Hassaine et al., 2014, Mowrey et al., 2013, Miller and Aricescu, 2014) indicate a number of conserved features, as well as notable differences, in this broad channel family. Critical future steps in this field may include the use of some or all of these structural templates to characterize TM sites involved in GlyR modulation, as well as optimization of GLIC or similar systems to better model allostery in Cys-loop receptors. For example, quantitative comparisons of apparent conducting and nonconducting states of a wide range of recent structures (Scott et al., 2015) revealed consistent changes in specific TM residues implicated in GlyR gating, supporting a broadly conserved mechanism among eukaryotic and prokaryotic models. Similarly, modeling of allosteric binding pockets in GABAARs (Jayakar et al., 2014) produced comparable dimensions and docking calculations based on either GLIC or the modified β3 GABAAR (Miller and Aricescu, 2014), supporting the generalizability of GLIC as a model system even for the binding of small modulators. In the present work, the quantitative transfer of ethanol potentiation via the TM domain of human GlyRs to a distantly related bacterial homolog demonstrates both the critical role of this region in allosteric modulation and the modularity of ligand-gated ion channels across species. The availability of crystallographic data for the GLIC/GlyR chimera itself (Moraga-Cid et al., 2015) offers new opportunities to bridge structure and function in this family of receptors, pending further validation of its physiological relevance.
Acknowledgments
Thanks to C.M. Borghese and G. Duret for helpful technical and conceptual discussions, and to Jody Mayfield for valuable editorial assistance.
Support: This work was supported by the National Institutes of Health National Institute on Alcohol Abuse and Alcoholism (R01 AA06399 to R.A.H., R01 AA013378 to J.R.T., and F32 AA019875-01 to R.J.H.) and by the Skidmore College Summer Faculty/Student Research Program.
Footnotes
The authors declare no competing financial interests.
REFERENCES
- Alqazzaz M, Thompson AJ, Price KL, Breitinger HG, Lummis SC. Cys-loop receptor channel blockers also block GLIC. Biophys J. 2011;101:2912–2918. doi: 10.1016/j.bpj.2011.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- Borghese CM, Blednov YA, Quan Y, Iyer SV, Xiong W, Mihic SJ, Zhang L, Lovinger DM, Trudell JR, Homanics GE, Harris RA. Characterization of two mutations, M287L and Q266I, in the α1 glycine receptor subunit that modify sensitivity to alcohols. J Pharmacol Exp Ther. 2012;340:304–316. doi: 10.1124/jpet.111.185116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Dillon GH, Huang R. Molecular determinants of proton modulation of glycine receptors. J Biol Chem. 2004;279:876–883. doi: 10.1074/jbc.M307684200. [DOI] [PubMed] [Google Scholar]
- Chiara DC, Gill JF, Chen Q, Tillman T, Dailey WP, Eckenhoff RG, Xu Y, Tang P, Cohen JB. Photoaffinity labeling the propofol binding site in GLIC. Biochemistry. 2014;53:135–142. doi: 10.1021/bi401492k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret G, Van Renterghem C, Weng Y, Prevost M, Moraga-Cid G, Huon C, Sonner JM, Corringer PJ. Functional prokaryotic-eukaryotic chimera from the pentameric ligand-gated ion channel family. Proc Natl Acad Sci. 2011;108:12143–12148. doi: 10.1073/pnas.1104494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman SA. Monod-Wyman-Changeux allosteric mechanisms of action and the pharmacology of etomidate. Curr Opin Anaesthesiol. 2012;25:411–418. doi: 10.1097/ACO.0b013e328354feea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassaine G, Deluz C, Grasso L, Wyss R, Tol MB, Hovius R, Graff A, Stahlberg H, Tomizaki T, Desmyter A, Moreau C, Li XD, Poitevin F, Vogel H, Nury H. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature. 2014;512:276–281. doi: 10.1038/nature13552. [DOI] [PubMed] [Google Scholar]
- Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilf RJ, Bertozzi C, Zimmermann I, Reiter A, Trauner D, Dutzler R. Structural basis of open channel block in a prokaryotic pentameric ligand-gated ion channel. Nat Struct Mol Biol. 2010;17:1330–1336. doi: 10.1038/nsmb.1933. [DOI] [PubMed] [Google Scholar]
- Hilf RJ, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- Howard RJ, Murail S, Ondricek KE, Corringer PJ, Lindahl E, Trudell JR, Harris RA. Structural basis for alcohol modulation of a pentameric ligand-gated ion channel. Proc Natl Acad Sci. 2011;108:12149–12154. doi: 10.1073/pnas.1104480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XQ, Hayrapetyan V, Gadhiya JJ, Rhubottom HE, Lovinger DM, Machu TK. Mutations of L293 in transmembrane two of the mouse 5-hydroxytryptamine3A receptor alter gating and alcohol modulatory actions. Br J Pharmacol. 2006;148:88–101. doi: 10.1038/sj.bjp.0706685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakar SS, Dailey WP, Eckenhoff RG, Cohen JB. Identification of propofol binding sites in a nicotinic acetylcholine receptor with a photoreactive propofol analog. J Biol Chem. 2013;288:6178–6189. doi: 10.1074/jbc.M112.435909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakar SS, Zhou X, Chiara DC, Dostalova Z, Savechenkov PY, Bruzik KS, Dailey WP, Miller KW, Eckenhoff RG, Cohen JB. Multiple propofol-binding sites in a ɣ-aminobutyric acid type A receptor (GABAAR) identified using a photoreactive propofol analog. J Biol Chem. 2014;289:27456–27468. doi: 10.1074/jbc.M114.581728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lido HH, Stomberg R, Fagerberg A, Ericson M, Soderpalm B. The glycine reuptake inhibitor org 25935 interacts with basal and ethanol-induced dopamine release in rat nucleus accumbens. Alcohol Clin Exp Res. 2009;33:1151–1157. doi: 10.1111/j.1530-0277.2009.00938.x. [DOI] [PubMed] [Google Scholar]
- Mascia MP, Mihic SJ, Valenzuela CF, Schofield PR, Harris RA. A single amino acid determines differences in ethanol actions on strychnine-sensitive glycine receptors. Mol Pharmcol. 1996;50:402–406. [PubMed] [Google Scholar]
- Mascia MP, Trudell JR, Harris RA. Specific binding sites for alcohols and anesthetics on ligand-gated ion channels. Proc Natl Acad Sci. 2000;97:9305–9310. doi: 10.1073/pnas.160128797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken LM, Trudell JR, Goldstein BE, Harris RA, Mihic SJ. Zinc enhances ethanol modulation of the α1 glycine receptor. Neuropharmacol. 2010;58:676–681. doi: 10.1016/j.neuropharm.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PS, Aricescu AR. Crystal structure of a human GABAA receptor. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PS, Da Silva HM, Smart TG. Molecular basis for zinc potentiation at strychnine-sensitive glycine receptors. J Biol Chem. 2005;280:37877–37884. doi: 10.1074/jbc.M508303200. [DOI] [PubMed] [Google Scholar]
- Moraga-Cid G, Sauguet L, Huon C, Malherbe L, Girard-Blanc C, Petres S, Murail S, Taly A, Baaden M, Delarue M, Corringer PJ. Allosteric and hyperekplexic mutant phenotypes investigated on an α1 glycine receptor transmembrane structure. Proc Natl Acad Sci U S A. 2015;112:2865–2870. doi: 10.1073/pnas.1417864112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowrey DD, Cui T, Jia Y, Ma D, Makhov AM, Zhang P, Tang P, Xu Y. Open-channel structures of the human glycine receptor α1 full-length transmembrane domain. Structure. 2013;21:1897–1904. doi: 10.1016/j.str.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito A, Muchhala KH, Asatryan L, Trudell JR, Homanics GE, Perkins DI, Davies DL, Alkana RL. Glycine and GABAA ultra-sensitive ethanol receptors as novel tools for alcohol and brain research. Mol Pharmacol. 2014;86:635–646. doi: 10.1124/mol.114.093773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nury H, Van Renterghem C, Weng Y, Tran A, Baaden M, Dufresne V, Changeux JP, Sonner JM, Delarue M, Corringer PJ. X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature. 2011;469:428–431. doi: 10.1038/nature09647. [DOI] [PubMed] [Google Scholar]
- Nys M, Kesters D, Ulens C. Structural insights into Cys-loop receptor function and ligand recognition. Biochem Pharmacol. 2013;86:1042–1053. doi: 10.1016/j.bcp.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Olsen RW, Li GD, Wallner M, Trudell JR, Bertaccini EJ, Lindahl E, Miller KW, Alkana RL, Davies DL. Structural models of ligand-gated ion channels: sites of action for anesthetics and ethanol. Alcohol Clin Exp Res. 2014;38:595–603. doi: 10.1111/acer.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Chen Q, Willenbring D, Mowrey D, Kong XP, Cohen A, Divito CB, Xu Y, Tang P. Structure of the pentameric ligand-gated ion channel GLIC bound with anesthetic ketamine. Structure. 2012;20:1463–1469. doi: 10.1016/j.str.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P, Vergnano AM, Barbour B, Casado M. Zinc at glutamatergic synapses. Neuroscience. 2009;158:126–136. doi: 10.1016/j.neuroscience.2008.01.061. [DOI] [PubMed] [Google Scholar]
- Perkins DI, Trudell JR, Asatryan L, Davies DL, Alkana RL. Charge and geometry of residues in the loop 2 β hairpin differentially affect agonist and ethanol sensitivity in glycine receptors. J Pharmacol Exp Ther. 2012;341:543–551. doi: 10.1124/jpet.111.190942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DI, Trudell JR, Crawford DK, Alkana RL, Davies DL. Molecular targets and mechanisms for ethanol action in glycine receptors. Pharmacol Ther. 2010;127:53–65. doi: 10.1016/j.pharmthera.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DI, Trudell JR, Crawford DK, Asatryan L, Alkana RL, Davies DL. Loop 2 structure in glycine and GABAA receptors plays a key role in determining ethanol sensitivity. J Biol Chem. 2009;284:27304–27314. doi: 10.1074/jbc.M109.023598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Prevost MS, Sauguet L, Nury H, Van Renterghem C, Huon C, Poitevin F, Baaden M, Delarue M, Corringer PJ. A locally closed conformation of a bacterial pentameric proton-gated ion channel. Nat Struct Mol Biol. 2012;19:642–649. doi: 10.1038/nsmb.2307. [DOI] [PubMed] [Google Scholar]
- Rienzo M, Lummis SC, Dougherty DA. Structural requirements in the transmembrane domain of GLIC revealed by incorporation of noncanonical histidine analogs. Chem Biol. 2014;21:1700–1706. doi: 10.1016/j.chembiol.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez A, Yévenes GE, San Martin L, Burgos CF, Moraga-Cid G, Harvey RJ, Aguayo LG. Control of ethanol sensitivity of the glycine receptor α3 subunit by transmembrane 2, the intracellular spliced cassette and C-terminus domain. J Pharmacol Exp Ther. 2015 doi: 10.1124/jpet.114.221143. doi:10.1124/jpet.114.221143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauguet L, Howard RJ, Malherbe L, Lee US, Corringer PJ, Harris RA, Delarue M. Structural basis for potentiation by alcohols and anaesthetics in a ligand-gated ion channel. Nat Commun. 2013;4:1697. doi: 10.1038/ncomms2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauguet L, Shahsavar A, Poitevin F, Huon C, Menny A, Nemecz A, Haouz A, Changeux JP, Corringer PJ, Delarue M. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc Natl Acad Sci U S A. 2014;111:966–971. doi: 10.1073/pnas.1314997111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberth SM, Chapman BE, Kuchel PW, Wittig RM, Grotendorst J, Jansen P, DeGraff AA. Ethanol transport in Zymomonas mobilis measured by using in vivo nuclear magnetic resonance spin transfer. J Bacteriol. 1996;178:1756–1761. doi: 10.1128/jb.178.6.1756-1761.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S, Lynch JW, Keramidas A. Correlating structural and energetic changes in glycine receptor activation. J Biol Chem. 2015;290:5621–5634. doi: 10.1074/jbc.M114.616573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurny R, Billen B, Howard RJ, Brams M, Debaveye S, Price KL, Weston DA, Strelkov SV, Tytgat J, Bertrand S, Bertrand D, Lummis SC, Ulens C. Multisite binding of a general anesthetic to the prokaryotic pentameric Erwinia chrysanthemi ligand-gated ion channel (ELIC). J Biol Chem. 2013;288:8355–8364. doi: 10.1074/jbc.M112.424507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderpalm B, Ericson M. Neurocircuitry involved in the development of alcohol addiction: the dopamine system and its access points. Curr Top Behav Neurosci. 2013;13:127–161. doi: 10.1007/7854_2011_170. [DOI] [PubMed] [Google Scholar]
- Talwar S, Lynch JW. Phosphorylation mediated structural and functional changes in pentameric ligand-gated ion channels: implications for drug discovery. Int J Biochem CellBiol. 2014;53:218–223. doi: 10.1016/j.biocel.2014.05.028. [DOI] [PubMed] [Google Scholar]
- Wang HL, Cheng X, Sine SM. Intramembrane proton binding site linked to activation of bacterial pentameric ion channel. J Biol Chem. 2012;287:6482–6489. doi: 10.1074/jbc.M111.305839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Yang L, Corringer PJ, Sonner JM. Anesthetic sensitivity of the Gloeobacter violaceus proton-gated ion channel. Anesth Analg. 2010;110:59–63. doi: 10.1213/ANE.0b013e3181c4bc69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Cromer BA, Harvey RJ, Parker MW, Lynch JW. A proposed structural basis for picrotoxinin and picrotin binding in the glycine receptor pore. J Neurochem. 2007;103:580–589. doi: 10.1111/j.1471-4159.2007.04850.x. [DOI] [PubMed] [Google Scholar]
- Yévenes GE, Moraga-Cid G, Peoples RW, Schmalzing G, Aguayo LG. A selective Gβɣ-linked intracellular mechanism for modulation of a ligand-gated ion channel by ethanol. Proc Natl Acad Sci U S A. 2008;105:20523–20528. doi: 10.1073/pnas.0806257105. [DOI] [PMC free article] [PubMed] [Google Scholar]