Summary
Ran is a small GTP binding protein that was originally identified as a regulator of nucleocytoplasmic transport [1], and subsequently found to be important for spindle formation [2-5]. In mitosis, a gradient of Ran-GTP emanates from chromatin and diminishes toward spindle poles [6]. Ran-GTP promotes spindle self-organization through the release of importin-bound spindle assembly factors (SAFs), which stimulate microtubule (MT) nucleation, organization and regulate MT dynamics [7-9]. Although many SAFs are non-motile MT-associated proteins, such as NuMA, TPX2, and HURP [7, 10-12], Ran also controls motor proteins, including Kid and HSET/XCTK2 [13, 14]. The Kinesin-14, XCKT2, is important for spindle assembly and pole organization [15-20], and Ran-GTP is proposed to promote XCKT2 MT crosslinking activity by releasing importin α/β from a bipartite nuclear localization signal (NLS) located in the tail domain [14]. Here we show that the Ran-GTP gradient spatially regulates XCTK2 within the spindle. A flattened Ran-GTP gradient blocked the ability of excess XCTK2 to stimulate bipolar spindle assembly and resulted in XCTK2-mediated bundling of free MTs. These effects required the XCTK2 tail, which promoted the motility of XCTK2 within the spindle independent of the Ran-GTP. In addition, the turnover kinetics of XCTK2 were spatially controlled: they were faster near the poles relative to the chromatin, but not with a mutant XCTK2 that cannot bind to importin α/β. Our results support a model in which the Ran-GTP gradient spatially coordinates motor localization with motility to ensure efficient spindle formation.
Results and Discussion
A Ran-GTP Gradient is Required for XCTK2-Mediated Stimulation of Spindle Assembly
Our previous work established that XCTK2 is a SAF whose MT crosslinking activity is regulated by Ran-GTP through importin α/β binding to the non-motor tail [14, 21]. However, it was unclear whether a physical gradient of Ran-GTP is required for XCTK2 function. To assess the role of the Ran-GTP gradient on XCTK2 activity, we flattened the gradient by adding a mixture of 10 μM RanQ69L [22] and 30 μM RanT24N [23], which generates high levels of Ran-GTP throughout the extract but eliminates the gradient around chromosomes [24]. Spindle formation was analyzed when the majority of structures in control reactions are spindle intermediates. Addition of GFP-XCTK2 increased the percentage of bipolar spindles relative to addition of GFP (Figures 1A and 1B, p < 0.001) [20]. Flattening the gradient by RanQ/T addition decreased the percentage of bipolar spindles in GFP control reactions (GFP + Q/T) compared to addition of GFP alone (p < 0.05), suggesting that spindle assembly is reduced in RanQ/T extracts. GFP-XCTK2 addition was not sufficient to stimulate spindle assembly in RanQ/T extracts relative to GFP +Q/T (p = 0.72), (Figure 1B). In addition, RanQ/T extracts with GFP-XCTK2 contained bundles of non-chromatin associated MTs throughout the extract (Figures 1A and S1A), suggesting that disruption of the Ran gradient perturbed the localization of XCTK2 and allowed XCTK2 to freely cross-link MTs throughout the extract rather than just within the spindle.
Figure 1. Stimulation of spindle assembly by XCTK2 requires a physical Ran-GTP gradient.
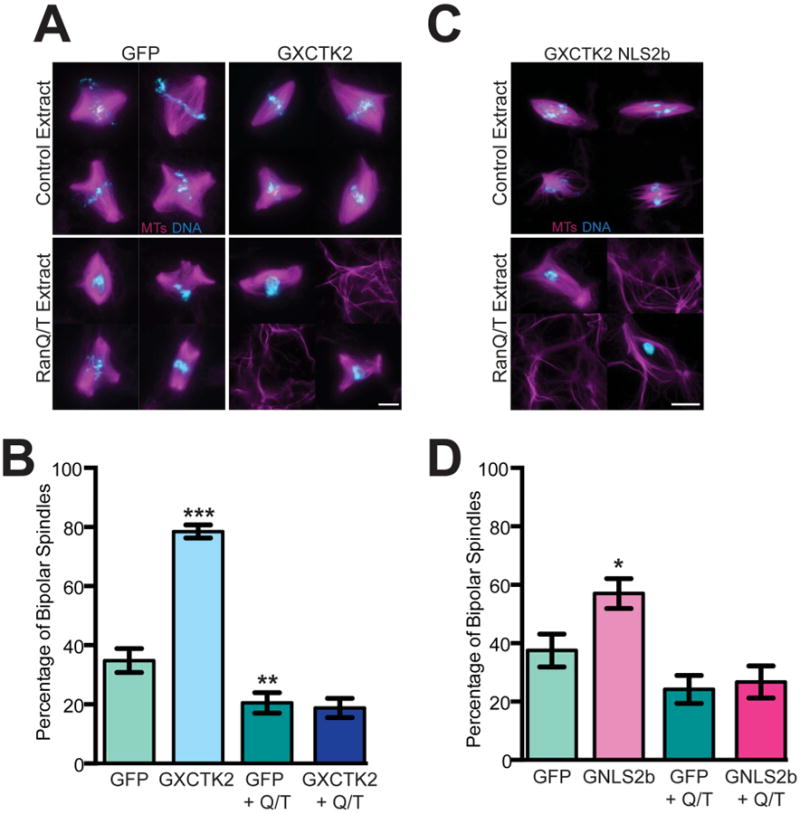
GFP, GFP-XCTK2, or GFP-XCTK2 NLS2b were added to cycled extracts at a five-fold molar excess relative to endogenous XCTK2 +/- 10 μM RanQ69L and 30 μM RanT24N (RanQ/T) 15 minutes post CSF addition. Representative image montages of (A) GFP and GFP-XCTK2 or (C) GFP-XCTK2 NLS2b reactions showing MTs (magenta) and DNA (blue). Scale bar, 10 μm. (B) Quantification of the percentage of bipolar spindles in GFP or GFP-XCTK2 extracts from 10 independent extracts (100 structures/experiment), and the mean ± SEM is graphed. (D) Quantification of the percentage of bipolar spindles in GFP-XCTK2 NLS2b extracts from six independent extracts (100 structures/experiment) where the mean ± SEM is reported. *, p < 0.05, **, p < 0.01, and ***, p < 0.001. (See also Figure S1).
It was previously reported that addition of RanQ/T to extracts liberated SAFs from importin α/β [24]. We therefore performed immunoprecipitations in which GFP-XCTK2 was added to extracts in the presence of RanQ69L and/or RanT24N. Addition of RanQ69L to extracts abolished the ability of importin β to co-immunoprecipitate with XCTK2 (Figure S1B, lanes 2 and 3); whereas addition of RanT24N did not inhibit importin β binding to XCTK2 (Figure S1B, lane 4) [14]. In the presence of RanQ/T, importin β still partially bound XCTK2 (Figure S1B, lane 5), suggesting that the high levels of Ran present in the RanQ/T extracts do not completely prevent importin α/β from binding to the XCTK2 tail. Analysis of GFP-XCTK2 localization (Figure S1C) showed that XCTK2 becomes slightly pole-enriched in the presence of RanQ/T. Together these results suggest that the Ran-GTP gradient may only spatially control XCTK2 when the importins can be released from the XCTK2 tail. In addition, these results demonstrate for the first time that the Ran-GTP gradient is necessary for the function of a SAF within the context of a spindle.
Release of Importins from the XCTK2 Tail Domain is not Sufficient to Stimulate Spindle Assembly in the Absence of a Ran-GTP Gradient
One hypothesis for the inability of XCTK2 to stimulate spindle assembly in RanQ/T extracts is that the XCTK2 tail needs to be completely free from importin α/β. To test this idea, we took advantage of an XCTK2 mutant, GFP-XCTK2 NLS2b, which contains a mutation in the NLS in the XCTK2 tail that prevents binding of the tail to importin α/β but not to MTs [14, 21]. As expected, GFP-XCTK2 NLS2b did not associate with importin β in the extracts (Figure S1C, lanes 6-10). Because this mutant increases the length of bipolar spindles [21], we hypothesized it is hyperactive and would increase spindle bipolarity similar to wt XCTK2. Addition of GFP-XCTK2 NLS2b to extracts caused an increase in the percentage of bipolar spindles compared to GFP addition (Figures 1C and 1D, p < 0.05). In contrast, addition of GFP-XCTK2 NLS2b to RanQ/T extracts did not increase spindle assembly relative to GFP + Q/T control (p = 0.74) and resulted in non-chromatin associated MT bundles similar to what we observed with wt GFP-XCTK2 in the RanQ/T extracts (Figures 1C, 1D and S1A). The localization of GFP-XCTK2 NLS2b on spindles and on MT bundles was highly variable and appeared cooperative (Figure S1B), similar to the cooperative binding observed with XCTK2 tail binding to MTs in vitro [14, 21]. Together, these results suggest that 1) XCTK2 needs a physical Ran gradient in order to stimulate spindle assembly and 2) because neither wt XCTK2 nor XCTK2-NLS2b can stimulate spindle assembly in RanQ/T extracts, other factors that influence spindle organization may also be perturbed in the RanQ/T extracts.
MT Binding by XCTK2 through the Tail and Motor Domains is Required for Stimulation of Spindle Assembly and MT Bundling
Because both the motor and tail domains of XCTK2 bind MTs, we reasoned that motor-tail MT crosslinking and sliding were critical for XCTK2 to stimulate spindle assembly. To test this idea, we generated a truncated XCTK2 construct (GFP-XCTK2 (122-643)) that lacks the tail domain, which includes the ATP-independent MT binding site and the NLS. Truncation of the tail domain abolished binding of importin β (Figure S2A), and when added in excess to extracts GFP-XCTK2 (122-643) had a faint localization on spindle MTs with enrichment at the spindle poles (Figure S2B). To test whether the tail was needed for stimulation of spindle assembly, we added a five-fold molar excess of GFP-XCTK2 (122-643) to extracts (Figure S2C) and assayed spindle formation (Figure 2A and B). Whereas both wt GFP-XCTK2 and GFP-XCTK2 NLS2b increased the percentage of bipolar spindles (Figure 2B, p < 0.001), GFP-XCTK2 (122-643) only modestly stimulated spindle assembly relative to GFP control (p < 0.05). This indicates that motor-tail MT crosslinking activity is needed to robustly stimulate spindle assembly.
Figure 2. The tail domain of XCTK2 is required to robustly stimulate spindle assembly.
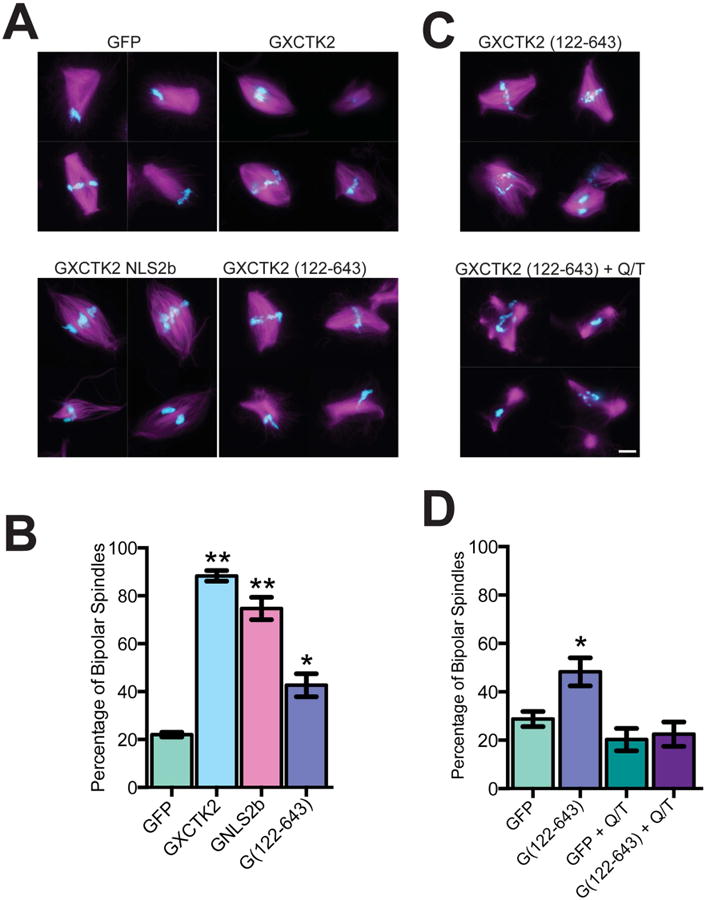
GFP, GFP-XCTK2, GFP-XCTK2 NLS2b, or GFP-XCTK2 (122-643) were added to extracts at a five-fold molar excess relative to endogenous XCTK2 and incubated for 30 min to allow for spindle assembly. (A) Image montages for the indicated protein additions in which the MTs are magenta and the DNA is blue. (B) Quantification of the percentage of bipolar spindles from three independent experiments (100 structures/experiment) with the mean ± SEM graphed. (C) GFP-XCTK2 (122-643) was added at a five-fold molar excess relative to endogenous XCTK2 +/- RanQ69L and RanT24N. Representative image montages of GFP-XCTK2 (122-643) extracts showing MTs (magenta) and DNA (blue). Scale bar, 10 μm. (D) Quantification of the percentage of bipolar spindles in each extract from four independent extracts (100 structures/experiment) with the mean ± SEM reported. *, p < 0.05, and **, p < 0.001 relative to control GFP. (See also Figure S2).
If crosslinking of MTs between the tail domain and the motor domain is responsible for the bundling of MTs in RanQ/T extracts, then XCTK2 (122-643) should not bundle MTs nor stimulate spindle assembly when added to RanQ/T extracts. Indeed, GFP-XCTK2 (122-643) did not stimulate spindle assembly in RanQ/T extracts, similar to what we observed with wt XCTK2 and XCTK2 NLS2b (Figures 2C and 2D). In addition, there was not excess bundling of non-chromatin associated MTs in RanQ/T extracts like that seen in RanQ/T extracts with wt XCTK2 or with XCTK2 NLS2b (Figure S1A). These results indicate that the excess MT bundling seen in the RanQ/T extracts is caused by motor-tail MT crosslinking.
XCTK2 Motility on the Spindle Requires both the Motor and the Tail Domains
One idea for how Ran-GTP controls the ability of XCTK2 to stimulate spindle assembly is that it may differentially regulate XCTK2 motility in the spindle. To test this idea, we imaged both GFP-XCTK2 and tubulin speckles and then used computer-based tracking to measure the velocity of speckles in the spindle [25]. GFP-XCTK2 speckles moved towards the poles at an average velocity of 5.87 ± 0.18 μm/min (Figures 3A and 3B, Video 1, Supplementary Table 1). Spatial analysis showed that there was a 24% reduction in XCTK2 velocity near the spindle poles (4.48 ± 0.34 μm/min, p < 0.01). The velocity of tubulin speckles and their distribution within the spindle was similar to previously published values, suggesting that XCTK2 addition is not disrupting spindle structure (Figures 3B and 3C). To determine whether the reduction of XCTK2 motility near the poles is due to the spatial regulation of the XCTK2 tail by Ran-GTP, we tracked GFP-XCTK2 NLS2b and GFP-XCTK2 (122-643) speckles over time. GFP-XCTK2 NLS2b speckles moved within the spindle at an average velocity of 6.08 ± 0.16 μm/min, with a reduction to 4.81 ± 0.32 μm/min at the poles (p < 0.01), similar to that of GFP-XCTK2 (p = 0.07), indicating that this differential velocity at the poles is likely not due to Ran-GTP regulation (Figure 3B, Video 1). In contrast, the overall velocity of GFP-XCTK2 (122-643) was significantly slower (3.95 ± 0.12 μm/min, p < 0.01) (Figure 3B, Video 1), and was only slightly faster than the rate of tubulin speckle movements toward the poles. These results suggest that both MT binding domains (motor and tail) are needed for robust motility.
Figure 3. XCTK2 motility is spatially regulated.
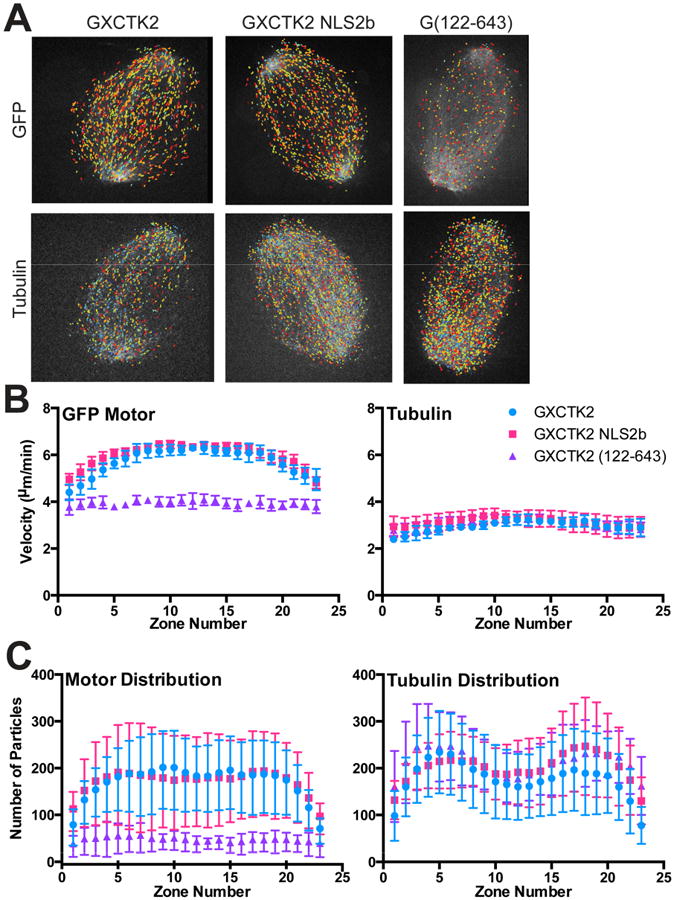
Cycled spindles containing GFP-XCTK2 (6 nM), GFP-XCTK2 NLS2b (6 nM) or GFP-XCTK2 (122-643) (20 nM) with 20 nM rhodamine-labeled tubulin were imaged by confocal microscopy. Images were collected in each channel every 2 s for 2.5 min and then analyzed by a custom MatLab algorithm (see methods), for speckle analysis. (A) Output images from the time-lapse movies with the speckles identified and color-coded according to their velocity with warmer colors indicating faster velocities. (B) Overall velocity of the indicated GFP proteins (left) or tubulin (right) in individual zones of the spindle. Data represent analysis of 8 movies under each condition and are the mean ± SD of the average values of the movies. (C) Average number of particles (GFP proteins, left or tubulin, right) moving in each zone of the spindle are graphed as the mean ± SD of the average values of eight movies. The number of tubulin speckles in one movie from GFP-XCKT2 NLS2b is not included because they were aberrantly high but did not have a different pattern of distribution. A more detailed data compilation is presented in Supplementary Table 1. (See also Video 1).
To ask whether there were regional variations in the number of motors moving, we analyzed the distribution of motors throughout the spindle and found that GFP-XCTK2 and GFP-XCTK2-NLS2b had a similar distribution of motors (Figure 3C). In contrast, there was a dramatic reduction in the number of motors on the spindle with GFP-XCTK2 (122-643) despite the addition of 5-fold more protein for the speckling analysis. These results show that the tail domain of XCTK2 is needed for both robust association of the motor to the spindle and for effective motility of the bound motors, suggesting that the XCTK2 tail anchors the motor to the spindle MTs. Ran-GTP likely modulates XCTK2 by controlling the strength of interaction of the tail with MTs in different regions of the spindle rather than by modulating its motility.
XCTK2 MT Turnover is Spatially Regulated by Ran-GTP in Different Regions of the Spindle
Because the XCTK2 tail is needed for robust association with the spindle and should be free from importin α/β in regions of high Ran-GTP, we postulated that XCTK2 dynamics might be spatially regulated by the Ran-GTP gradient. To test this idea, we analyzed XCTK2 turnover using fluorescence recovery after photobleaching (FRAP) to measure the kinetics of fluorescence recovery of XCTK2 in regions of the spindle where the Ran gradient is the highest (near chromatin) and where the Ran gradient is lowest (near poles) (Figures 4A and S3, Video 2). The t1/2 of XCTK2 recovery around chromatin (29.6 ± 13.0s) was ~30% slower than at the poles (21.0 ± 11.1s, p < 0.01). One possible explanation is that tubulin fluxes toward the poles, which could result in differences in tubulin dynamics in these regions. However, tubulin recoveries near chromatin and at the pole regions were indistinguishable (t1/2 of chromatin, 67.5 ± 25.9; t1/2 of poles, 61.9 ± 29.0; p = 0.31) (Figure 4A). There was no significant difference in the percent recovery of the fluorescence in the different regions, suggesting that there is not an immobile pool of motor (Figure S3). These results show that XCTK2 MT association dynamics are spatially regulated in different regions of the spindle and that XCTK2 turnover kinetics are faster than that of tubulin.
Figure 4. XCTK2 turnover is spatially regulated by Ran-GTP.
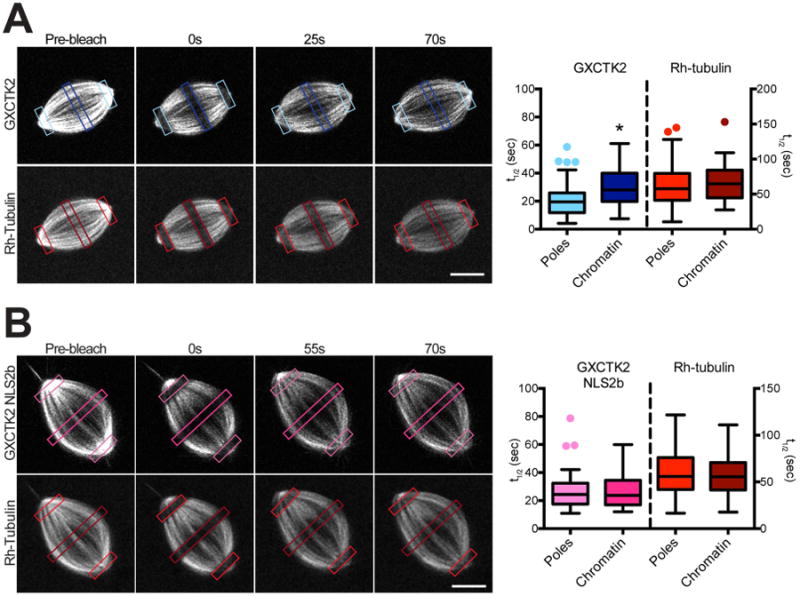
Cycled spindles containing 100 nM excess GFP-XCTK2 (A) or 50 nM GFP-XCTK2 NLS2b (B) and 0.5 μM X-rhodamine-labeled tubulin were photobleached by specifying a 3 μm width box (colored boxes) in the regions of the chromatin and poles and imaged at 0.7 s intervals in the green and red channels for at least 70 s post-bleach. Scale bar, 10 μm. The t1/2 values in each region for GFP-XCTK2, GFP-XCTK2 NLS2b or rhodamine-tubulin were calculated for at least 30 spindles and are plotted as box and whisker plots. *, p < 0.001. (The fluorescence recovery curves for the representative regions are shown in Figure S3. Also see Video 2).
To test the idea that the spatial regulation of XCTK2 turnover was due to regulation by Ran-GTP, we measured the turnover of GFP-XCTK2 NLS2b. Because addition of a five-fold excess of the XCTK2 NLS mutants to extracts perturbs spindle morphology [21], we added only a 2.5-fold excess of GFP-XCTK2 NLS2b for FRAP analysis, which did not perturb spindle structure. Interestingly, the recovery of GFP-XCTK2 NLS2b near chromatin and at the poles was not significantly different (Figure 4B, Video 2) (t1/2 of chromatin, 26.1 ± 11.6s; t1/2 of poles, 26.4 ± 12.0s; p = 0.92) and was similar to the recovery of GFP-XCTK2 near the chromatin, suggesting that the spatial differences in the recovery of GFP-XCTK2 are due to Ran-GTP regulation. The recovery times of tubulin in both regions were not different (t1/2 of chromatin, 59.3 ± 24.0s; t1/2 of poles, 59.3 ± 32.7s; p = 0.99) (Figure 4B), and the recovery of GFP-XCTK2 NLS2b was faster than rhodamine-tubulin (Figure S3B). These results suggest that Ran-regulated importin α/β binding to the tail of XCTK2 provides a mechanism to spatially regulate XCTK2 within the spindle.
Our study is the first to demonstrate that Ran regulation is not simply due to the high levels of Ran-GTP, but rather it is the physical gradient of Ran-GTP that is necessary for SAF function. A slope of association dynamics between the chromatin and poles provides a mechanism whereby different levels of Ran-GTP spatially control spindle organization by generating a secondary gradient of SAF function. When Ran-GTP levels drop near the poles, importins α/β are able to compete more effectively with MTs for binding of the XCTK2 tail domain, thus increasing the turnover kinetics and reducing the anchoring of the protein to spindle MTs, consistent with studies showing reduced MT affinity of the XCTK2 tail in the presence of importins α/β [14]. Our current data show that Ran-GTP does not directly regulate the motility of XCTK2 in the spindle, but suggest that Ran-GTP indirectly regulates motility by modulating the affinity of the XCTK2 tail for MTs [26-30]. This idea is consistent with the observation that Kinesin-14 proteins are dynamic cross-linkers in the spindle, which slide anti-parallel MTs and lock together parallel MTs [31-33]. If the motors only slide MTs effectively when the tail domain is bound between cross-linked MTs, then the ability of Ran to modulate this cross-link by regulating tail MT affinity would be a powerful mechanism to control motor activity in the spindle. In addition, our work is the first to demonstrate the consequences of disrupting the Ran-GTP gradient on a SAF activity. In the future it will be critical to understand which other SAFs are regulated by Ran-GTP gradient as well as to elucidate how the activities of different SAFs may be specifically modulated in regions of lower and higher Ran-GTP. These studies will provide new insight to how cues within the spindle coordinate motor activity to ensure proper spindle formation and maintenance.
Supplementary Material
Highlights.
XCTK2 requires a gradient of Ran-GTP to stimulate spindle assembly
The Ran-GTP gradient regulates XCTK2 microtubule binding affinity
XCTK2 turnover kinetics are spatially regulated in the spindle
Acknowledgments
This work was supported by NIH grant R01-GM059618 to CEW, grant F31GM099309 from the NIGMS to LNW, and NSF MCB 1157982 to SLS. GY was supported by an NSF Faculty Early Career Award DBI-1149494. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NSF, NIGMS or the NIH. The IU Light Microscopy Imaging Center is supported in part by the Office of Vice President for Research at Indiana University. We thank the members of the Walczak lab for support and for comments on the manuscript.
Footnotes
Author Contributions: LNW developed the project, and was responsible for the majority of the experiments and the writing. SEM contributed reagents, experimental design, and edited the paper. SRC and GY developed and implemented algorithms for speckle analysis. SLS proposed the model for XCTK2 turnover and developed the FRAP analysis code. CEW led the overall project, aided in experimental design, analysis, and interpretation, and wrote the paper with LNW.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- 2.Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- 3.Kalab P, Pu RT, Dasso M. The Ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- 4.Ohba T, Nakamura M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- 5.Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- 6.Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- 7.Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 8.Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin β is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- 9.Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. Role of importin-β in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- 10.Wittmann T, Wilm M, Karsenti E, Vernos I. TPX2, A novel Xenopus MAP involved in spindle pole organization. J Cell Biol. 2000;149:1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 12.Sillje HH, Nagel S, Korner R, Nigg EA. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol. 2006;16:731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 13.Trieselmann N, Armstrong S, Rauw J, Wilde A. Ran modulates spindle assembly by regulating a subset of TPX2 and Kid activities including Aurora A activation. J Cell Sci. 2003;116:4791–4798. doi: 10.1242/jcs.00798. [DOI] [PubMed] [Google Scholar]
- 14.Ems-McClung SC, Zheng Y, Walczak CE. Importin α/β and Ran-GTP regulate XCTK2 microtubule binding through a bipartite nuclear localization signal. Mol Biol Cell. 2004;15:46–57. doi: 10.1091/mbc.E03-07-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goshima G, Nedelec F, Vale RD. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatsumi M, Endow SA. Mutants of the microtubule motor protein, nonclaret disjunctional, affect spindle structure and chromosome movement in meiosis and mitosis. J Cell Sci. 1992;101:547–559. doi: 10.1242/jcs.101.3.547. [DOI] [PubMed] [Google Scholar]
- 17.Matuliene J, Essner R, Ryu J, Hamaguchi Y, Baas PW, Haraguchi T, Hiraoka Y, Kuriyama R. Function of a minus-end-directed kinesin-like motor protein in mammalian cells. J Cell Sci. 1999;112:4041–4050. doi: 10.1242/jcs.112.22.4041. [DOI] [PubMed] [Google Scholar]
- 18.Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton DA. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol. 1999;147:351–366. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharp DJ, Yu KR, Sisson JC, Sullivan W, Scholey JM. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat Cell Biol. 1999;1:51–54. doi: 10.1038/9025. [DOI] [PubMed] [Google Scholar]
- 20.Walczak CE, Verma S, Mitchison TJ. XCTK2: A kinesin-related protein that promotes mitotic spindle assembly in Xenopus laevis egg extracts. J Cell Biol. 1997;136:859–870. doi: 10.1083/jcb.136.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai S, Weaver LN, Ems-McClung SC, Walczak CE. Kinesin-14 family proteins HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol Biol Cell. 2009;20:1348–1359. doi: 10.1091/mbc.E08-09-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci U S A. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dasso M, Seki T, Azuma Y, Ohba T, Nishimoto T. A mutant form of the Ran/TC4 protein disrupts nuclear function in Xenopus laevis egg extracts by inhibiting the RCC1 protein, a regulator of chromosome condensation. EMBO J. 1994;13:5732–5744. doi: 10.1002/j.1460-2075.1994.tb06911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maresca TJ, Groen AC, Gatlin JC, Ohi R, Mitchison TJ, Salmon ED. Spindle assembly in the absence of a RanGTP gradient requires localized CPC activity. Curr Biol. 2009;19:1210–1215. doi: 10.1016/j.cub.2009.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang G, Cameron LA, Maddox PS, Salmon ED, Danuser G. Regional variation of microtubule flux reveals microtubule organization in the metaphase meiotic spindle. J Cell Biol. 2008;182:631–639. doi: 10.1083/jcb.200801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuta K, Toyoshima YY. Minus-end-directed motor Ncd exhibits processive movement that is enhanced by microtubule bundling in vitro. Curr Biol. 2008;18:152–157. doi: 10.1016/j.cub.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 27.Chandra R, Salmon ED, Erickson HP, Lockhart A, Endow SA. Structural and functional domains of the Drosophila ncd microtubule motor protein. J Biol Chem. 1993;268:9005–9013. [PubMed] [Google Scholar]
- 28.Karabay A, Walker RA. Identification of microtubule binding sites in the Ncd tail domain. Biochemistry. 1999;38:1838–1849. doi: 10.1021/bi981850i. [DOI] [PubMed] [Google Scholar]
- 29.Walker RA. ncd and kinesin motor domains interact with both alpha- and beta-tubulin. Proc Natl Acad Sci U S A. 1995;92:5960–5964. doi: 10.1073/pnas.92.13.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao L, Mogilner A, Civelekoglu-Scholey G, Wollman R, Evans J, Stahlberg H, Scholey JM. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol. 2006;16:2293–2302. doi: 10.1016/j.cub.2006.09.064. [DOI] [PubMed] [Google Scholar]
- 31.Braun M, Drummond DR, Cross RA, McAinsh AD. The kinesin-14 Klp2 organizes microtubules into parallel bundles by an ATP-dependent sorting mechanism. Nat Cell Biol. 2009;11:724–730. doi: 10.1038/ncb1878. [DOI] [PubMed] [Google Scholar]
- 32.Fink G, Hajdo L, Skowronek KJ, Reuther C, Kasprzak AA, Diez S. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat Cell Biol. 2009;11:717–723. doi: 10.1038/ncb1877. [DOI] [PubMed] [Google Scholar]
- 33.Hentrich C, Surrey T. Microtubule organization by the antagonistic mitotic motors kinesin-5 and kinesin-14. J Cell Biol. 2010;189:465–480. doi: 10.1083/jcb.200910125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


