Abstract
Metformin is a widely prescribed drug for the treatment of type-2 diabetes. Although epidemiological data have provided a strong rationale for investigating the potential of this biguanide for use in cancer prevention and control, uncertainty exists whether metformin should be expected to have an impact in non-diabetic patients. Furthermore, little attention has been given to the possibility that other biguanides may have anticancer activity. In this study, the effects of clinically relevant doses of metformin (9.3mmol/kg diet), buformin (7.6 mmol/kg diet), and phenformin (5.0 mmol/kg diet) were compared to rats fed control diet (AIN93-G) during the post initiation stage of 1-methyl-1-nitrosourea-induced (50 mg/kg body weight) mammary carcinogenesis (n = 30/group). Plasma, liver, skeletal muscle, visceral fat, mammary gland, and mammary carcinoma concentrations of the biguanides were determined. In comparison to the control group, buformin decreased cancer incidence, multiplicity, and burden; whereas, metformin and phenformin had no statistically significant effect on the carcinogenic process relative to the control group. Buformin did not alter fasting plasma glucose or insulin. Within mammary carcinomas, evidence was obtained that buformin treatment perturbed signaling pathways related to energy sensing. However, further investigation is needed to determine the relative contributions of host systemic and cell autonomous mechanisms to the anticancer activity of biguanides such as buformin.
Keywords: Biguanides, mammary carcinogenesis, metformin, buformin, phenformin
Introduction
Considerable attention is being given to using metformin, one of the most widely prescribed drugs for the treatment of type-2 diabetes, for the prevention and control of a number of types of cancer, including breast cancer (1–3). While epidemiologic studies, coupled with in vitro mechanistic evidence, have propelled metformin into clinical trials as a potential chemopreventive and adjuvant, preclinical observations have raised concerns about metformin efficacy (4–8). Magnifying the problem are conflicting reports regarding mechanism of action, particularly in the non-diabetic state (3, 9, 10).
Metformin is prescribed for the treatment of type-2 diabetes, but understanding of its mechanism of action in this context is limited and has recently been challenged (11,12). The prevailing view has been that metformin induces activation of AMP activated protein kinase (AMPK) in the liver, presumably by reducing intracellular energy charge via partial inhibition of complex I activity. However, evidence from mice has been inconsistent, with the absence of either LKB1 or AMPK activity failing to block the glucose lowering effects of metformin (12). Rather, new evidence has been published indicating that metformin, as well as a related biguanide, phenformin, induce AMP accumulation resulting in inhibition of adenyl cyclase and down regulation of protein kinase A activity (11). It is hypothesized that reduced activity of protein kinase A decreases the expression of genes that regulate hepatic gluconeogenesis, thereby decreasing hepatic glucose output. Nonetheless, this work failed to clarify the mechanism that accounts for AMP accumulation. One possible alternative to the proposed inhibition of complex I is that metformin and other biguanides inhibit the activity of AMP deaminase, the enzyme that regulates the conversion of AMP to IMP (13–15). If AMP deaminase is inhibited, AMP would accumulate intracellularly, not only activating AMPK but also inhibiting adenyl cyclase and leading to decreases in cyclic AMP and the activity of protein kinase A.
Recognizing that other biguanides differ from metformin in their biological activities and that phenformin and buformin have been reported to inhibit mammary carcinogenesis in various experimental models (7, 16, 17), the experiments reported herein were conducted to compare effects of these biguanides. Based on the results of the carcinogenesis study, additional experiments were conducted to generate hypotheses about potential targets of the most effective anticancer biguanide, buformin, and about the likely involvement of host systemic and cell autonomous mechanisms in accounting for cancer inhibitory activity.
Materials and Methods
Chemicals
Primary antibodies used in this study were purchased from Cell Signaling Technology (Beverly, MA): anti-pPKAThr198/PKA, anti-pCREBSer133/CREB, anti-pSTAT3Tyr705/STAT3, anti-pSrcSer17/Src, anti-pACC Ser79/ACC, anti-pAktSer473/Akt, anti-pAMPK Thr172/AMPK, anti-p4EBP1Thr37/46/4EBP1, anti-pmTORSer2448/mTOR, anti-pP70S6KThr389/P70S6K, anti-EPAC-1, anti-BEATA2_AR, anti-FAK, anti-FASN, anti-GLUT4, anti-OCT3, anti-NOTCH, anti-PGC1, anti-pPRAS40Thr246, anti-PRAS40, anti-pRAPTORSer792, anti-RAPTOR, and anti-PI3K110α. Anti-BCL-2 was purchased from BD Bioscience (San Jose, CA). Anti-P27 was purchased from Thermo Fisher Scientific (Waltham, MA). Anti-rabbit immunoglobulin-horseradish peroxidase-conjugated secondary antibody, LumiGLO reagent with peroxide and cAMP assay kit were also purchased from Cell Signaling Technology, Inc. (Beverly, MA). Anti-IGF1Rα, anti-HMGCR, anti-P21, anti-SCD1, and anti-SCEBP1 were obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX); mouse anti-β-actin primary antibody was obtained from Sigma Aldrich (St. Louis, MO). The 1-methyl-1-nitrosourea (MNU) was obtained from Ash Stevens (Detroit, MI) and stored at −80°C prior to use. Metformin and buformin were obtained from Waco Pure Chemical Industries, (Waco, TX); phenformin was obtained from Sigma Aldrich (St. Louis, MO).
Biguanides
Biguanides were incorporated into an AIN-93G purified diet formulation (18–20). The dietary concentration of metformin, (9.3mmol/kg diet), provided the rat with a dose equivalent to a 50 kg women receiving 800 mg metformin/d which is within the range typically given for the control of type-2 diabetes. The concentrations of buformin (7.6 mmol/kg diet) or phenformin (5.0 mmol/kg diet) were determined to be the maximum tolerated dose in the rat based on a preliminary feeding study. Here, maximum tolerated dose is defined as the highest dose that causes no weight loss, slows growth rate by less than 10%, or induces no external signs of toxicity (21). The human dose equivalent for buformin and phenformin is also 800 mg/d which is higher than what has been given in clinical studies of these compounds. Buformin is still used clinically in Romania (Silubin, sustained release) where a common dose is 50 to 300 mg/d (22). Before being withdrawn from the market, the recommended dose of phenformin was 400 to 800 mg/d (23, 24). Biguanide concentrations in each diet were confirmed by HPLC analysis.
HPLC analysis of biguanides
Diet and tissue were homogenized (20% w/v) using ultrasound and then extracted with acetonitrile/methanol (2/1 v/v) for 30 min at room temperature. Following extraction, samples were centrifuged at 17,000 × G for 10 min and the supernatant fraction was directly analyzed via HPLC using a Waters C18 reverse phase column (μbondapak C18 10μm column, 3.9×300mm, equipped with a μbondapak C18 guard) with UV detection (235 nm). Run conditions were: isocratic elution using a 10mM KH2PO4/K2HPO4 pH 6.8 : ACN (37:63) mobile phase; flow rate was 1.5mL/min, the injection volume was 20uL and the run time was 8 min.
Carcinogenesis experiment
Female Sprague Dawley rats were obtained (Charles River, Wilmington, MA) at 20 days of age. At 21 days of age, 120 rats were injected with 50 mg MNU/kg body weight, i.p. as previously described (25). Rats were housed in solid bottomed polycarbonate cages equipped with food cups. At 28 days of age, 1 week after carcinogen injection, rats were assigned by stratified randomization using body weight to one of four groups (30 rats/group) and were ad libitum fed either control diet (AIN 93-G) or AIN 93-G diet into which metformin, buformin, or phenformin had been incorporated. Throughout the experiment, animal rooms were maintained at 22 ± 1°C with 50% relative humidity and a 12-h light/12-h dark cycle. Rats were weighed weekly and were palpated for the detection of mammary tumors twice weekly starting from 21 days post carcinogen. At necropsy, rats were skinned and the skin to which mammary gland chains were attached was examined under translucent light for detectable mammary pathologies at 5× magnification. All detectable mammary gland pathologies were excised and prepared for histological classification according to published criteria (26, 27). Only confirmed mammary carcinomas are reported. The experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee and conducted according to the committee guidelines.
Blood collection and plasma biomarker analyses
Blood collection
Following an overnight fast, rats were euthanized over a 3-hour time interval, between 8 and 11 a.m., via inhalation of gaseous carbon dioxide. The sequence in which rats were euthanized was stratified across groups to minimize the likelihood that order effects would masquerade as treatment associated effects. After the rat lost consciousness, blood was directly obtained from the retro-orbital sinus and gravity fed through heparinized capillary tubes (Fisher Scientific, Pittsburgh, PA) into EDTA coated tubes (Becton Dickinson, Franklin Lakes, NJ) for plasma. The bleeding procedure took approximately 1 min/rat. Thereafter, the unconscious rat was euthanized by cervical dislocation. Plasma was isolated by centrifugation at 1000 × g for 10 min at room temperature.
Assessment of circulating molecules
Insulin-like growth factor 1 (IGF-1), IGF binding protein 3 (IGFBP-3), adiponectin, insulin, leptin, and glucagon in plasma or cAMP in tissue lysate were determined using commercially available ELISA assays as previously described. Glucose in plasma was determined enzymatically using a commercially available kit (Pointe Scientific, Inc., Canton, MI.).
Western blotting
Each mammary carcinoma was homogenized in lysis buffer [40 mM Tris-HCl (pH 7.5), 1% Triton X-100, 0.25 M sucrose, 3 mM EGTA, 3 mM EDTA, 50 μM β-mercaptoethanol, 1 mM phenyl-methylsulfonyl fluoride, and complete protease inhibitor cocktail (Calbiochem, San Diego, CA)]. The lysates were centrifuged at 7500 × g for 10 min at 4 °C and supernatant fractions collected and stored at −80 °C. Supernatant protein concentrations were determined by the Bio-Rad protein assay (Bio-Rad, Hercules, CA). Western blotting was performed as described previously. Briefly, 40 μg of protein lysate per sample was subjected to 8–16% sodium dodecyl sulfate-polyacrylamide gradient gel electrophoresis (SDS-PAGE) after being denatured by boiling with SDS sample buffer [63 mM Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 50 mM DTT, and 0.01% bromophenol blue] for 5 min. After electrophoresis, proteins were transferred to a nitrocellulose membrane. The levels of pPKAThr198/PKA, pCREBSer133/CREB, pSTAT3Tyr705/STAT3, pSrcSer17/Src, pACC Ser79/ACC, pAktSer473/Akt, pAMPK Thr172/AMPK, p4EBP1Thr37/46/4EBP1, pmTORSer2448/mTOR, pP70S6KThr389/P70S6K, EPAC-1, IGF1Rα, PI3K110α and β-actin were determined using specific primary antibodies, followed by treatment with the appropriate peroxidase-conjugated secondary antibodies and visualized by LumiGLO reagent western blotting detection system. The chemiluminescence signal was captured using a ChemiDoc densitometer (Bio-Rad) that was equipped with a CCD camera having a resolution of 1300 × 1030. Quantity One software (Bio-Rad) was used in the analysis. The Quantity One software has a warning algorithm that notifies the user if pixel density is approaching saturation so that all signals used for analysis are in the linear range. All Western blot signals were within a range where the signal was linearly related to the mass of protein and actin-normalized scanning density data were used for analysis.
Statistical Analyses
Differences among groups were evaluated as follows: incidence of mammary carcinomas by the Fischer exact test, the number of mammary carcinomas per rat (multiplicity) by Poisson regression, and cancer burden by the nonparametric Kruskal-Wallis test. Palpable cancer latency was evaluated by survival analysis using the Mantel-Haenszel method. P-values were adjusted for multiple comparisons using a Bonferroni correction. Differences in final body weight and circulating analytes were evaluated by ANOVA with post hoc comparisons by the method of Tukey (28). For Western blots, the data were either the actin-normalized scanning data for proteins or the ratio of the actual scanning units derived from the densitometry analysis of each Western blot for the phospho-proteins. For statistical analyses, the actin-normalized scanning density data obtained from the ChemiDoc scanner using Quantity One (Bio-Rad, Hercules, CA) were rank transformed, an approach that is particularly suitable for semi-quantitative measurements that are collected as continuously distributed data, as is the case with Western blots (29). Ratio data were computed from the scanning units derived from the densitometry analysis, i.e. the arbitrary units of optical density, and then the ratios were rank transformed and medians ± the interquartile ranges were computed and shown using scatter plots (Supplementary Fig. S1). All analyses were performed using Systat statistical analysis software, version 13 (Systat Software, Inc., Chicago, IL) or Prism for Windows, version 6 (GraphPad Software Inc.). All P values are 2-sided and statistical significance was set a priori at P < 0.05.
Multivariate Analysis
Supervised and unsupervised multivariate techniques were employed to evaluate and visualize the data per our previously published approach (30–33).
Unsupervised Analysis
Principal components analysis (PCA) was used to analyze plasma analytes and Western data (34). The PCA model can be written:
| (eq 1) |
where X is the matrix of measured variables, Xbar is a vector of means (all 0 when the data are centered), T is a matrix of scores that summarize the X variables, P is a matrix of loadings, and E is a matrix of residuals.
Supervised Multivariate Analysis
Orthogonal projections to latent structures for discriminant analysis (OPLD-DA) was used as a supervised, class-based method (30–32, 35) to study plasma analytes and Western data. The OPLS-DA model can be written:
| (eq 2) |
where the interpretation of equation 2 is similar to that for the PCA model; however, an additional rotation has been applied using the class information to partition TP′ into a predictive, TpPp′, and an orthogonal, ToPo′, component. The number of predictive and orthogonal components in the models was determined by cross-validation.
Three key statistics describe the fit of each model: R2X(cum), the total amount of explained variation in X; R2Y(cum), the total amount of variation explained in Y; and Q2Y(cum), the total amount of predicted variability in Y, estimated by 7-fold cross validation. The contribution of each component partitioned into between (predictive) and within (orthogonal) class is also estimated, and summarized as R2Xp and R2Xo, respectively. R2Xp and R2Xo sum to R2X(cum).
Visualization of PCA and OPLS-DA
Scatter plots of the first two score vectors for the PCA models were drawn, along with 95% confidence ellipses based on Hotelling’s multivariate T2, to identify outliers that might bias the results of OPLS-DA. For OPLS-DA, class separation was shown in several ways. The first predictive score was plotted against the first orthogonal score to visualize the within- and between-class variability associated with the first principal component, and dendrograms were drawn using the first (or first and second) predictive scores, by the single linkage method, and sorted by size. In the single linkage method, observations were merged by proximity to neighbors based on Euclidean distance, building the hierarchy from individual observations by progressively merging clusters until all observations are merged into one parent cluster (36).
S-plots were constructed to identify influential proteins in the separation of treatment groups. S-plots based on the first principal component show reliability (modeled correlation) plotted against feature magnitude (loadings or modeled covariance). If protein concentrations have variation in correlation and covariance between classes, this plot will assume an S-shape, with heavily influential features separating from other features at the upper right and lower left tails of the feature cloud within the model space (31, 32). All multivariate analyses were done using SIMCA-P+ v.12.0.1 (Umetrics, Umea, Sweden).
Results
Effect of biguanides on the carcinogenic response
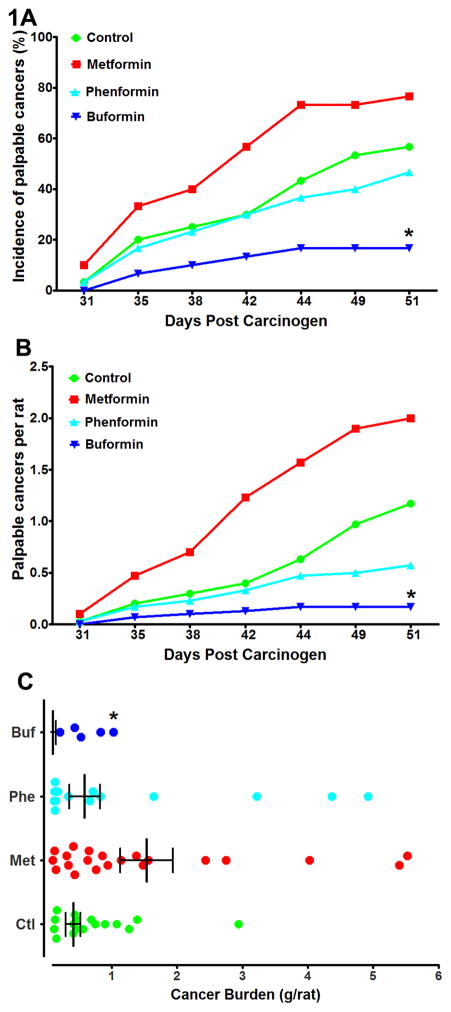
A total of 106 histopathologically confirmed mammary carcinomas were detected by palpation in this study and 11 additional carcinomas with a mass > 100 mg were found at necropsy. The effects of biguanide treatment were determined not only on palpable mammary pathologies, but also small pathologies that were identified by 5× magnification at necropsy and that we refer to as microcarcinomas (< 100 mg in size) (Table 1). In comparison to the control group, only buformin treatment reduced the incidence and multiplicity of palpable carcinomas and microcarcinomas at levels of statistical significance that were significant with adjustment for multiple comparisons (P < 0.008). In addition, the carcinogenic response was lower in buformin treated rats in comparison to rats treated with metformin or phenformin, although the magnitude and statistical significance of these differences varied by cancer endpoint (Supplementary Table S1 for detailed statistical summary).
Table 1.
Effect of biguanides on final mammary cancer incidence and multiplicity
| Treatmentb | Palpable carcinomasa | Microcarcinomas | Total | |||
|---|---|---|---|---|---|---|
| Incidence % | Multiplicity Number/rat | Incidence % | Multiplicity Number/rat | Incidence % | Multiplicity Number/rat | |
| Control | 56.7c | 1.2 ± 0.3c,d | 80.0c | 1.6 ± 0.3c | 83.3c | 2.7 ± 0.4c,d |
| Metformin | 76.7c | 2.0 ± 0.3c | 86.7c | 1.6 ± 0.2c | 93.3c | 3.6 ± 0.4c |
| Buformin | 16.7d | 0.2 ± 0.1e | 36.7d | 0.6 ± 0.2d | 43.3d | 0.7 ± 0.2e |
| Phenformin | 46.7c,d | 0.6 ± 0.1d,e | 63.3c,d | 1.0 ± 0.2c,d | 76.7c,d | 1.6 ± 0.2d,e |
| Overall P value | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
Values are means ± SEM for cancer multiplicity, n = 30 rats/group. Cancer incidence was analyzed by the Fisher exact test and cancer multiplicity by Poisson regression.
Values within a column with different superscripts were statistically different by post hoc multiple comparisons, P < 0.008.
Eleven (9%) of these carcinomas were detected at necropsy. This 9% error rate was not statistically different among groups (P = 0.261).
Biguanide was provided in the diet at concentrations of: 9.3mmol metformin/kg diet, 7.6 mmol buformin/kg diet, or 5.0 mmol phenformin/kg diet.
The emergence of carcinomas over time (cancer latency, Fig. 1) was most rapid in the metformin group whether the endpoint was the occurrence of the first carcinoma in an animal (cancer incidence, Fig. 1A) or the average number of cancers per rat (cancer multiplicity, Fig. 1B). In comparison to the control group, metformin shortened cancer latency (P = 0.025), whereas buformin prolonged cancer latency (P = 0.003). Phenformin was without effect on cancer latency relative to the control group (P = 0.265). Buformin prolonged cancer latency relative to either metformin (P < 0.001) or phenformin (P = 0.015). However, these P-values are not significant after adjustment for multiple comparisons. Thus, only buformin significantly affected cancer latency. Fig. 1C shows the effect of biguanide treatment on cancer mass per rat, with only buformin reducing cancer burden.
Figure 1.
Effects of biguanide treatment on various aspects of the carcinogenic response. A, the incidence of palpable mammary cancer as a function of days post carcinogen injection. B, the average number of palpable cancers per rat as a function of days post carcinogen injection. C, the cancer burden in grams per rat determined at necropsy. The graph is a scatter plot showing the median and interquartile range. The computation includes tumor free rats (not shown in the graph). Note that only 5 buformin treated rats had palpable carcinomas, a fact that limited the number of carcinomas that could be Western blotted in this group.
*Significantly different from the control group when adjusted for multiple comparisons.
Tissue concentrations of biguanides
Biguanide concentrations were determined in blood and tissue obtained at necropsy following an overnight fast (Supplementary Table S2). Plasma and tissue concentrations of metformin and buformin were similar and not statistically different with the exception of skeletal muscle in which metformin concentration was three times higher for metformin than buformin. Phenformin was only detected in skeletal muscle and mammary gland following the overnight fast. In mammary gland, the concentrations of the three biguanides were similar. In mammary carcinomas, metformin and buformin were found to have similar concentrations.
Effects of biguanides on circulating analytes
Blood from overnight fasted rats was collected at necropsy and all plasma samples were evaluated for the analytes shown in Table 2. Of the analytes measured, only the plasma concentrations of IGF-1 (P < 0.001), glucagon (P = 0.002), and adiponectin (P = 0.003) were affected. In the buformin and phenformin treated groups, IGF-1 was 30.4% and 22.8% lower than the control group and glucagon was 63.0% and 57.5%, lower than the control group, respectively. In the buformin group, adiponectin was 19.1% higher than in the control group.
Table 2.
Effect of biguanides on plasma analytes
| Analyte | Control | Metformina | Phenformin | Buformin | Overall P value |
|---|---|---|---|---|---|
| Glucose (mg/dL) | 125.4 ± 12.9 | 128.5 ± 11.7 | 128.7 ± 17.4 | 130.7 ± 23.9 | 0.697 |
| Insulin (ng/mL) | 1.2 ± 0.3 | 1.1 ± 0.2 | 1.1 ± 0.3 | 1.1 ± 0.4 | 0.879 |
| IGF-l (ng/mL) | 328.7 ± 94.8b | 332.5 ± 74.4b | 254.2 ± 62.4c | 229.4 ± 61.5c | < 0.001 |
| IGFBP3 (ng/mL) | 112.9 ± 29.9 | 113.0 ± 18.8 | 107.2 ± 24.3 | 106.3 ± 18.5 | 0.542 |
| Glucagon (pg/mL) | 73.5 ± 59.9b | 63.9 ± 40.7b,d | 26.8 ± 12.1c | 31.1 ± 9.5c,d | 0.002 |
| Leptin (ng/mL) | 1.9 ± 0.7 | 1.8 ± 0.7 | 1.5 ± 0.4 | 1.5 ± 0.6 | 0.087 |
| Adiponectin (ng/mL) | 18.8 ± 3.7b | 18.6 ± 4.0b | 20.8 ± 3.3b,c | 22.4 ± 6.2c | 0.003 |
| Total Cholesterol (mg/dL) | 101.7 ± 8.1 | 104.5 ± 14.9 | 100.2 ± 16.5 | 100.3 ± 13.0 | 0.564 |
| HDL Cholesterol (mg/dL) | 50.6 ± 9.1b | 55.2 ± 10.0b,c | 55.5 ± 11.1b,c | 60.4 ± 12.0c | 0.0068 |
| LDL Cholesterol (mg/dL) | 31.6 ± 11.4b,d | 30.6 ± 8.2b,d | 25.9 ± 9.3b,c,d | 21.1 ± 9.1c | 0.0001 |
| Triglyceride (mg/dL) | 97.6 ± 17.1 | 93.7 ± 20.6 | 94.0 ± 18.1 | 94.2 ± 11.9 | 0.7973 |
Values are means ± SD, n = 30/group. Data were evaluated by ANOVA with post hoc comparison by the method of Tukey. Values within a row with different superscripts were statistically different by post hoc multiple comparisons, P < 0.05.
Biguanide was provided in the diet at concentrations of: 9.3mmol metformin/kg diet, 7.6 mmol buformin/kg diet, or 5.0 mmol phenformin/kg diet.
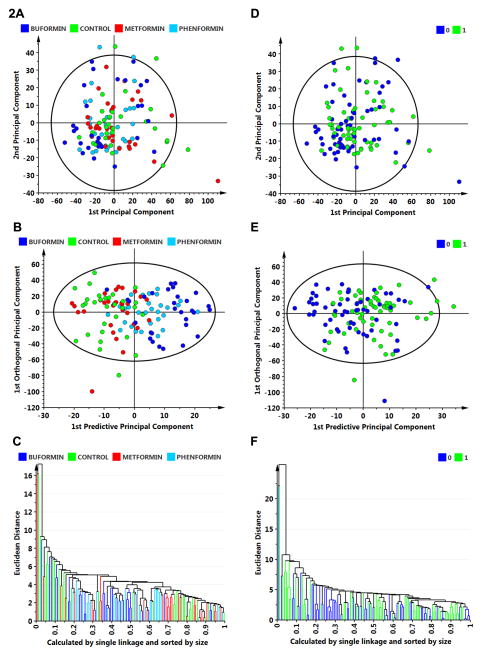
For hypothesis generation concerning the role of host systemic factors in accounting for the effects of biguanide treatment, plasma analyte data were subjected to multivariate analysis of variance using unsupervised and supervised clustering techniques as implemented in SIMCA-P. Those analyses (Fig. 2) failed to reveal a pattern of analytes that distinguished among treatment groups (Fig. 2A) using unsupervised principle components analyses (Fig. 2A) or a supervised technique, OPLS-DA (Fig. 2B). The poor classification rate (overall rate is 65% correct) is illustrated in the dendrogram from the supervised analysis (Fig. 2C) with the poorest classification occurring between the control group and the metformin group. This approach also determined whether the plasma analyte profile could distinguish between cancer bearing and cancer free rats irrespective of the treatment group in which they occurred. Neither unsupervised (Fig. 2D) or supervised (Fig. 2E) analyses distinguished between these classes as illustrated in the dendrogram from the supervised analysis (Fig. 2F).
Figure 2.
Multivariate discriminant analysis was used to determine whether plasma analyte data could distinguish among treat groups (A–C) or whether an animal was cancer bearing versus cancer free (D–F). A, to visualize inherent clustering patterns, the scatter plot represents unsupervised analysis through the PCA 4-class model. Poor separation of treatment groups is observed. Model fit: R2X(cum)= 0.437, and Q2(cum)= 0.093. B, to determine contributing sources of variation, the scatter plot represents supervised analysis of the 4-class OPLS-DA model, which rotates the model plane to maximize separation due to class assignment. Separation is still poor with an overall misclassification rate of 45%. Model fit: R2Y(cum)= 0.199, Q2Y(cum)= 0.132. C, to visualize the misclassification rate, the dendrogram depicts hierarchical clustering patterns among treatment groups using single linkage and size. D, to visualize inherent clustering patterns, the scatter plot represents unsupervised analysis through the PCA 2-class model. Poor separation of treatment groups is observed for the categories: cancer free = 0 versus cancer bearing = 1. E, to determine contributing sources of variation, the scatter plot represents supervised analysis of the 2-class OPLS-DA model, which rotates the model plane to maximize separation due to class assignment. Separation is still poor with an overall misclassification rate of 55%. Model fit: R2Y(cum) = 0.124, Q2Y(cum) = 0.080. F, to visualize the misclassification rate, the dendrogram depicts hierarchical clustering patterns among treatment groups using single linkage and size.
Between-class discrimination by Western blots
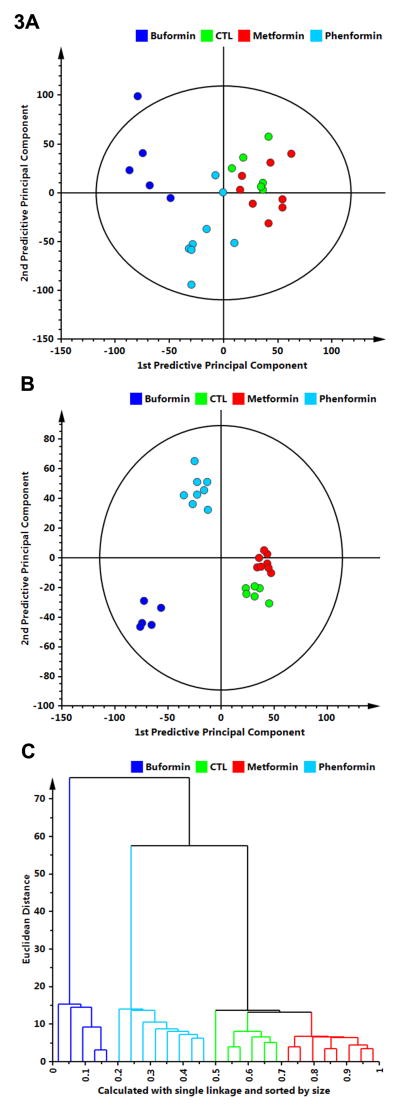
To determine if treatments had distinct effects on cell signaling, all Western blot data were first evaluated using PCA on all 4 treatment groups. PCA identified two significant components that explained a total of 47.7% of the variance in protein concentration. The first 2 component scores of the model are shown in Fig. 3A. Buformin and phenformin separated well from control and metformin which were not separated from one another. Only one buformin tumor fell outside the 95% confidence ellipse.
Figure 3.
Multivariate discriminant analysis was used to determine whether Western blot data for 26 proteins assessed in mammary carcinomas (Supplementary Fig. S1 and 2) could distinguish among treat groups. A, to visualize inherent clustering patterns, the scatter plot represents unsupervised analysis through the PCA 4-class model. Separation of treatment groups is observed. Model fit: R2X(cum) = 0.747, with 5 components, and Q2(cum) = 0.155. B, to determine contributing sources of variation, the scatter plot represents supervised analysis of the 4-class OPLS-DA model, which rotates the model plane to maximize separation due to class assignment. Complete separation of the 4 classes was observed. Model fit: R2Y(cum) = 0.984, Q2Y(cum) = 0.963. C, to visualize the misclassification rate, the dendrogram depicts hierarchical clustering patterns among the treatment groups using single linkage and size. Two main clusters comprise 1) buformin versus 2) phenformin, control, metformin.
OPLS-DA was then used to refine the model fit and partition the variance into predictive (protein expression differences related to treatment) and orthogonal (protein expression differences unrelated to treatment) sources. The first predictive and orthogonal components are plotted in Fig. 3B; 24.1% of the variance in protein concentration was related to treatment (first predictive component), whereas 14.4% of the variance was unrelated to treatment (first orthogonal component); the remaining variation is attributed to noise. The overall fit of the model was good (R2XP1-3 = 80.9%, R2XO1 = 37.2%, R2Y(cum) = 96.3%, Q2(cum) = 81.1%). The dendrogram (Fig. 3C), constructed using the first 2 score vectors form the OPLS-DA model, illustrates the classification accuracy. Two main clusters were defined: 1) buformin and 2) all other treatment groups, which subsequently split into clusters comprising phenformin, control and metformin, which mirrored the ordering observed in the carcinogenic response (Table 1 and Fig. 1).
Identification of influential proteins
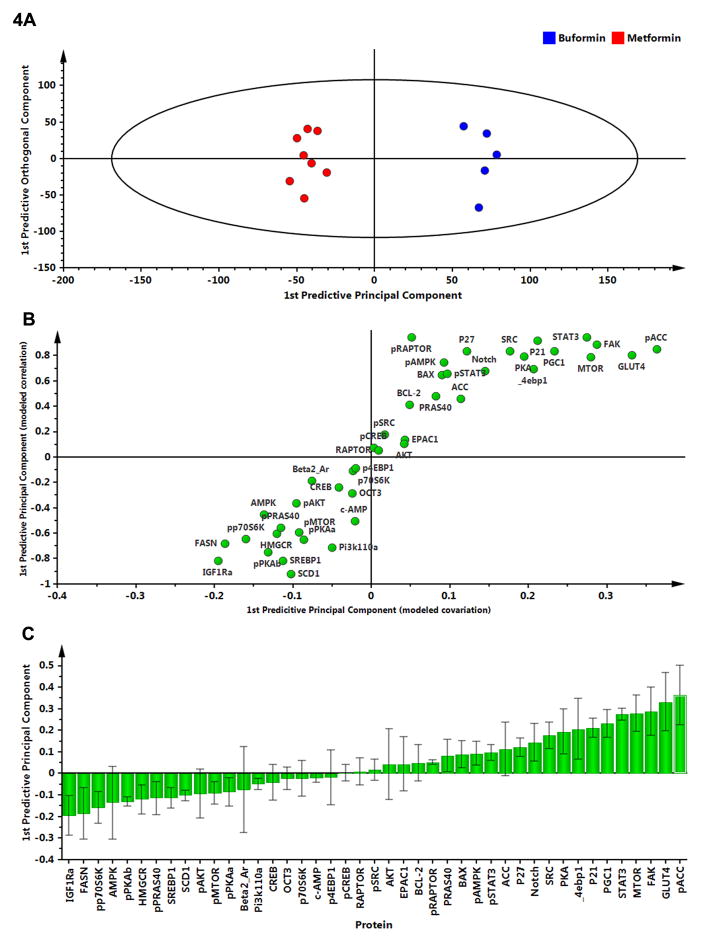
To determine the proteins responsible for the distinctions observed among treatment groups and provide insight into differences in the carcinogenic response, two-class OPLS-DA modeling was performed. Since buformin and metformin were in similar concentrations in mammary carcinomas and mammary gland, but metformin was ineffective in inhibiting the carcinogenic process, these two treatments were compared. A two-class OPLS-DA model comparing buformin and metformin (Fig. 4A) was used to generate the S-plot shown in Fig. 4B with magnitude and variation in each protein shown in Fig. 4C. Among the most influential proteins accounting for separation are those involved in the regulation of de novo lipid synthesis (pACC and FASN) and known to be regulated at least in part by components of the mTOR signaling pathway which also contributed significantly to observed separation between groups.
Figure 4.
To determine the proteins responsible for class separation, multivariate analysis was extended to identify influential proteins responsible for the separation between classes. A, a supervised OPLS-DA model was created to compare buformin to metformin; complete separation was observed. B, an S-plot was constructed by plotting modeled correlation in the first predictive principal component against the modeled covariance in the first predictive component. Upper right and lower left regions of the S-plots contain candidate proteins with both high reliability and high magnitude. C, to determine the statistical reliability of the proteins shown in 4B, jack-knifed confidence intervals (JKCI) were created on the magnitude of covariance in the first component for the 26 proteins and sorted in ascending order based on expression in the buformin group; proteins with JKCIs including 0 were not considered to account for separation.
Discussion
Many compounds exist that have biguanide related activity, and two of these compounds, buformin and phenformin, were synthesized in the same era that metformin was developed (9). In general, they are not used in the treatment of diabetes because they have a greater potential to induce lactic acidosis. Nonetheless, the question has been raised about whether metformin is the best biguanide with which to answer questions about the anti-cancer activity of this class of compounds (10). To address this question relative to breast cancer, the effects of metformin, buformin, and phenformin were evaluated. Buformin, but not metformin or phenformin, was found to have strong cancer inhibitory activity. In this regard, the biguanide concentration data (Supplementary Table S2) merit comment. Buformin and metformin were present in tissue at similar concentrations yet buformin was highly effective in inhibiting mammary cancer; whereas, metformin was not. This situation established a rationale for the post hoc analyses reported in Fig. 3 and Fig. 4, the goal of which was hypothesis generation about the targets of effective biguanides for breast cancer prevention and control. Relative to phenformin, which was fed at the lowest dietary concentration of the three biguanides, it was not detectable in any tissue after an overnight fast other than mammary gland and skeletal muscle. Given that 5 mmol/kg diet appears to be a maximal tolerated dose of this compound, it is worth considering that phenformin’s ineffectiveness against breast cancer might be associated with either rapid metabolism or the need to bolus dose rather than feed it in the diet in order to achieve effective doses of the compound in target tissue(s).
The main finding for metformin was a lack of effectiveness against the carcinogenic response, compared to control and other biguanides. The results of this study showed that administration of a clinically relevant dose of metformin in the diet, based on allometric dose conversion, may have accelerated the carcinogenic response in this non-diabetic model for breast cancer. However, the effects of metformin on multiple cancer endpoints were judged not to be statistically different from the control group following adjustment for multiple comparisons (Supplementary Table S1). Metformin was detected in liver, mammary gland, and mammary carcinomas suggesting that absence of the compound in potential target tissues is unlikely to account for lack of cancer inhibitory activity (Supplementary Table S2). Additionally, the effect of metformin compared to control was qualitatively in the wrong direction. Though existing literature has supported an indirect action by metformin through its activity in the liver, differences in plasma analytes, associated with hepatic metabolism and thought to play a role in breast carcinogenesis, were not detected in metformin versus the control group (Table 2).
Studies of mechanism
The understanding of how biguanides exert protective activity against cancer in vivo is limited. One question of interest is whether anticancer activity is driven indirectly through effects on host systemic factors that impact cellular activity in the breast or whether the effects are direct. In order to generate further hypotheses about this question for investigation in future experiments, we used multivariate analysis of plasma analytes (Fig. 2) and mammary carcinoma Western blot data (Fig. 3 and Fig. 4) to determine whether either dataset would distinguish among treatment groups. The hypothesis generating experiments on plasma analytes were insufficient to distinguish among treatment groups or cancer bearing versus cancer free animals (Fig. 2); whereas, the patterns of protein expression in carcinomas resulted in 100% classification accuracy by treatment group (Fig. 3).
The multivariate analyses that resulted in high classification accuracy were based on the evaluation of the effects of biguanide treatment on cell signaling events in mammary carcinomas. The measured proteins represent multiple mechanisms proposed to explain the anti-diabetic effects of biguanides. In terms of anticancer activity, these proteins are associated with energy sensing pathways, cell proliferation, and cell death. Western blot data (Supplementary Fig. S1) were subject to unsupervised and supervised multivariate clustering analyses. This approach revealed that mammary carcinoma patterns of protein expression distinguished among treatment groups with the groups ordering in a manner consistent with treatment response (Fig. 4 dendrogram; buformin < phenformin < control < metformin). This finding is consistent with biguanides mediating direct effects in the target tissue.
Whether biguanides act indirectly, directly, or a combination, there is considerable discussion about the specific targets and cellular processes mediating biguanide effects. While some evidence indicates that biguanides act as weak mitochondrial poisons by reducing activity of complex I of the electron transport system, there is an alternative hypothesis that biguanides inhibit AMP deaminase (13, 14). In either case, intracellular concentrations of AMP would increase, activating AMP activated protein kinase and suppressing protein kinase A activity. The multivariate analysis of protein expression emphasized the negative regulation of mTOR activity via AMPK activation. This observation is supported by: a) elevated levels of activated ACC and RAPTOR which are direct targets of activated AMPK (Fig. 4 and Supplementary Fig. S1); and b) lower levels of the activated targets of mTOR, e.g., p70S6K. Further, changes in ACC activation, FASN, SREBP1, and SCD1 indicate alterations in intra-tumoral lipid synthesis may be occurring in carcinomas from buformin-treated rats. These findings, which are considered hypothesis generating, will permit further experiments to be designed to identify causal mechanisms.
Limitations
The studies carried out in this work used a non-diabetic animal model in which metformin had less anti-cancer activity than other biguanides. Though non-diabetic animal models are relevant to anticipating outcomes in ongoing clinical trials that exclude diabetics and individuals taking metformin, the biguanides buformin and phenformin are no longer used clinically due to risk for lactic acidosis. Though buformin and phenformin are unlikely to see clinical use, understanding biguanide drug targets in preclinical animal models will help develop new agents.
Human subjects are generally administered metformin once or twice a day; whereas, it was administered in the diet in this study. This could result in differences in tissue concentrations that might impact target protein activity. This underscores a critical need for more data on the pharmacokinetics and pharmacodynamics of metformin if its current indications are extended to cancer prevention at various organ sites.
Implications
A lack of effect for metformin in non-diabetic preclinical rodent models of breast cancer has been recently reported (37). These findings are confirmed and extended in this work by evaluation of additional biguanides. These preclinical studies raise many questions about the use of biguanides such as metformin in cancer prevention and control. In the context of having an impact in vivo, this study indicates that in non-diabetic individuals, the effects of an anti-diabetic therapeutic dose of metformin may have limited benefit. In contrast, other biguanides outperform metformin in the preclinical model. As the mechanisms that account for the observed protection are established, the goal will be to identify agents that render protection in the absence of dose limiting toxicities, e.g. lactic acidosis, and such agents would represent the next generation of compounds with biguanide-related anti-cancer activity.
Supplementary Material
Acknowledgments
Funding: Supported by PHS grant R01-CA52626 from the National Cancer Institute, awarded to Henry J. Thompson.
The authors thank Elizabeth Neil, Angela Neil, Shawna Matthews, and Yijun Wang for their excellent technical assistance.
Footnotes
Conflicts of Interest: The authors disclose no potential conflicts of interest.
References
- 1.Azvolinsky A. Repurposing to fight cancer: the metformin-prostate cancer connection. J Natl Cancer Inst. 2014;106:dju030. doi: 10.1093/jnci/dju030. [DOI] [PubMed] [Google Scholar]
- 2.Yue W, Yang CS, Dipaola RS, Tan XL. Repurposing of metformin and aspirin by targeting AMPK-mTOR and inflammation for pancreatic cancer prevention and treatment. Cancer Prev Res (Phila) 2014 doi: 10.1158/1940-6207.CAPR-13-0337. [DOI] [PubMed] [Google Scholar]
- 3.Quinn BJ, Kitagawa H, Memmott RM, Gills JJ, Dennis PA. Repositioning metformin for cancer prevention and treatment. Trends Endocrinol Metab. 2013;24:469–80. doi: 10.1016/j.tem.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Alimova IN, Liu B, Fan Z, Edgerton SM, Dillon T, Lind SE, et al. Metformin inhibits breast cancer cell growth, colony formation and induces cell cycle arrest in vitro. Cell Cycle. 2009;8:909–15. doi: 10.4161/cc.8.6.7933. [DOI] [PubMed] [Google Scholar]
- 5.Liu B, Fan Z, Edgerton SM, Deng XS, Alimova IN, Lind SE, et al. Metformin induces unique biological and molecular responses in triple negative breast cancer cells. Cell Cycle. 2009;8:2031–40. doi: 10.4161/cc.8.13.8814. [DOI] [PubMed] [Google Scholar]
- 6.Thompson MD, Thompson HJ. A systems pharmacokinetic and pharmacodynamic approach to identify opportunities and pitfalls in energy stress-mediated chemoprevention: the use of metformin and other biguanides. Curr Drug Targets. 2012;13:1876–84. doi: 10.2174/138945012804545614. [DOI] [PubMed] [Google Scholar]
- 7.Anisimov VN, Egormin PA, Piskunova TS, Popovich IG, Tyndyk ML, Yurova MN, et al. Metformin extends life span of HER-2/neu transgenic mice and in combination with melatonin inhibits growth of transplantable tumors in vivo. Cell Cycle. 2010;9:188–97. doi: 10.4161/cc.9.1.10407. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Z, Jiang W, Thompson MD, McGinley JN, Thompson HJ. Metformin as an energy restriction mimetic agent for breast cancer prevention. J Carcinog. 2011;10:17. doi: 10.4103/1477-3163.83043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollak M. Metformin and other biguanides in oncology: advancing the research agenda. Cancer Prev Res (Phila) 2010;3:1060–5. doi: 10.1158/1940-6207.CAPR-10-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollak MN. Investigating metformin for cancer prevention and treatment: the end of the beginning. Cancer Discov. 2012;2:778–90. doi: 10.1158/2159-8290.CD-12-0263. [DOI] [PubMed] [Google Scholar]
- 11.Miller RA, Chu Q, Xie J, Foretz M, Viollet B, Birnbaum MJ. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature. 2013;494:256–60. doi: 10.1038/nature11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foretz M, Hebrard S, Leclerc J, Zarrinpashneh E, Soty M, Mithieux G, et al. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J Clin Invest. 2010;120:2355–69. doi: 10.1172/JCI40671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouyang J, Parakhia RA, Ochs RS. Metformin activates AMP kinase through inhibition of AMP deaminase. J Biol Chem. 2011;286:1–11. doi: 10.1074/jbc.M110.121806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vytla VS, Ochs RS. Metformin increases mitochondrial energy formation in L6 muscle cell cultures. J Biol Chem. 2013;288:20369–77. doi: 10.1074/jbc.M113.482646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanaspa MA, Cicerchi C, Garcia G, Li N, Roncal-Jimenez CA, Rivard CJ, et al. Counteracting roles of AMP deaminase and AMP kinase in the development of fatty liver. PLoS One. 2012;7:e48801. doi: 10.1371/journal.pone.0048801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anisimov VN. Effect of buformin on the life span, estrous cycle, and spontaneous tumors. Vop Onkol. 1980;26:42–8. [PubMed] [Google Scholar]
- 17.Anisimov VN, Dilman VM. Inhibitory effect of phenformin in the development of mammary tumors induced by N-nitrosomethylurea. Vop Onkol. 1980;26:58. [Google Scholar]
- 18.Reeves PG. Components of the AIN-93 diets as improvements in the AIN-76A diet. J Nutr. 1997;127:838S–41S. doi: 10.1093/jn/127.5.838S. [DOI] [PubMed] [Google Scholar]
- 19.Reeves PG, Nielsen FH, Fahey GC., Jr AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr. 1993;123:1939–51. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- 20.Reeves PG, Rossow KL, Lindlauf J. Development and testing of the AIN-93 purified diets for rodents: results on growth, kidney calcification and bone mineralization in rats and mice. J Nutr. 1993;123:1923–31. doi: 10.1093/jn/123.11.1923. [DOI] [PubMed] [Google Scholar]
- 21.Rao CV, Steele VE, Swamy MV, Patlolla JM, Guruswamy S, Kopelovich L. Inhibition of azoxymethane-induced colorectal cancer by CP-31398, a TP53 modulator, alone or in combination with low doses of celecoxib in male F344 rats. Cancer Res. 2009;69:8175–82. doi: 10.1158/0008-5472.CAN-09-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beckmann R, Lintz W, Schmidt-Bothelt E. Evaluation of a sustained release form of the oral antidiabetic butylbiguanide (Silubin-retard) European journal of clinical pharmacology. 1971;3:221–8. doi: 10.1007/BF00565010. [DOI] [PubMed] [Google Scholar]
- 23.Hunt JA, Catellier C, Dupre J, Gardiner RJ, McKendry JB, Toews CJ, et al. The use of phenformin and metformin. Can Med Assoc J. 1977;117:429–30. [PMC free article] [PubMed] [Google Scholar]
- 24.McKendry JB, KUWAYTI K, RADO PP. Clinical experience with DBI (phenformin) in the management of diabetes. Can Med Assoc J. 1959;80:773–8. [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson HJ, McGinley JN, Rothhammer K, Singh M. Rapid induction of mammary intraductal proliferations, ductal carcinoma in situ and carcinomas by the injection of sexually immature female rats with 1-methyl-1-nitrosourea. Carcinogenesis. 1995;16:2407–11. doi: 10.1093/carcin/16.10.2407. [DOI] [PubMed] [Google Scholar]
- 26.Singh M, McGinley JN, Thompson HJ. A comparison of the histopathology of premalignant and malignant mammary gland lesions induced in sexually immature rats with those occurring in the human. Lab Invest. 2000;80:221–31. doi: 10.1038/labinvest.3780025. [DOI] [PubMed] [Google Scholar]
- 27.Thompson HJ, Singh M, McGinley J. Classification of premalignant and malignant lesions developing in the rat mammary gland after injection of sexually immature rats with 1-methyl-1-nitrosourea. J Mammary Gland Biol Neoplasia. 2000;5:201–10. doi: 10.1023/a:1026495322596. [DOI] [PubMed] [Google Scholar]
- 28.Sokal RR, Rohlf FJ. Biometry the principles and practice of statistics in biological research. 3. New York: W.H. Freeman; 1995. [Google Scholar]
- 29.Conover WJ, Iman RL. Rank Transformations as a Bridge between Parametric and Nonparametric Statistics. The American Statistician. 1981;35:124–9. [Google Scholar]
- 30.Trygg J, Wold S. Orthogonal projections to latent structures (OPLS) Journal of Chemometrics. 2002;16:119–28. [Google Scholar]
- 31.Wiklund S. Multivariate Data Analysis and Modeling in “omics”. Umetrics; Umea, Sweden: 2008. [Google Scholar]
- 32.Wiklund S, Johansson E, Sjostrom L, Mellerowicz EJ, Edlund U, Shockcor JP, et al. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem. 2008;80:115–22. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- 33.User’s Guide to Simca-P, Simca-P+ 11. Umea, Sweden: Umetrics AB; 2005. [Google Scholar]
- 34.Morrison DF. Multivariate Statistical Methods. 3. New York: McGraw-Hill Publishing Co; 1990. [Google Scholar]
- 35.Gabrielsson J, Jonsson H, Airiau C, Schmidt B, Escott R, Trygg J. OPLS methodology for analysis of pre-processing effects on spectroscopic data. Chemometrics and Intelligent Laboratory Systems. 2006;84:153–8. [Google Scholar]
- 36.Tushar Roy SS, Pratihar DK. Determination of optimal clusters using a genetic algorithm. In: Taniar D, editor. Data mining and knowledge discovery technologies. Hershey, PA: IGI Publishing; 2008. pp. 98–117. [Google Scholar]
- 37.Thompson MD, Grubbs CJ, Bode AM, Reid JM, McGovern R, Bernard PS, et al. Lack of effect of metformin on mammary carcinogenesis in nondiabetic rat and mouse models. Cancer Prevention Research. 2015 doi: 10.1158/1940-6207.CAPR-14-0181-T. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.