Abstract
Current methodologies used for mitochondrial motility analysis tend to either overlook individual mitochondrial tracks or analyze only peripheral mitochondria instead of mitochondria in all regions of the cell. Furthermore, motility analysis of an individual mitochondrion is usually quantified by establishing an arbitrary threshold for “directed” motion. In this work, we created a custom, publicly available computational algorithm based on a previously published approach (Giedt et al., 2012) in order to characterize the distribution of mitochondrial movements at the whole-cell level, while still preserving information about single mitochondria. Our technique is easy to use, robust and computationally inexpensive. Images are first pre-processed for increased resolution, and then individual mitochondria are tracked based on object connectivity in space and time. When our method is applied to microscopy fields encompassing entire cells, we reveal that the mitochondrial net distances in fibroblasts follow a lognormal distribution within a given cell or group of cells. The ability to model whole-cell mitochondrial motility as a lognormal distribution provides a new quantitative paradigm by which to compare mitochondrial motility in naïve and treated cells. We further demonstrate that microtubule and microfilament depolymerization shift the lognormal distribution in directions which indicate decreased and increased mitochondrial movement, respectively. These findings advance earlier work on neuronal axons (Morris and Hollenbeck, 1993) by relating them to a different cell type, applying them on a global scale, and automating measurement of mitochondrial motility in general.
Keywords: cytoskeleton, microfilaments, microtubules, mitochondrial motility, single particle tracking
Introduction
Mitochondrial motility has become a subject of increasing study in recent years. This focus was borne out of the observation that mitochondria localize to subcellular areas of high metabolic activity, including the growth cones of neuronal axons (Morris and Hollenbeck, 1993), virus assembly sites in cells infected with African swine fever virus (Rojo et al., 1998), uropods in migrating lymphocytes (Campello et al., 2006) and the leading edge of migrating cancer epithelial cells (Desai et al., 2013). Such localization implies that mitochondria in healthy cells exhibit characteristic motility which may be impaired in diseased cells. Accordingly, many studies now focus on analysis of mitochondrial motility using live cell imaging, attempting to elucidate the physiological (Morris and Hollenbeck, 1995; Yi et al., 2004) and pathological (Du et al., 2010) factors which affect it.
Different strategies have been employed in order to track mitochondrial movement, with a vast majority of studies investigating mitochondrial transport in neuronal axons. Neuronal mitochondria are individually well-resolved and also migrate towards the growth cone, so this system is by nature uniquely suitable for particle tracking. De Vos and Sheetz (De Vos and Sheetz, 2007) describe two methods to quantify mitochondrial motility in more generalized cell types. In the first, the non-overlapping regions of two successive images of a cell with fluorescently stained mitochondria are measured as a whole-cell index of mitochondrial motility, and is used by the Hajnóczky group (Yi et al., 2004), among others. Rintoul et al. take a similar approach to subcellular analysis of mitochondrial movement by measuring changes in fluorescent intensity of individual pixels (Rintoul et al., 2003). While this general approach might consider the whole cell, the biological interpretation of the motility index is unclear. Additionally, results can be drastically skewed by movement of the cell substrate or the cell itself, and information about individual mitochondrial tracks is lost. De Vos and Sheetz also discuss the use of a kymograph, which analyzes space vs. time images for individual mitochondria. Kymographs are used by some (Du et al., 2010), while others have analogously employed the NIH image analysis software ImageJ to analyze individual or groups of mitochondria within cells (e.g. (Chen et al., 2012)). Several groups have employed advanced image processing strategies in order to track groups of mitochondria (e.g. (Gerencser and Nicholls, 2008)) but these strategies remain limited to subcellular areas.
Despite the intense research focus on mitochondrial motility, analysis of individual mitochondrial tracks on the whole-cell level remains elusive. Kymographs are ill-suited for large scale analysis of mitochondrial motility, since they only consider one-dimensional movement and do not represent an automated method for motility detection. A more appropriate approach would be along the lines of Giedt et al. (Giedt et al., 2012) who first pre-processed time-lapse images and then automatically tracked mitochondria using object connectivity in space and time. Their technique is simpler in concept than most single particle tracking (SPT) algorithms, and is well-suited to non-spherical particles which undergo fusion and fission. However, like many others, Giedt et al. only utilized this strategy to analyze groups of mitochondria in subcellular regions instead of at the whole-cell level. We provide a major advancement in mitochondrial motility analysis by characterizing motion throughout the entire cell. In constructing our algorithm, we build on Geidt et al.'s basic approach but implement multiple changes to the overall methodology, technical processing and conditions for data inclusion. The resulting “whole-cell” analysis we describe refers to a two-dimensional slice of the cell as captured with our fluorescence microscopy setup, which optimizes visualization of most mitochondria in the cell. In addition to eliminating bias potentially introduced by only studying peripheral mitochondria, our technique critically allowed us to establish a whole-cell depiction of mitochondrial motility. This in turn enables the distinction between baseline motility in healthy cells and motility in cells characterized by acute or chronic dysfunction. We additionally made our algorithm publicly available and accessible with a user-friendly graphical interface.
Ultimately, our goal was to use this method to assess the effects of microtubule and microfilament depolymerization on mitochondrial motility in fibroblasts on the whole-cell level. Studies point to the structural association between mitochondria and the cytoskeleton (Boldogh and Pon, 2006; Heggeness et al., 1978). This connection may extend to a functional one, as a higher concentration or greater polarization of mitochondria is often observed in cellular regions associated with mechanosensing (Campello et al., 2006). In the pathological sense, many diseases are characterized by both mechanical and mitochondrial dysfunction. Cancer cells, for instance, abnormally rely on glycolysis for ATP production (Warburg, 1925), and also exhibit decreased elastic moduli and traction force generation as compared to normal cells (Munevar et al., 2001; Paszek et al., 2005). While others have investigated the effects of cytoskeletal impairment on mitochondrial motility in neurons (Morris and Hollenbeck, 1995) and other limited settings, studying this connection on the whole-cell level in generalized cell types will shed light on the general integration between mitochondrial health and cell mechanics.
Materials and Methods
Cell Culture and Reagents
Adult human dermal fibroblasts (Lifeline Cell Technology, Walkersville, MD) were cultured in FibroLife cell culture media (Lifeline Cell Technology) as previously described (Kandel et al., 2014). MatTek 35-mm glass-bottom dishes (MatTek, Ashland, MA) were coated for 30-40 minutes with 5 μg/mL fibronectin (BD Biosciences, San Jose, CA) dissolved in PBS, and then plated with cells between passages 1 and 5 at a density of approximately ∼25,000 cells/plate. The day before experiments, cells were transfected with CellLight Mitochondria-GFP, BacMam 2.0 (Life Technologies, Grand Island, NY) at a concentration of 40 particles per cell, and kept in the dark at 37°C. Several hours before experiments, some cells were also dye-loaded with 20 nM Tetramethylrhodamine, methyl ester (TMRM, Life Technologies), a mitochondrial potential dye. Cells were placed in Recording HBSS (HBSS pH 7.4 with 1.3 mmol/L CaCl2, 0.9 mmol/L MgCl2, 2 mmol/L glutamine, 0.1 g/L heparin, 5.6 mmol/L glucose, and 1% FBS) for imaging. Changes in mitochondrial potential are not related in this work.
Immediately before imaging, cells were incubated for 30 minutes in either replacement Recording HBSS, cytochalasin D (cytD; Sigma Aldrich, St. Louis, MO; 200 nM, 1 μM, or 5 μM, taken from a 1 mM stock dissolved in DMSO and diluted using Recording HBSS), nocodazole (noco; Sigma; 2 μM, 10 μM, or 50 μM, taken from a 10 mM stock dissolved in DMSO), both 50 μM noco and 5 μM cytD, or carbonyl cyanide 3-chlorophenylhydrazone (CCCP; Sigma; 5 μM, taken from a 10 μM stock dissolved in DMSO). Data shown from each group represent mitochondria pooled from 5 or more cells over multiple days.
The cytoskeletal inhibitors were confirmed to be working in preliminary experiments as follows: cells were incubated in a given concentration of noco (for microtubule depolymerization) or a given concentration of cytD (for microfilament depolymerization) for 30 minutes. Noco-treated cells were then incubated in 250 nM TubulinTracker Green (Life Technologies) for 30 minutes at 37°C, then rinsed 3 times and placed in recording HBSS for fluorescence imaging with a standard TRITC filter. Fig. S3 in the supplementary data shows that increasing concentrations of noco result in progressive breakdown of microtubules. CytD-treated cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA; diluted with HBSS from a 16% stock solution) for 10 minutes, then permeabilized for 3 minutes with 0.1% Triton, followed by 20 minutes of dye-loading with 100 nM Acti-stain 488 phalloidin (Cytoskeleton, Inc, Denver, CO) taken from a 14 μM stock prepared according to the manufacturer's instructions. Actin fiber visualization using a standard FITC filter showed that in a similar manner to noco, the concentrations used of cytD affected the actin cytoskeleton in accordance with the level of concentration used (Fig. S1).
CCCP was confirmed to be working by dyeing CellLight Mitochondria GFP transfected cells with 20 nM tetramethylrhodamine methyl ester perchlorate (TMRM, Sigma), a mitochondrial inner membrane potential dye. Incubation was performed for 1 hour at room temperature in the dark. Cells were illuminated in both the FITC and TRITC channels before treatment with CCCP, and the two dyes showed nearly perfect overlap for a successfully transfected cell. Imaging cells ten minutes after 5 μM CCCP was added showed that the TMRM dye had left the mitochondria and leaked into the cytoplasm (data not shown), indicating inner membrane depolarization.
Fluorescence Microscopy
Fluorescence microscopy was performed similarly to our previous methods (Sobolewski et al., 2011). Cells were imaged using a QImaging QIClick camera (QImaging, Surrey, BC, Canada) (1×1 binning, 1392×1040 pixels) attached to Olympus IX70 microscope (Olympus, Melville, NY) with an Olympus 40x oil immersion objective lens (Olympus) and Photofluor light source (89 North, Burlington, VT). Computer control of the microscope was facilitated by LUDL programmable filter wheels, shutters, and focus control (LUDL Electronic Products, Hawthorne, NY) and images were collected using IPL 3.7 software (BD, Rockville, MD). For each experiment, cells were visualized using a standard FITC filter, and isolated cells with well-resolved mitochondria were selected. Cells were imaged for 5-10 minutes every 1.5 or 3 seconds, eventually to be sampled every 3 seconds for a total of 5 minutes.
ImageJ Image Pre-Processing
We preprocessed raw image files in ImageJ using an approach previously presented (Giedt et al., 2012)Briefly, time-lapse images were first convolved using the 5×5 edge-detection filter given in (Giedt et al., 2012). Next, images were converted to the frequency domain using a Fast Fourier Transform (FFT) in ImageJ, and subjected to a bandpass filter ranging from 2 pixels (∼0.3 μm with our resolution) to 100 pixels (∼16 μm). Finally, the resulting images were thresholded by eye to best eliminate noise, with the results saved as a sequence of individual binary images. Fig. 1 shows the evolution of a single frame through the ImageJ processing steps. Video S1 shows a raw image sequence of a control cell and Video S2 shows the same video after pre-processing in ImageJ.
Figure 1. Image processing of a sample cell image.
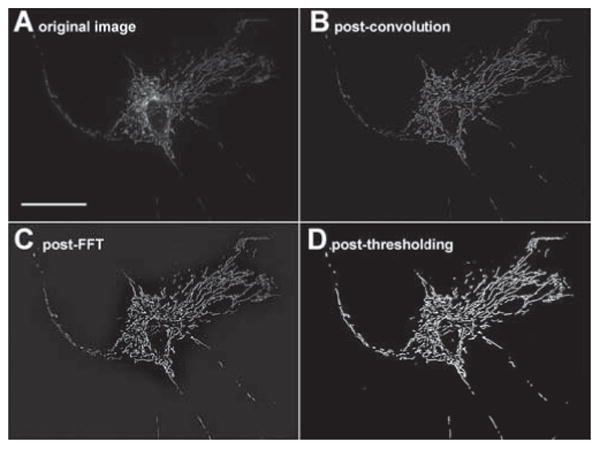
A Original image. Scale bar is 50 μm and scale remains the same across all four images.
B The same image after convolution.
C The same image after FFT.
D The final image after thresholding.
Matlab Algorithm
Stacks of images were analyzed by a custom Matlab script designed to track the movement of individual mitochondria. Fig. 2A displays a flowchart describing the basic structure of the program, and Fig. 2B shows how the algorithm treats a 4-frame sequence of sample data. The scripts and instructions of our algorithm are included online on Github (www.github.com/kandelj/MitoSPT), a freely available software sharing repository, and are intended for public use with proper citation. The Matlab processing is done via a user-friendly interface with inputs such file names, image speed, image resolution, and desired lifetime minimum, and is usually completed in a few minutes. Data can be viewed in the interface in the formats shown in Fig. 3A and Fig. 4A.
Figure 2. Matlab based algorithm tracks centroid locations of all objects as they travel throughout a time-lapse recording.
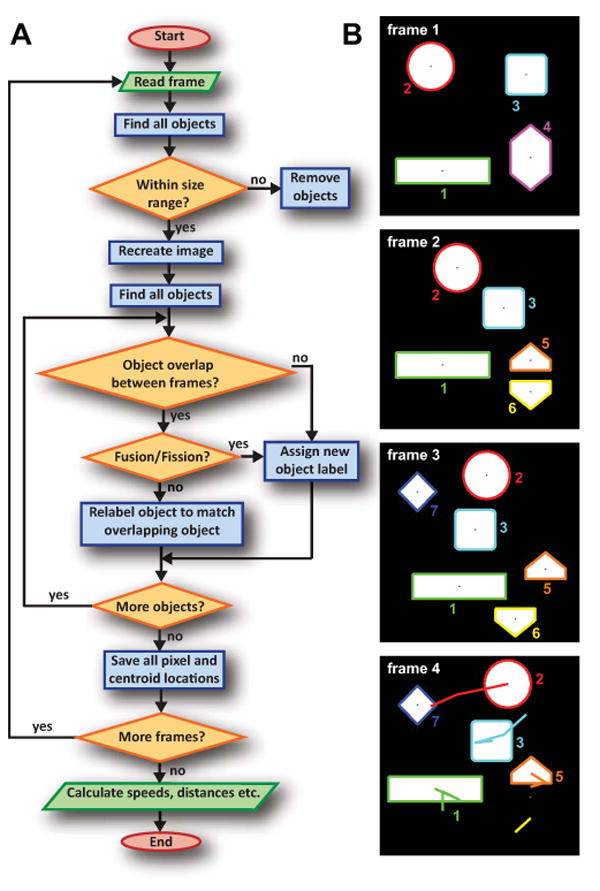
A Flowchart of algorithm used to calculate net distances traveled by mitochondria over the course of a 5 minute video.
B Illustration of the algorithm's results for artificial sample data. A comparison of frame 1 to frame 2 shows that object 4 has split into objects 5 and 6, necessitating new labels. In addition, all other objects have moved. Object 7 newly appears in frame 3, and object 6 has disappeared in frame 4. The centroid paths from the first frame are traced in the final frame. The path shown below object 5 is from object 6 (sample data lifetime is not limited to 4 frames as true data are).
Figure 3. Net distances traveled by mitochondria are lognormally distributed.
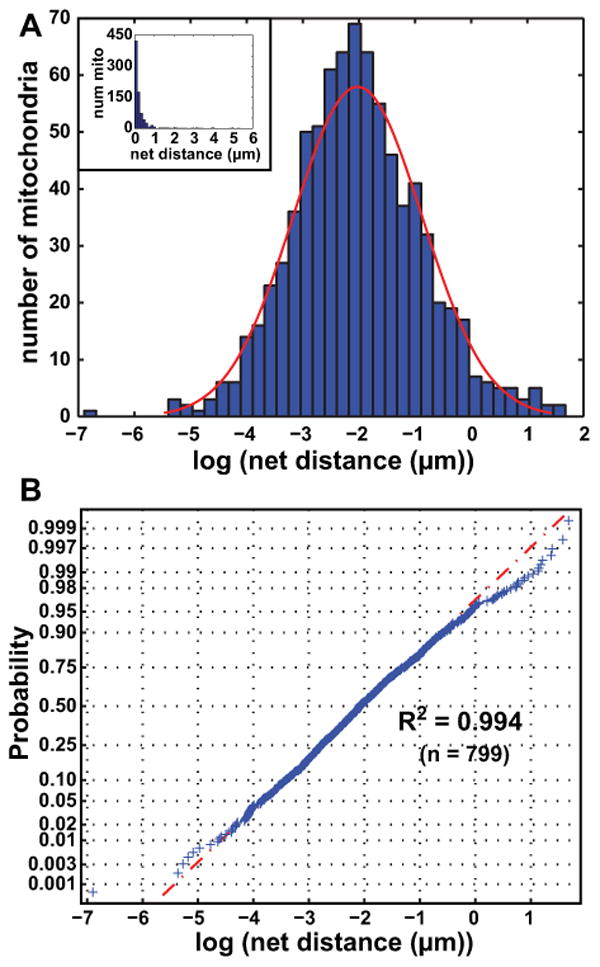
A Histogram of log values of net distances traveled by all mitochondrial objects satisfying our size and lifetime inclusion criteria in a given control cell over the video. A normal distribution (red) has been overlaid onto the histogram, with a mean of -2.023 (=135.3 nm) and standard deviation of 1.150. Inset: histogram of net distances traveled by mitochondria without the log transformation.
B Normal probability plot of the logs of net mitochondrial distances traveled. The data give an R2 of 0.994 when fit to a lognormal distribution.
Figure 4. CCCP and nocodazole lower the distribution of net distances traveled by mitochondria while cytochalasin D raises it.
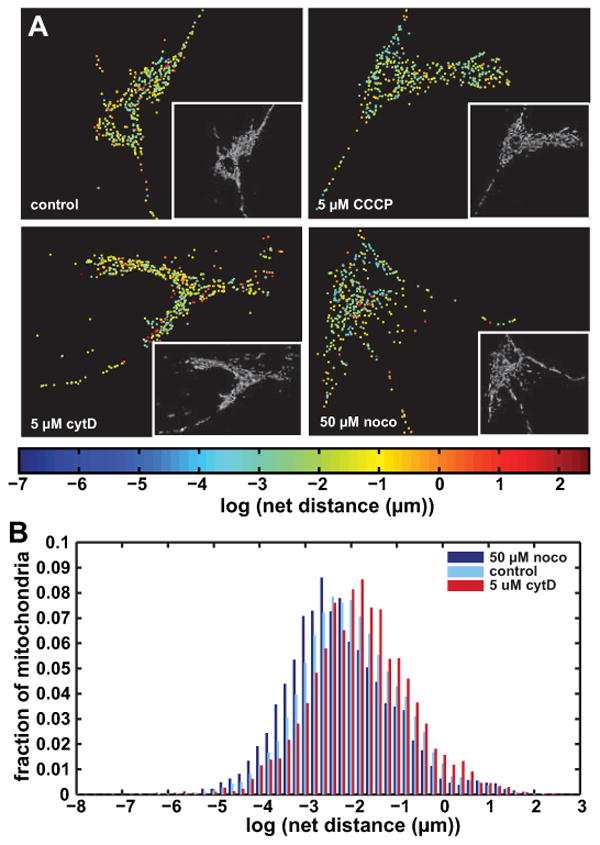
A Images of representative cells from the control, 5 μM CCCP, 5 μM cytD and 50 μM noco groups. Each dot represents a mitochondrial object in its final location after tracking, and is colored by the log of the net distance traveled. Inset into each figure is the first pre-processed binary image from the corresponding video.
B Grouped histograms of net distances traveled by mitochondria in control, 50 μM noco and 5 μM cytD groups. Mitochondria are pooled together from 5-17 cells per group, with a total mitochondrial number of 4323 (cytD), 5530 (noco) and 19334 (control). HIstograms are normalized to reflect percentages of objects rather than absolute numbers.
Here, we relate a brief conceptual outline of the program. First, the algorithm reads the current frame into Matlab, and the built-in function bwconncomp is used to find the connected white objects in the image (using 8-point connectivity rather than Giedt's 4-point connectivity). The Matlab function regionprops is used to find object sizes so the image can be recreated to contain only those objects whose area lies within specified limits. Bwconncomp is used to automatically number these objects and save their pixel locations, while regionprops finds the objects' centroid locations. Beginning with the second frame of the program, the object pixel locations in the current frame (“frame n+1”) are compared with those of the previous frame (“frame n”), yielding four possibilities for each object:
Simple overlap between a single object from frame n+1 and a single object from frame n. We conclude that the two objects are identical, and relabel the object in frame n+1 to match the corresponding object in frame n.
Fusion: An object from frame n+1 overlaps with multiple objects from frame n. We assume a fusion event has occurred, and we permanently retire the labels of the corresponding objects in frame n. The fused object in frame n+1 gets a new, unique label. Our approach here differs from that of Giedt et al., which identically labels mitochondrial objects which fuse at some point.
Fission: Several objects from frame n+1 overlap with one object in frame n. We infer that a fission event has occurred. As in the case of fusion, we introduce a new label for this object and retire the old label.
New object: an object appears which exhibits no overlap with any other object. This may occur e.g. in the case of an object which is hovering around our assigned object area constraints, and suddenly falls into the specified range when it did not previously. We also use new unique labels for such objects.
Once all objects are relabeled so that they are trackable, the new object pixel locations are stored so that the program can then compare the next frame, n+2, to frame n+1. After the program has cycled through all frames, the stored centroid locations are used to calculate distances traveled by each object at each interval, as well as net distances traveled by each object using the simple distance formula:
| (1) |
where dnet is the net distance traveled by the object, xlast and ylast are the x- and y-positions, respectively, of the centroid in the last frame in which the object appears, and x0 and y0 are the x- and y-positions of the centroid in the first frame in which the object appears.
In order to maximize our signal-to-noise ratio (SNR), we have subjected our algorithm to several restrictions. First, we limit object size to be between 20 and 500 pixels (∼0.5 μm2 to ∼13 μm2 with our microscope's resolution). We interpret smaller objects as noise. Larger objects represent extremely integrated mitochondrial networks whose movement will be difficult to track, and generally represent <2% of objects from a given cell (see Fig. S2A). We also established a minimum lifetime of 4 or more consecutive frames (≥ 9 seconds) per object (similar to (Giedt et al., 2012) and (Jaqaman et al., 2008) in at least one application) in order to increase our SNR, assuming that more temporary objects are often due to imperfect image processing. For example, a fused object which immediately undergoes fission probably never fused in the first place, and including its track in our analysis may result in underestimated values of net distances. A histogram showing object lifetimes for an individual cell (Fig. S2B) demonstrates that a majority of objects last very temporarily due to fusion, fission, and low SNR resulting from limited resolution. The lifetime cutoff definitely affects the net distance distribution, as shorter net distances are generally associated with shorter lifetimes (see Fig. S2C). However, avoiding setting a lifetime threshold likely decreases the SNR. We have chosen our sampling frequency to be every 3 seconds, observing that more frequent sampling increases noise, but less frequent sampling increases our probability of missing true signal.
Statistics
For computing the coefficient of determination (R2) of the probability plots, a probability plot was first made by applying the Matlab normplot function to the log values of the net distances traveled by mitochondria in a given cell or cells. X and y values were extracted from the plot, with each data point represented as (xi,yi). The backslash operator was used in Matlab to fit a line to this data, giving expected y values (fi) based on this model. The total sum of squares (SStot) was calculated as:
| (2) |
The residual sum of squares (SSresid) was calculated as:
| (3) |
R2 was calculated as:
| (4) |
The Matlab function normfit gave the mean and standard deviation of the normal distribution best fitting the distribution of logs of netdistances. P-values comparing log distributions were computed using two-sample Kolmogorov-Smirnov tests (K-S test; kstest2 function in Matlab). This statistical method tests the null hypothesis that two sets of data are taken from the same continuous distribution. Since acceptance or rejection of the null hypothesis based on p-value is by definition affected by dataset sizes, we merely report the p-values in supplemental data instead of establishing a threshold for significance when comparing our large datasets.
Results
Algorithm
A description of the image pre-processing performed prior to the particle tracking algorithm is given in the Materials and Methods section and is illustrated in Fig. 1. In addition, Video S1 (sped up 20x, such that 1 second of video equals 1 minute in real time) displays an unprocessed video of a control cell, with Video S2 showing the corresponding processed video. Fig. 2A displays a flowchart representing the algorithm, and Fig. 2B shows how the algorithm treats a sample 4-frame sequence of data. The algorithm is discussed in greater detail in the Materials and Methods section.
Net distances traveled by mitochondria follow a lognormal distribution
After applying our method to several cells, we performed a log-transformation on the net distances traveled by mitochondria. We found that on a cell-by-cell basis, the data appear to follow a continuous lognormal distribution. Fig. 3A shows a histogram of log values of the net distances traveled by all objects in a given control cell over the 5 minutes recorded, with the inset graph showing untransformed data. A normal distribution with a mean of -2.023 (= 132.3 nm) and standard deviation of 1.15 is overlaid onto the log-transformed histogram. The mean value corresponds to the geometric mean of the original distribution (Limpert et al., 2001); thus, means of data groups are henceforth reported as geometric means of original data. Since the standard deviations of all groups in this study were similar and exhibited no obvious trends (see Table I), we focus on the geometric means of various groups as their defining parameter. Limpert et al. (Limpert et al., 2001) gives a more thorough description regarding the spatial interpretation of standard deviations in lognormal distributions. In general, we found that a vast majority of mitochondria traveled very small net distances (< 1 μm), which is consistent with what has been previously reported in the literature (Giedt et al., 2012). Additionally, the net distances traveled by individual mitochondria had no obvious connection to their subcellular spatial location. This is shown in Fig. 4A, which displays an image of a representative control cell at the upper left. Mitochondrial objects are positioned in their final locations, with each mitochondrion being color coded by the log value of its net distances traveled. The lack of dependence of mitochondrial motility on subcellular spatial location persisted when cells were subjected to various mitochondrial and cytoskeletal toxins (presented below). It is possible that this absence of correlation occurs only in resting cells, as it is likely that cells which are metabolically stimulated in some spatially asymmetrical manner will exhibit spatially dependent mitochondrial motility.
Table I. Parameters of all groups and p-values resulting from pairwise K-S tests.
| control | 5 μM CCCP | 200 nM cytD | 1 μM cytD | 5 μM cytD | 2 μM noco | 10 μM noco | 50 μM noco | cytD + noco | n | geo mean | stdev | R2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| control | x | -91.86 | -2.17 | -1.70 | -71.49 | -82.95 | -118.71 | -116.95 | -4.77 | 19334 | 135.53 | 1.11 | 0.994 |
| 5 μM CCCP | x | x | -77.03 | -67.53 | -199.83 | -0.31 | -2.08 | -3.89 | -40.00 | 5522 | 106.92 | 1.10 | 0.992 |
| 200 nM cytD | x | x | x | -0.70 | -43.39 | -75.58 | -95.79 | -95.42 | -8.38 | 6441 | 141.06 | 1.15 | 0.993 |
| 1 μM cytD | x | x | x | x | -42.11 | -67.28 | -86.27 | -87.79 | -6.34 | 5653 | 141.57 | 1.10 | 0.995 |
| 5 μM cytD | x | x | x | x | x | -190.93 | -228.71 | -216.87 | -70.01 | 4323 | 167.91 | 1.08 | 0.997 |
| 2 μM noco | x | x | x | x | x | x | -3.95 | -2.69 | -45.20 | 6188 | 108.27 | 1.11 | 0.991 |
| 10 μM noco | x | x | x | x | x | x | x | -4.38 | -55.40 | 5996 | 102.86 | 1.05 | 0.993 |
| 50 μM noco | x | x | x | x | x | x | x | x | -67.85 | 5530 | 105.54 | 1.15 | 0.988 |
| cytD + noco | x | x | x | x | x | x | x | x | x | 5113 | 134.47 | 1.12 | 0.987 |
P-value cells are color coded by log value, with red cells indicating most significant, and blue cells indicating least significant. Additional columns give number of objects per group (n), the geometric mean, the standard deviation (in log scale), and the coefficient of determination to the normal probability plot for each group.
We performed a Shapiro-Wilk (S-W) test (using SigmaPlot v11.0) in order to test the goodness of fit of the motility data for a given group to a lognormal distribution, The resulting W-statistics were very high, often above 0.99, but large datasets led the p-values to fall below the standard significance threshold of 0.05. We instead opted to visually assess whether the data followed a normal distribution using normal probability plots. Fig. 3B shows the probability plot corresponding to the cell shown in Fig. 3A. The calculated R2 is 0.994, demonstrating an excellent fit. In general, after converting net distances to the log scale, R2 values usually exceeded 0.98 for individual cells and groups of cells. We thus conclude that net distances traveled by mitochondria follow a lognormal distribution and should be quantified as such on the whole-cell level.
While different cells within each group displayed individual geometric means, combining them into a group maintained the lognormal shape of the distribution. Fig. S3 shows an example of this, with the probability plot of the entire group (10 μM noco) giving an R2 value of 0.993. Despite the high R2 values, we opted to use the non-parametric K-S test instead of the Student's t-test (which assumes a normal distribution) when comparing the net distances traveled by mitochondria in different groups, on account of the failed S-W tests. Overall, mitochondria in control cells traveled a geometric mean net distance of 135.5 nm during the 5-minute recording period.
Cyanide decreases mitochondrial motility
For our first experiment comparing mitochondrial motility in normal and diseased cells, we used 5 μM CCCP in order to verify that this uncoupler decreases mitochondrial motility, as shown by others (Giedt et al., 2012; Safiulina et al., 2006). The geometric mean of the mitochondrial net distances traveled by the treated cells was 106.9 nm, compared with 135.5 nm of all mitochondria in control cells. Fig. 4A compares the logs of the net distances in a representative control cell and CCCP-treated cell. The CCCP-treated cell appears bluer overall, illustrating the shift of its distribution towards decreased motility. Video S3 shows the pre-processed movie of this particular cell, and Fig. 5 plots the parameters of the control and CCCP distributions among other groups. Table I shows a matrix comparing logs of p-values across different groups using the two-sample K-S test, with cells color coded by the log of their p-value value to illustrate relative significance. Warmer colors indicate smaller, more significant p-values, and cooler colors represent larger, less significant ones. Absolute significance is highly affected by our large sample sizes, resulting in nearly any group comparison falling below the standard threshold of p = 0.05. As such, we present logs of p-values in order to highlight their relative, rather than absolute, significance, and colored them according to this scale. A comparison of the control group to the CCCP treated group gives a relatively low p-value, indicating a significant difference.
Figure 5. Box plots showing data from all groups.
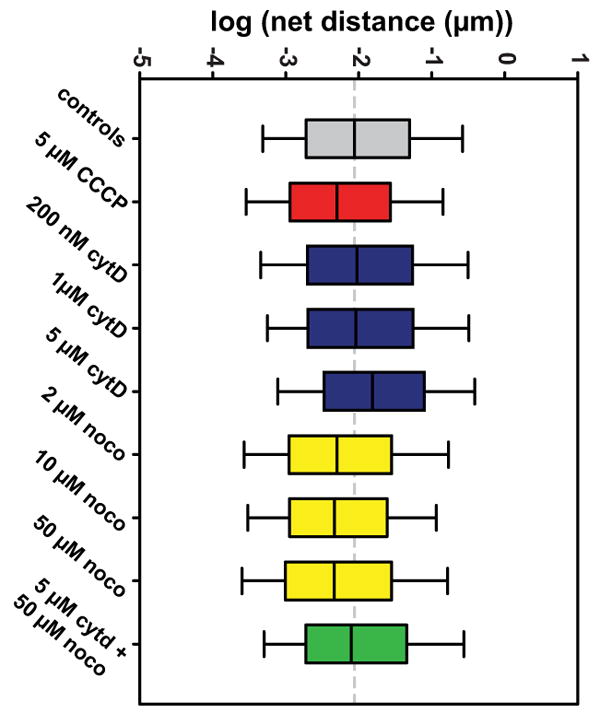
Whiskers extend to 10th and 90th percentiles of data. The median of the control group is extended throughout the plot as a dashed gray line.
Others have shown that high concentrations of CCCP can result in microtubule depolymerization (Maro et al., 1982), which itself impairs mitochondrial motility as shown next. However, we verified with fluorescence microscopy that at the concentration and treatment length used, microtubule structure was preserved (Fig. S4), and appears similar to that of the control cells shown in Fig S1. This suggests that the mitochondrial depolarization induced by CCCP primes them for motility dysfunction in a manner which is independent of microtubule structure, ultimately suggesting an underlying link between mitochondrial function and motility.
Microtubule depolymerization decreases net distance traveled by mitochondria, while microfilament depolymerization increases it
Cells treated with various concentrations of noco in order to depolymerize their microtubules exhibited decreased mitochondrial motility. 2 μM, 10 μM and 50 μM concentrations were used, resulting in geometric means of 108.3 nm, 102.9 nm, and 105.5 nm, respectively. The sample cell from the 50 μM group shown in Figure 4A appears overall more blue than the control cell, indicating shorter net distances traveled by the corresponding mitochondria. The pre-processed movie of this cell is given in Video S4. The group geometric means did not differ significantly among different noco concentrations, as plotted in Fig. 5 and shown in the p-value matrix in Table I. Thus, while these concentrations appear to affect the microtubule network in a somewhat dose-dependent manner (Fig. S1), the affect on mitochondrial transport does not seem to vary by dose and is exhibited with even partial disruption of the network.
As opposed to noco, cytD increased mitochondrial motility. For the 200 nM and 1 μM concentrations, this effect was hardly pronounced with geometric means of 141.1 and 141.6 nm, respectively. However, at 5 μM the lognormal distribution of net distances exhibited a geometric mean of 167.9 nm as compared to 135.5 nm in the control group. Like in the case of noco treatment, our results show that this effect is not dose-dependent. However, as opposed to the effects of noco on microtubule-mediated mitochondrial transport, only extreme disruption of the actin cytoskeleton affects mitochondrial motility. A sample cell from the 5 μM cytD group is shown in Fig. 4A, with the object colors tending towards yellow and orange values when compared to control cells. Video S5 shows the pre-processed movie of this cell. Fig. 4B shows overlaid histograms of the 5 μM cytD treated group, the control group, and the 50 μM noco group, with the effect of each drug on the motility distribution appearing quite evident.
Finally, we were interested in whether the noco or cytD effect on mitochondrial motility would dominate when both drugs were used. We treated cells for 30 minutes with 50 μM noco and 5 μM cytD. Resulting motility was virtually the same as control cells, with a geometric mean of 134.5 nm (see Fig. 5 and Table I). Thus, the effects of cytD and noco act in opposition to restore mitochondrial motility to its baseline level. Fig. 5 shows box plots of all groups studied and Table I shows the relative p-values resulting from the K-S tests comparing all groups.
Discussion
We have created a publicly available, easy-to-use method for mitochondrial tracking in whole cells based on fluorescent live imaging. The algorithm is formulated based on the approach described in (Giedt et al., 2012), and uses ImageJ for image pre-processing and Matlab for object recognition and tracking. We used our method in order to easily compare the whole-cell distribution of mitochondrial motility between untreated cells and cells subjected to mitochondrial and cytoskeletal toxins.
One major insight provided by our work is that whole-cell mitochondrial movement follows a lognormal distribution. Until now, it has become standard to categorize mitochondrial movement as either small-scale Brownian-like motion or brief spurts of directed motion (e.g. (Giedt et al., 2012; Nekrasova et al., 2011)), with one group even adding an intermediate category of small-scale directed motion (Saunter et al., 2009). We show, by contrast, that mitochondrial motility follows a continuous distribution, an observation which is also made by Gerencser and Nicholls (Gerencser and Nicholls, 2008). We provide further insight into their observation by showing that the distribution is of a particular type; namely, a lognormal distribution.
Our data slightly deviates from the lognormal distribution at extremely high and low net distances, indicating fatter tails than expected. This is especially pronounced at the upper extremes (see Fig. 3B and Fig. S3), which lends some support to the idea that certain mitochondria undergo bursts of directed motion. However, setting an arbitrary threshold for such bursts within a continuous distribution can make comparison between studies difficult. More problematically, the threshold value heavily affects the resulting analysis. For example, Fig. 4B shows that control, cytD and noco treated cells all have similar percentages of mitochondria which undergo net distance changes above ∼2.1 μm (∼0.75 on the x-axis), but a lower cutoff might accurately capture differences between these groups. In contrast to the arbitrary nature of mitochondrial motility categorization, quantification of net distances using a lognormal distribution, as is descriptive of many biological phenomena (Limpert et al., 2001), highlights the substantial shift between different groups even at a glance. Motor-driven events are particularly interesting in their ability to provide insight into mechanisms of mitochondrial translocation, but an accurate understanding of mitochondrial motility at the whole-cell level includes many small-scale movements which reflect mitochondrial drifting or even possibly fluctuations due to microscopy limitations. The lognormal distribution successfully encapsulates all types of mitochondrial movement occurring within a cell, and the pharmacological agents we used (noco, cytD, CCCP) affect the entire range of mitochondrial motility across the cell. Considering the geometric mean of the data rather than the arithmetic mean as is more commonly used enables the determination of a whole-cell index of motility which reflects true biological parameters emanating from individual mitochondria data. This can easily be used to compare different groups of cells subjected to a variety of experimental conditions.
Overall, our findings are consistent with previous research showing that microtubule depolymerization hinders mitochondrial motility in mammalian cells (Morris and Hollenbeck, 1995; Yi et al., 2004; Ligon and Steward, 2000; Müller et al., 2005). Our ability to demonstrate the same effect across entire microscopy fields encompassing whole cells lends support to our algorithm's ability to characterize mitochondrial motility, but further expands this finding to fibroblasts on the whole-cell level. Prior results on the role of actin filaments in this regard has been divided, with some studies reporting impaired mitochondrial motility resulting from microfilament depolymerization (Müller et al., 2005; Quintero et al., 2006), and others finding, like us, that it was enhanced (Ligon and Steward, 2000; Morris and Hollenbeck, 1995). One recent study even found that actin depolymerization enhanced mitochondrial motility in neuronal dendrites and axons, but had little effect on mitochondrial motility in the cell body (Narayanareddy et al., 2014). A general perspective on organelle motility presents microtubules and actin as tracks for long-range and short-range movement, respectively(Langford, 1995). With specific regard to mitochondria in eukaryotic cells, some investigators suggest that intermediate filaments impair mitochondrial motility (Nekrasova et al., 2011), while others have proposed that actin filaments are responsible for mitochondrial docking in neurons in particular (Chada and Hollenbeck, 2004). Our data showing that actin depolymerization increases mitochondrial transport supports the extension of this latter theory to fibroblasts. Frederick and Shaw (Frederick and Shaw, 2007) review mitochondrial motility in greater detail, including some discussion of the motor proteins used by mitochondria to travel along cytoskeletal tracks. Our finding that combined effective doses of noco and cytD restore baseline mitochondrial motility are at odds with others' results in neurons (Morris and Hollenbeck, 1995), and perhaps suggests that neither actin nor tubulin dominates mitochondrial motility in fibroblasts. Much remains to be elucidated regarding the mechanisms of interaction between mitochondria and the actin cytoskeleton in generalized cell types.
We believe that increased optical resolution can further improve our algorithm. Our current limits in microscopy resolution probably lead to increased variability in successive binary images, which may lead to some of the small-scale movements we observe. For example, a one-pixel fluctuation in a large object can lead to a centroid translocation of <10 nm, far less than the ∼160 nm length of a pixel in our system. It is impossible to tell whether these fluctuations originate in light detection limitations or truly reflect small-scale mitochondrial movement, and improved resolution might better allow for this differentiation. Greater resolution would also sharpen the appearance of the perinuclear mitochondria. In this work, we were careful to only select cells with well-resolved perinuclear mitochondria following the pre-processing steps. Our resulting analysis shows that these mitochondria exhibit motility on a scale similar to peripheral mitochondria (see Fig. 4A), suggesting that we are detecting true perinuclear mitochondrial motility rates. In the Materials and Methods section, we describe other strategies we used to curb other effects of limited resolution, including image pre-processing and imposing a minimum object lifetime.
As a final strategy to maximize SNR, objects resulting from fusion or fission are newly labeled, rather than having their tracks continue as in other studies (Giedt et al., 2012; Jaqaman et al., 2008). Continuing tracks rather than starting new ones in these cases can exaggerate the actual motion traveled by creating jumps in centroid location, especially if the fusion or fission is illusory. Whether this approach is logically justifiable in the case of true fusion and fission is open to debate. One reason to continue tracks of fused or split objects is that poor SNR can cause that illusory fusion, fission, or momentary object disappearance. Since shorter tracks of several such objects may in reality comprise the lengthier track of a single object, prematurely ending the recording of such objects may lead to underestimating their net distance traveled. This phenomenon was observed by Jaqaman et al. (Jaqaman et al., 2008) using a global optimization technique in order to fuse tracks of various subcellular particles over a time course video and thus reduce such error. While possible application of Jaqaman's technique may benefit our algorithm in the future, we are reluctant to apply it to mitochondria since fusion and fission can only be detected by connecting tracks. This will lead to jumps in centroid location as discussed above. We would prefer to develop a hybrid approach in which unique identities of mitochondria preceding and following fusion and fission are preserved, but possibly employs track linking to enable more accurate detection of true fusion and fission and disregards momentary disappearance events due to limited resolution. This approach will help us preserve as much data as possible about mitochondrial motility as well as fusion and fission rates.
Another limitation in this work is the possibility of confounding mitochondrial movement with cell movement, which is particularly of concern with the use of cytD. In this work, we were careful to discard any time-lapse images with significant cell movement, as frequently occurred in cytD-treated cells. Video S6 (which also exhibits poor resolution) shows an example of one such cell, with a corresponding color-coded plot of the cell shown in Fig. S5. Despite our meticulousness, it is possible that imperceptible cellular motility can still artificially increase the range of mitochondrial motility in a given cell. Future experiments may be able to account for this by introducing a cellular dye and then correcting for cell movements, in both magnitude and direction, at each time point. This correction may help elucidate whether actin in fact plays a role in mitochondrial docking, as this previously unmentioned confound may relate to conflicting data in this regard.
This foundational study prompts a variety of additional experiments relating to many different scientific areas of research. In addition to the computational improvements suggested above, some follow-up mechanistic studies may be interesting as well. One obvious question to explore is the mechanism by which microfilaments normally “impair” mitochondrial transport. Since actin filaments are principally involved in the well-studied field of cell mechanics (Janmey and McCulloch, 2007), we additionally intend to investigate general connections between cell mechanics and bioenergetics.
Conclusions
Despite a plethora of research focused on mitochondrial motility, methods commonly used to analyze mitochondrial movement fail to provide a whole-cell view of individual mitochondrial trajectories. Here, we achieve this synthesis by adapting a previously reported SPT method for whole-cell analysis and additionally making it publicly available online for general use. The approach first improves resolution by pre-processing images and then uses Matlab for object recognition and tracking. We use our method to demonstrate that on a whole-cell level, mitochondrial net distances excellently follow a lognormal distribution in fibroblasts. The continuous distribution followed by small-scale diffusion-like movements and bursts of directed motion challenges the widely held categorization of mitochondrial motion as either undirected or motor-driven. We further show that the lognormal distribution is shifted to lower net distances when microtubules are depolymerized, and shifted to higher net distances when microfilaments are depolymerized. These findings point to a general connection between mitochondria and the cytoskeleton which may extend beyond structural association into cellular function. We aim to further elucidate the mechanisms behind these interactions as well as refine our algorithm to optimize our detection of mitochondrial motility.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. Martin Picard, Alessia Angelin, Marni Falk and Douglas C. Wallace for helpful discussions regarding mitochondrial behavior; Ben Kandel for help with statistical assessments, image processing, and general MATLAB support; and Eric Abhold for helpful discussions and writing evaluation. The following funding sources are acknowledged for supporting this work: NIH grant T32 HL007954, Office of Naval Research grant N000141410538, and the Horatio C. Wood Endowment at the University of Pennsylvania.
References
- Boldogh IR, Pon LA. Interactions of mitochondria with the actin cytoskeleton. Biochim Biophys Acta BBA - Mol Cell Res. 2006;1763:450–462. doi: 10.1016/j.bbamcr.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Campello S, Lacalle RA, Bettella M, Manes S, Scorrano L, Viola A. Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J Exp Med. 2006;203:2879–2886. doi: 10.1084/jem.20061877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chada SR, Hollenbeck PJ. Nerve Growth Factor Signaling Regulates Motility and Docking of Axonal Mitochondria. Curr Biol. 2004;14:1272–1276. doi: 10.1016/j.cub.2004.07.027. [DOI] [PubMed] [Google Scholar]
- Chen HK, Liu Z, Meyer-Franke A, Brodbeck J, Miranda RD, McGuire JG, Pleiss MA, Ji ZS, Balestra ME, Walker DW, Xu Q, Jeong De, Budamagunta MS, Voss JC, Freedman SB, Weisgraber KH, Huang Y, Mahley RW. Small Molecule Structure Correctors Abolish Detrimental Effects of Apolipoprotein E4 in Cultured Neurons. J Biol Chem. 2012;287:5253–5266. doi: 10.1074/jbc.M111.276162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai SP, Bhatia SN, Toner M, Irimia D. Mitochondrial Localization and the Persistent Migration of Epithelial Cancer cells. Biophys J. 2013;104:2077–2088. doi: 10.1016/j.bpj.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Guo L, Yan S, Sosunov AA, McKhann GM, ShiDu Yan S. Early deficits in synaptic mitochondria in an Alzheimer's disease mouse model. Proc Natl Acad Sci. 2010;107:18670–18675. doi: 10.1073/pnas.1006586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick RL, Shaw JM. Moving Mitochondria: Establishing Distribution of an Essential Organelle. Traffic. 2007;8:1668–1675. doi: 10.1111/j.1600-0854.2007.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerencser AA, Nicholls DG. Measurement of Instantaneous Velocity Vectors of Organelle Transport: Mitochondrial Transport and Bioenergetics in Hippocampal Neurons. Biophys J. 2008;95:3079–3099. doi: 10.1529/biophysj.108.135657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedt RJ, Pfeiffer DR, Matzavinos A, Kao CY, Alevriadou BR. Mitochondrial Dynamics and Motility Inside Living Vascular Endothelial Cells: Role of Bioenergetics. Ann Biomed Eng. 2012;40:1903–1916. doi: 10.1007/s10439-012-0568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heggeness MH, Simon M, Singer SJ. Association of mitochondria with microtubules in cultured cells. Proc Natl Acad Sci U S A. 1978;75:3863–3866. doi: 10.1073/pnas.75.8.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA, McCulloch CA. Cell Mechanics: Integrating Cell Responses to Mechanical Stimuli. Annu Rev Biomed Eng. 2007;9:1–34. doi: 10.1146/annurev.bioeng.9.060906.151927. [DOI] [PubMed] [Google Scholar]
- Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel J, Lee HS, Sobolewski P, Tomczyk N, Composto RJ, Eckmann DM. Chemically grafted fibronectin for use in QCM-D cell studies. Biosens Bioelectron. 2014 doi: 10.1016/j.bios.2014.02.053. http://linkinghub.elsevier.com/retrieve/pii/S0956566314001419. [DOI] [PMC free article] [PubMed]
- Langford GM. Actin- and microtubule-dependent organelle motors: interrelationships between the two motility systems. Curr Opin Cell Biol. 1995;7:82–88. doi: 10.1016/0955-0674(95)80048-4. [DOI] [PubMed] [Google Scholar]
- Ligon LA, Steward O. Role of microtubules and actin filaments in the movement of mitochondria in the axons and dendrites of cultured hippocampal neurons. J Comp Neurol. 2000;427:351–361. doi: 10.1002/1096-9861(20001120)427:3<351::aid-cne3>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Limpert E, Stahel WA, Abbt M. Log-normal Distributions across the Sciences: Keys and Clues. BioScience. 2001;51:341. [Google Scholar]
- Maro B, Marty MC, Bornens M. In vivo and in vitro effects of the mitochondrial uncoupler FCCP on microtubules. EMBO J. 1982;1:1347–1352. doi: 10.1002/j.1460-2075.1982.tb01321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RL, Hollenbeck PJ. The regulation of bidirectional mitochondrial transport is coordinated with axonal outgrowth. J Cell Sci. 1993;104(Pt 3):917–927. doi: 10.1242/jcs.104.3.917. [DOI] [PubMed] [Google Scholar]
- Morris RL, Hollenbeck PJ. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J Cell Biol. 1995;131:1315–1326. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Mironov SL, Ivannikov MV, Schmidt J, Richter DW. Mitochondrial organization and motility probed by two-photon microscopy in cultured mouse brainstem neurons. Exp Cell Res. 2005;303:114–127. doi: 10.1016/j.yexcr.2004.09.025. [DOI] [PubMed] [Google Scholar]
- Munevar S, Wang Y, Dembo M. Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys J. 2001;80:1744–1757. doi: 10.1016/s0006-3495(01)76145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanareddy BRJ, Vartiainen S, Hariri N, O'Dowd DK, Gross SP. A Biophysical Analysis of Mitochondrial Movement: Differences Between Transport in Neuronal Cell Bodies Versus Processes: A Biophysical Analysis of Mitochondrial Movement. Traffic. 2014;15:762–771. doi: 10.1111/tra.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasova OE, Mendez MG, Chernoivanenko IS, Tyurin-Kuzmin PA, Kuczmarski ER, Gelfand VI, Goldman RD, Minin AA. Vimentin intermediate filaments modulate the motility of mitochondria. Mol Biol Cell. 2011;22:2282–2289. doi: 10.1091/mbc.E10-09-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Quintero M, Colombo SL, Godfrey A, Moncada S. Mitochondria as signaling organelles in the vascular endothelium. Proc Natl Acad Sci U S A. 2006;103:5379–5384. doi: 10.1073/pnas.0601026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintoul GL, Filiano AJ, Brocard JB, Kress GJ, Reynolds IJ. Glutamate decreases mitochondrial size and movement in primary forebrain neurons. J Neurosci Off J Soc Neurosci. 2003;23:7881–7888. doi: 10.1523/JNEUROSCI.23-21-07881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo G, Chamorro M, Salas ML, Viñuela E, Cuezva JM, Salas J. Migration of mitochondria to viral assembly sites in African swine fever virus-infected cells. J Virol. 1998;72:7583–7588. doi: 10.1128/jvi.72.9.7583-7588.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safiulina D, Veksler V, Zharkovsky A, Kaasik A. Loss of mitochondrial membrane potential is associated with increase in mitochondrial volume: Physiological role in neurones. J Cell Physiol. 2006;206:347–353. doi: 10.1002/jcp.20476. [DOI] [PubMed] [Google Scholar]
- Saunter CD, Perng MD, Love GD, Quinlan RA. Stochastically determined directed movement explains the dominant small-scale mitochondrial movements within non-neuronal tissue culture cells. FEBS Lett. 2009;583:1267–1273. doi: 10.1016/j.febslet.2009.02.041. [DOI] [PubMed] [Google Scholar]
- Sobolewski P, Kandel J, Klinger AL, Eckmann DM. Air bubble contact with endothelial cells in vitro induces calcium influx and IP3-dependent release of calcium stores. Am J Physiol Cell Physiol. 2011;301:C679–686. doi: 10.1152/ajpcell.00046.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos KJ, Sheetz MP. Visualization and quantification of mitochondrial dynamics in living animal cells. Methods Cell Biol. 2007;80:627–682. doi: 10.1016/S0091-679X(06)80030-0. [DOI] [PubMed] [Google Scholar]
- Warburg O. The metabolism of carcinoma cells. J Cancer Res. 1925;9:148–163. [Google Scholar]
- Yi M, Weaver D, Hajnóczky G. Control of mitochondrial motility and distribution by the calcium signal: a homeostatic circuit. J Cell Biol. 2004;167:661–672. doi: 10.1083/jcb.200406038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


