Abstract
Elucidating mechanisms of hepatitis B virus (HBV)-mediated hepatocarcinogenesis is needed to gain insights into the etiology and treatment of liver cancer. Cells where HBV is replicating exhibit increased expression of Plk1 kinase and reduced levels of two transcription repression factors, SUZ12 and ZNF198. SUZ12 is an essential subunit of the transcription repressive complex PRC2. ZNF198 stabilizes the transcription repressive complex composed of LSD1, Co-REST and HDAC1. These two transcription repressive complexes are held together by binding the long noncoding RNA HOTAIR. In this study we linked these regulatory events mechanistically, by showing that Plk1 induces proteasomal degradation of SUZ12 and ZNF198 by site-specific phosphorylation. Plk1-dependent ubiquitination of SUZ12 and ZNF198 was enhanced by expression of HOTAIR, significantly reducing SUZ12 and ZNF198 stability. In cells expressing the HBV X protein (HBx) downregulation of SUZ12 and ZNF198 mediated global changes in histone modifications. In turn, HBx-expressing cells propagated an altered chromatin landscape after cell division, as exemplified by changes in histone modifications of the EpCAM promoter, a target of PRC2 and LSD1/Co-REST/HDAC1 complexes. Notably, liver tumors from X/cmyc bitransgenic mice exhibited downregulation of SUZ12 and ZNF198 along with elevated expression of Plk1, HOTAIR, and EpCAM. Clinically, similar effects were documented in a set of HBV-related liver tumors consistent with the likelihood that downregulation of SUZ12 and ZNF198 leads to epigenetic reprogramming of infected hepatocytes. Since both Plk1 and HOTAIR are elevated in many human cancers, we propose that their combined effects are involved in epigenetic reprogramming associated broadly with oncogenic transformation.
Keywords: Hepatitis B virus X protein (HBx), SUZ12/PRC2, ZNF198/LSD1/Co-REST/HDAC1 complex, Polo-like kinase 1, lncRNA HOTAIR, hepatocarcinogenesis
INTRODUCTION
Chronic infection by the Hepatitis B virus is a major etiologic factor in the development of hepatocellular carcinoma (HCC) (1). The World Health Organization estimates that 250 million people globally are chronically infected with HBV. Despite the availability of the HBV vaccine, the vaccine is not always protective and children born of infected mothers become chronically infected. Current treatments include antiviral nucleoside analogs, but eventually this treatment results in viral resistance (2). In advanced stage HCC, targeted therapies such as sorafenib (anti-MAPK) are of modest but significant benefit (3). Thus, new and effective mechanism-based therapies are needed, along with new prognostic markers for molecular staging of the disease (4).
Pathogenesis of HBV-mediated HCC involves effects of chronic inflammation of the liver (5) and effects of the HBV X protein (HBx), which acts as a weak oncogene (6) or co-factor in hepatocarcinogenesis (7). HBV DNA integrates into the host genome at early steps of clonal tumor expansion and the majority of tumors display continued expression of HBx (8). HBx is a multifunctional protein (9, 10), is required for the HBV life cycle (11), and is implicated in HCC pathogenesis by a mechanism not yet understood. Regarding this mechanism, our studies (12, 13) discovered an inverse relationship between the protein levels of two transcription repressive factors, SUZ12 and ZNF198, and the mitotic polo-like-kinase1 (Plk1) (14). This intriguing relationship between Plk1, SUZ12 and ZNF198 was observed in cellular and animal models of HBx- and HBV-mediated oncogenic transformation as well as in HBV replicating cells (15).
SUZ12 is one of the three essential subunits of the transcription repressive PRC2 complex (polycomb repressive complex 2). The other two core subunits of PRC2 include EED protein and the histone methyltransferase EZH2 (16). This chromatin modifying complex epigenetically regulates lineage selection during embryonic development and stem cell differentiation (17, 18) by trimethylating histone3 on lysine 27 (H3K27me3), a transcription silencing modification (16). PRC2 forms multi-protein complexes with other chromatin modifying proteins (19, 20), and associates with noncoding RNAs (21). PRC2 binds the long noncoding RNA (lncRNA) HOTAIR which also binds another transcription repressive complex composed of lysine demethylase1 (LSD1), Co-REST, and histone deacetylase 1(HDAC1) (22). Interestingly, the LSD1/Co-REST/HDAC1 complex which enzymatically removes histone acetylations and H3K4 methylations that activate transcription, is stabilized by the chromosomal protein ZNF198 (23). Thus, HOTAIR couples the transcription repressive activity of PRC2 and LSD1/Co-REST/HDAC1 complexes (22). In addition to the involvement of these chromatin modifying complexes in cell fate determination during embryonic development, accumulating evidence links deregulated expression of polycomb proteins (24) and ZNF198 (25, 26) to cancer pathogenesis.
In liver tumors of X/c-myc bitransgenic mice and those induced by chronic infection with Woodchuck hepatitis virus (WHV), increased protein levels of Plk1 correlated with reduced protein levels of SUZ12 and ZNF198 (15). Our studies suggested that Plk1, activated by HBx in late G2 phase (27), is responsible for down-regulating the protein levels, but not the transcription of SUZ12 and ZNF198 (12, 13, 27). Studies by others have shown that Plk1 phosphorylation induces proteasomal degradation of its substrates involved in cell cycle progression (28, 29). Elevated Plk1 occurs in various types of human cancers, including liver cancer (30). The function of Plk1 substrates ranges from proteins involved in mitotic progression, chromosome segregation, DNA replication, and signal transduction (28-30). In this study we show that Plk1 signals by phosphorylation the proteasomal degradation of the chromatin modifying proteins SUZ12 and ZNF198. Clinically, elevated expression of HOTAIR and Plk1 was quantified in aggressive liver tumors derived from chronically infected HBV patients, supporting the involvement of this novel mechanism in HBV-mediated hepatocarcinogenesis.
MATERIALS AND METHODS
Cell culture, transfections, plasmids, siRNAs and synchronization protocols
Tetracycline regulated HBx-expressing 4pX-1 cells, derived from mouse hepatocyte AML12 cell line (31), was grown as described (32), in the presence of tetracycline (5μg/ml) or without tetracycline for 16-18h to allow HBx expression. HBx expression was confirmed by RT-PCR. Synchronization of 4pX-1 cells in G1/S by the double thymidine block (dTB) was performed as described (27). Synchronization of 4pX-1 cells in G2/M was by incubation with 100 nM nocodazole for 12h, and release from the block for 4h. HepAD38 cells, derived from HepG2 cell line, were grown as described (33). Transient transfections in HEK293T cells or 4pX-1 cells were performed employing Lipofectamine 2000 (Invitrogen) as described by manufacturer, with 4μg each of the following mammalian expression plasmids: SUZ12-HA, ZNF198-GFP (34), Plk1CA-GFP, Plk1KD-GFP, Ubiquitin-FLAG (35, 36), and pcDNA3-HOTAIR (22). HOTAIR siRNA and scrambled control siRNA were transfected in 4pX-1 cells using Lipofectamine® RNAiMAX (Invitrogen).
All cell lines used in this study are routinely tested for mycoplasma (employing commercially available kits), and retention of described properties and growth characteristics. 293HEK and AML12 cell lines were purchased from ATCC. HepAD38 cell line (33) was obtained from Dr. C. Seeger, Fox Chase Cancer Center in 2010. Expression of HBV pregenomic RNA in HepAD38 cells is tested by PCR at regular intervals, less than six months. HBx expression in tetracycline regulated HBx expressing cell lines (4pX-1, 4pX-1-SUZ12kd) is confirmed by PCR at regular intervals, less than six months. Knockdown of SUZ12 in 4pX-1-SUZ12kd cell line is confirmed by immunoblots compared to 4pX-1 cells. Selection of knockdown cells is carried out by cell sorting based on GFP fluorescence, should the expected knockdown of SUZ12 is lost. This is performed as needed. Last cell sorting for GFP+ cells was performed in 2014.
In vitro Plk1 kinase assays
performed exactly as described (35, 36) employing affinity purified SUZ12-HA and ZNF198-GFP following transient expression in HEK293 cells. Site-directed mutagenesis of putative Plk1 phosphorylation sites in SUZ12-HA and ZNF19-GFP was performed employing the QuickChange site-directed mutagenesis kit (Stratagene). Mutations were confirmed by DNA sequencing.
Immunoblots, immunoprecipitations and immunofluorescence microscopy were performed employing standard protocols (13). Antibodies used: HA (Sigma), FLAG (Sigma), GFP (Santa Cruz), Plk1 (Abcam), phospho- Plk1T210 (BD Biosciences), SUZ12 (Abcam), ZNF198 (Thermo Fisher Scientific), HBc (Abcam) LSD1 (Abcam), HDAC1 (Abcam), EZH2 (Cell Signaling), H3 (Active Motif), H3K27me3 (Abcam), H3K4me3 (Cell Signaling), H3K4me2 (Cell Signaling), H3K4me1 (Cell Signaling).
Chromatin immunoprecipitation assays (ChIP)
performed using 4pX-1 cells synchronized by nocodazole (100ng/ml) in G2/M for 12h, followed by release from nocodazole block for 4h, with or without HBx expression by tetracycline removal for 16h. The Millipore ChIP Assay Kit (Cat. No. 17-295) and the Mono-Methyl-Histone H3 (Lys4) Antibody #9723S (Cell Signaling) and Anti-Histone H3 (tri methyl K27) antibody-ChIP Grade (ab6002) were employed.
Reverse Transcription and Quantitative Real-Time PCR
RNA isolated from normal liver and liver tumors of X/c-myc mice (6) and human HBV-related HCCs employing PureLink RNA Mini Kit (12183018A, Invitrogen). Liver tissues from chronic HBV patients with HCC, tumor and peritumoral tissue, were obtained from the French National Biological Resources Centre following approved consent from the French Liver Tumor Network Scientific Committee. cDNA was synthesized from 2.0 μg total RNA isolated using iScript™ cDNA Synthesis Kit (170-8891, Bio-Rad). Quantitative real-time PCR reactions were performed in triplicates and normalized to GAPDH employing FastStart Essential DNA Green Master (06924204001, Roche), SYBR green (Roche), and Roche LightCycler 96. The 2-ΔΔCt method was used for analysis. Primer sequences are listed in Supplementary Data section.
RESULTS
HBx and Plk1 regulate protein levels of SUZ12 and ZNF198
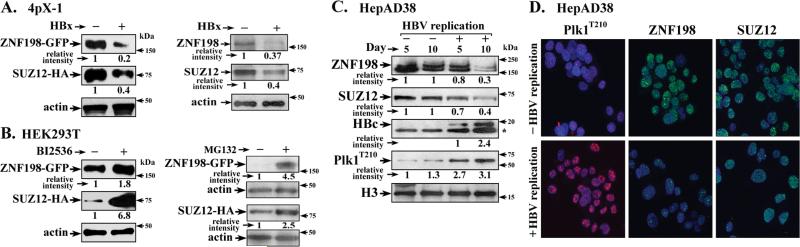
To investigate whether Plk1, which becomes activated by HBx expression (27), downregulates SUZ12 and ZNF198 proteins (15), we transfected mammalian expression vectors encoding SUZ12-HA and ZNF198-GFP in tetracycline-regulated HBx-expressing 4pX-1 cells (32). Expression of HBx decreased by > 50% protein levels of transfected SUZ12-HA and ZNF198-GFP as well as endogenous SUZ12 and ZNF198 proteins (Fig. 1A). To link this HBx-mediated downregulation of SUZ12 and ZNF198 to Plk1, HEK293T cells transfected with SUZ12-HA and ZNF198-GFP were treated with Plk1 inhibitor BI2536 or proteasomal inhibitor MG132 (Fig. 1B). Treatment with BI2536 or MG132 restored SUZ12-HA and ZNF198-GFP protein levels (Fig. 1B), suggesting SUZ12 and ZNF198 are down-regulated via Plk1 mediated phosphorylation.
Figure 1. Plk1 down-regulates SUZ12 and ZNF198 proteins.
A. (left panel) 4pX-1 cells, transiently transfected with SUZ12-HA and ZNF198-GFP encoding plasmids, were grown with (+) or without (−) HBx expression by tetracycline removal for 48h; whole cell extracts (WCE) were immunoblotted with GFP or HA antibodies. (Right panel) WCE from 4pX-1 cells treated with nocodazole (100 nM) for 12h and grown with (+) or without (−) HBx expression for 16h, were immunoblotted with ZNF198 or SUZ12 antibodies; B. HEK293T cells transiently transfected with SUZ12-HA and ZNF198-GFP encoding plasmids, were grown with (+) or without (−) BI2536 (250 nM) for 12h or MG132 (2.5 μM). (Left panel) DMSO serves as vehicle control. (Right panel) Transfected cells were synchronized in G2/M by addition of nocodazole (100 nM) for 12 hr. Lysates were immunoblotted with HA or GFP antibodies C. Immunoblots of indicated proteins using WCE from HepAD38 cells grown with (+) or without (−) HBV replication by tetracycline removal for 5 and 10 days. D. Immunofluorescence microscopy of indicated proteins, +/- HBV replication in HepAD38 cells.
These effects of HBx on SUZ12 and ZNF198 proteins were also observed in HBV replicating HepAD38 cells (13, 15). In the HepAD38 cellular model of HBV replication the integrated HBV genome is under control of the Tet-off promoter (33). HBV replication is initiated by tetracycline removal, allowing transcription of the pregenomic RNA, the template of viral replication. In the presence of HBV replication, using the viral core protein HBc as marker for HBV replication, endogenous SUZ12 and ZNF198 protein levels were reduced (Fig. 1C). Importantly, employing the phospho-T210 Plk1 (Plk1T210) antibody that detects active Plk1, we observe that in HBV replicating cells Plk1 was activated (Fig. 1C) and localized to the nucleus (Fig. 1D).
Plk1 phosphorylates SUZ12 and ZNF198 in vitro
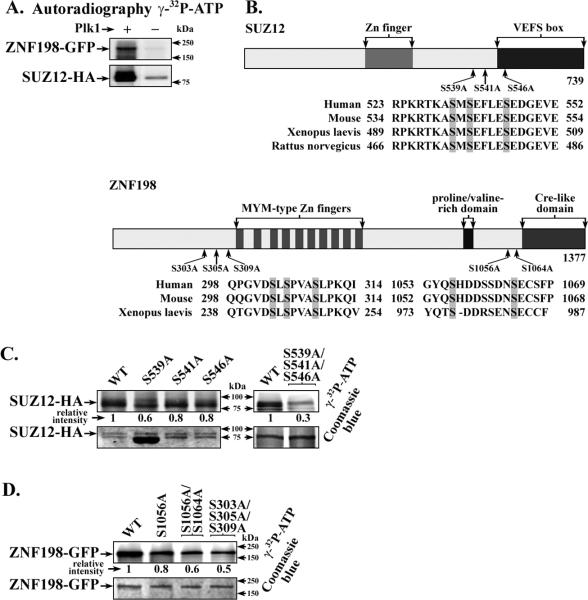
Since SUZ12 and ZNF198 are nuclear proteins and active Plk1 was in the nucleus of HBV replicating cells (Fig. 1D), these results suggest SUZ12 and ZNF198 could serve as Plk1 substrates. To test this hypothesis, we performed in vitro Plk1 assays (Fig. 2). SUZ12-HA and ZNF198-GFP proteins were expressed by transient transfection in HEK293T cells and purified by affinity chromatography. In vitro kinase assays with affinity purified substrates and Plk1 (36) were performed in the presence of γ-P32-ATP, analyzed by SDS PAGE, and autoradiography (35, 36). Indeed, wild-type (WT) SUZ12 and ZNF198 proteins were phosphorylated in vitro by Plk1 (Fig. 2A).
Figure 2. In vitro Plk1 kinase assays of SUZ12 and ZNF198 and site-directed mutants.
A. Affinity purified SUZ12-HA and ZNF198-GFP isolated from transfected HEK293T cells and Plk1 (35, 36) were used in in vitro Plk1 kinase assays. Reactions were performed with (+) or without (−) Plk1, in the presence of γ-32P-ATP and analyzed by SDS PAGE and autoradiography (35, 36). B. Functional regions of SUZ12 (40) and ZNF198 (23) and putative Plk1 phosphorylation sites are shown. Ser to Ala substitutions constructed at highlighted residues. C. and D., in vitro kinase assays of wildtype (WT) SUZ12-HA (C.) and ZNF198-GFP (D.) and site-directed mutants, as indicated. Commassie blue staining shows the same gel used for autoradiography. Relative intensity quantified by ImageJ software is ratio of signal from in vitro kinase reaction vs. corresponding commassie blue stained band, expressed relative to WT. (n=3)
Next, we examined the protein sequence of SUZ12 and ZNF198 for conserved, consensus Plk1 phosphorylation sites (D/E-X-S/T-Φ-D/E), and constructed Ser to Ala substitutions at those sites (Fig. 2B). Phosphoproteomic studies have shown SUZ12 is phosphorylated in vivo at S546 using nocodazole-arrested Hela cells (37) and S539 in human embryonic stem cells, hESCs (38); on the other hand, ZNF198 is phosphorylated in vivo at S305 using nocodazole-arrested Hela cells (39). However, the kinase(s) mediating these phosphorylations are unknown.
To determine if these sites in SUZ12 and ZNF198 are phosphorylated by Plk1, we expressed each mutant protein in HEK293T cells, purified them by affinity chromatography, and performed in vitro Plk1 kinase assays (Fig. 2C-D). SUZ12 proteins with Ser to Ala substitutions at residues 539, 541 and 546 (single and triple site mutants) were used in in vitro kinase assays. The results show that all three sites were phosphorylated by Plk1, the triple mutant showing 70% reduction in phosphorylation (Fig. 2C), and identify Plk1 as the kinase mediating the in vivo detected phosphorylations of SUZ12 at S539 and S546 (37, 38).
Similar analyses with ZNF198 identified two clusters of putative Plk1 phosphorylation sites in vitro. A cluster of serine residues at the N-terminus of ZNF198, S303, S305, and S309, and a cluster at the C-terminus, S1056 and S1064. The triple Ser to Ala mutant, S303A/S305A/S309A, consistently exhibited the lowest level of phosphorylation in vitro, in comparison to the double S1056A/S1064A mutant (Fig. 2D). Importantly, in vivo phosphorylation of S305 of ZNF198 was detected by phosphoproteomic studies (39), suggesting that Plk1 is the kinase that phosphorylates S305 of ZNF198 in vivo.
Plk1 phosphorylation dissociates SUZ12- and ZNF198-containing complexes
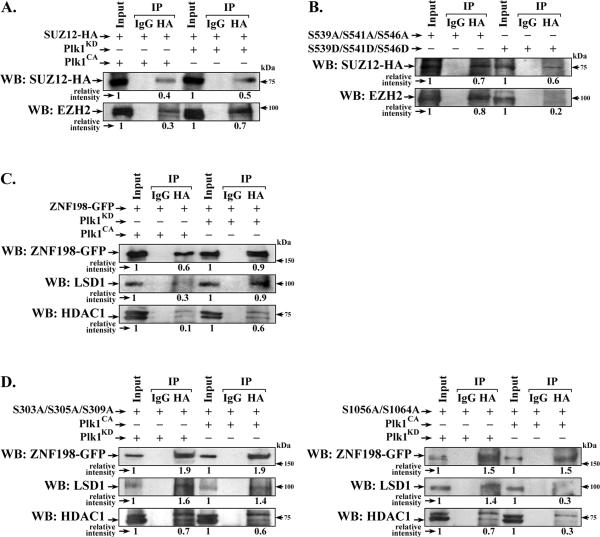
In SUZ12, residues 539, 541 and 546 phosphorylated by Plk1 in vitro (Fig. 2C), are adjacent (S539 and S541) and within (S546) the VEFS box (Fig. 2B) that interacts with EZH2 (40), suggesting that phosphorylation at those sites will disrupt SUZ12 interaction with EZH2. Accordingly, we examined the effect of expression of constitutively active (CA) Plk1CA on the interaction of SUZ12 with EZH2. Expression of active Plk1CA decreased the amount of EZH2 that co-immunoprecipitated with SUZ12, in comparison to the kinase dead (KD) Plk1KD (Fig. 3A). To confirm these results we examined endogenous EZH2 interaction with SUZ12 mutants containing Ser to Ala or Ser to Asp substitutions at all three sites (S539, S541 and S546). The phospho-mimetic substitutions (Ser to Asp) significantly reduced interaction of SUZ12 with EZH2 (Fig. 3B).
Figure 3. Plk1-mediated phosphorylations of SUZ12 and ZNF198 disrupt interaction with respective chromatin modifying complexes.
A. Transient co-expression of SUZ12-HA and Plk1CA or Plk1KD in HEK293T cells. WCE of transfected cells immunoprecipitated with IgG or HA antibodies; immunoprecipitates (IP) were immunoblotted (WB) with EZH2 or HA antibodies. Relative intensity quantified vs. Input. B. Transient co-expression of ZNF198-GFP and Plk1CA or Plk1KD in HEK293T cells. WCE of transfected cells immunoprecipitated with IgG or GFP antibodies; IPs were immunoblotted for GFP, HDAC1 or LSD1. C. and D., transient co-expression of indicated site-directed mutants of SUZ12 (C.) and ZNF198 (D.) in HEK293T cells. IPs immunoblotted with indicated antibodies.( n=3)
We also examined whether Plk1-mediated phosphorylation of ZNF198 disrupts interaction with LSD1/Co-REST/HDAC1 complex (23). ZNF198-GFP was co-transfected with either Plk1CA or Plk1KD. Upon expression of inactive Plk1KD, ZNF198-GFP co-immunoprecipitated both LSD1 and HDAC1 (Fig. 3C). By contrast, active Plk1CA reduced co-immunoprecipitation of these proteins (Fig. 3C). Cotransfection of active Plk1CA with N-terminal S303A/S305A/S309A or C-terminal S1056A/S1064A ZNF198 mutants demonstrated that only the N-terminal mutant maintained interaction with LSD1 and HDAC1 (Fig. 3D, compare left vs. right panel). We conclude that Plk1 phosphorylation at the N-terminal sites dissociates ZNF198 from the LSD1/Co-REST/HDAC1 complex.
Plk1 phosphorylation promotes ubiquitin-mediated degradation of SUZ12 and ZNF198
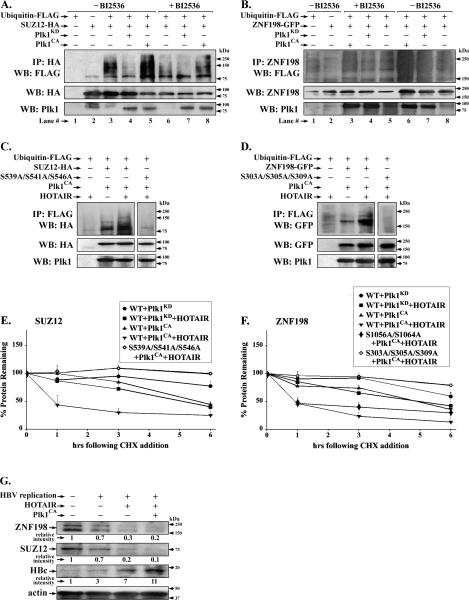
Since phosphorylation by Plk1 is often associated with ubiquitin-mediated proteolysis of its substrates (29), we investigated whether Plk1 phosphorylation of SUZ12 and ZNF198 regulates their ubiquitination. Expression vectors encoding SUZ12-HA or ZNF198-GFP were co-transfected in HEK293T cells with plasmids encoding ubiquitin-FLAG, and either Plk1CA or Plk1KD. All transfections assays were performed in the presence of MG132, an inhibitor of proteasomal degradation. Ubiquitinated WT SUZ12-HA, detected by immunoblots with FLAG antibody, was observed upon expression of active Plk1CA (Fig. 4A, lane 5), while the Plk1 inhibitor BI2536 suppressed ubiquitination (Fig. 4A, lane 8). Similar results were observed with WT ZNF198-GFP (Fig. 4B); compare lane 6 without BI2536 vs. lane 3 with BI2536. Since ubiquitination of SUZ12 and ZNF198 depends on active Plk1 (Fig. 4A and B), we examined the ubiquitination potential of their Plk1 phosphorylation mutants in transfected HEK293T cells. The triple Ser to Ala mutants of SUZ12 and ZNF198 (Fig. 2) lacked ubiquitination (Fig. S1A and S1B), supporting that Plk1-mediated phosphorylations at those sites are necessary for ubiquitination.
Figure 4. Plk1 dependent ubiquitination of SUZ12 and ZNF198 facilitated by lncRNA HOTAIR.
Ubiquitination assays of SUZ12-HA (A.) and ZNF198-GFP (B.) in HEK293T cells with addition (+) of BI2536 (500 nM for 20h) or (−) addition of DMSO. HEK293T cells co-transfected with 4.0μg each of indicated plasmids (+) or pCDNA3 empty vector (−). In A., WCE (0.5mg) immunoprecipitated with HA antibody and immunoblotted with FLAG antibody. In B., WCE (1.0 mg) immunoprecipitated with ZNF198 antibody and immnunobloted with FLAG. C. and D., ubiquitination assays of WT SUZ12-HA (C.) and WT ZNF198-GFP (D.), and the respective Ser to Ala mutants, in HEK293T cells transfected with 4.0μg each of plasmid DNA encoding Plk1CA and HOTAIR (+) or pcDNA3 empty vector (−). WCE (0.5mg) utilized for immunoprecipitations. (n=3) E. Quantification of transfected protein (SUZ12 and ZNF198) remaining, in a time course (0-6h) after cyclohexamide (CHX) addition. The indicated WT and Ser to Ala mutants of SUZ12-HA (A.) and ZNF198-GFP (F.) were co-transfected with Plk1CA and HOTAIR expression vectors; Plk1KD and pcDNA3 served as negative controls, respectively. WCE were harvested at 0-6h following addition of 20μg/ml of CHX to HEK293T cells; CHX was added 24h after transfection. Results represent the average from two independent experiments. Error bars are standard deviation. G. HepAD38 cells grown (−) HBV replication by addition of 5 μg/ml tetracycline or with (+) HBV replication by tetracycline removal. HOTAIR and Plk1 CA expression vectors were transfected on day-1 after tetracycline removal and cells were harvested 48h later. WCE were innumoblotted with indicated antibodies. (n=3)
HOTAIR increases ubiquitination of SUZ12 and ZNF198, reducing their stability
HOTAIR functions as scaffold in ubiquitination by interacting with RNA binding E3 ubiquitin ligases (41). Since HOTAIR bridges PRC2 to the LSD1/Co-REST/HDAC1 complex, stabilized by ZNF198 (23), we investigated whether HOTAIR has a role in ubiquitination of SUZ12 and ZNF198. We co-transfected in HEK293T cells expression vectors encoding HOTAIR and WT SUZ12-HA or ZNF198-GFP, together with active Plk1CA. Increased ubiquitination of SUZ12 and ZNF198 was observed with overexpression of transfected HOTAIR (Fig. S1C and D, and Fig. 4C and D). By contrast, the triple mutants of SUZ12 and ZNF198 that cannot be phosphorylated by Plk1 (Fig. 2) lacked ubiquitination, even with HOTAIR overexpression (Fig. 4C and D).
To directly demonstrate the role of Plk1 and HOTAIR on the stability of SUZ12 and ZNF198 proteins, we quantified their half-life (t1/2) following treatment with cyclohexamide (CHX) (Fig. 4E-F, and Fig. S2). Co-expression of active Plk1CA and HOTAIR, reduced the half-life of WT SUZ12 and ZNF198 to 1 h, in comparison to t1/2 > 6h with expression of inactive Plk1KD and HOTAIR. Conversely, the triple SUZ12 and ZNF198 mutants that cannot be phosphorylated by Plk1 (Fig. 2), exhibited enhanced stability, even in the presence of active Plk1CA and HOTAIR. By contrast, the t1/2 of C-terminal ZNF198 mutant (S1056A/S1064A) resembled WT ZNF198 (Fig. 4F). We interpret these results to mean that HOTAIR facilitates ubiquitination of Plk1-phosphorylated SUZ12 and ZNF198 proteins, thereby accelerating their proteasomal degradation.
To confirm that this mechanism is operational with endogenous SUZ12 and ZNF198 proteins, we co-expressed Plk1CA and HOTAIR in HBV-replicating HepAD38 cells. Expression plasmids were tranfected on day-1 after tetracycline removal, and cells were harvested 48h later. Co-expression of Plk1CA and HOTAIR significantly reduced endogenous levels of ZNF198 and SUZ12 (Fig. 4G). Interestingly, under these conditions HBc levels increased by 11-fold, in agreement with earlier results showing that knockdown of SUZ12 and ZNF198 enhances HBV replication (13).
HBx downregulates SUZ12 and ZNF198 during cell cycle progression in untransformed hepatocytes and alters the chromatin landscape of daughter cells
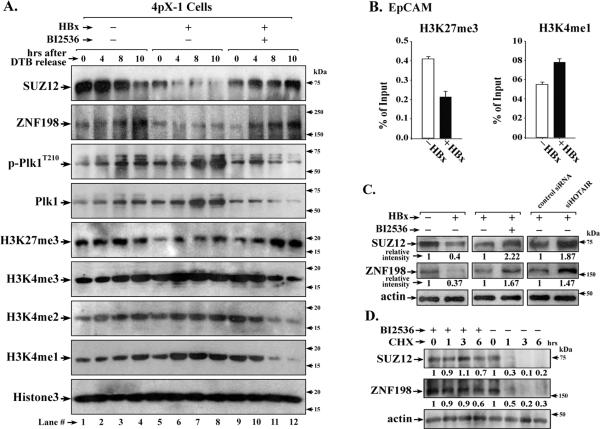
To investigate whether this mechanism occurs inHBx-expressing cells, we employed tetracycline-regulated HBx-expressing 4pX-1 cells (32). HBx activates Plk1 in late G2 phase (27). Employing double thymidine block (dTB) to synchronize 4pX-1 cells in G1/S, with or without HBx expression (27), we monitored protein levels of SUZ12 and ZNF198 as a function of cell cycle progression. Samples were collected at 4h (S phase), 8h (G2) and 10h (late G2/M) after release from dTB, as a function of HBx expression and BI2536 treatment (Fig. S3A). Immunoblots demonstrate reduction in SUZ12 and ZNF198 levels with HBx expression (Fig. 5A, lanes 5-8) that parallels activation of Plk1 (p-Plk1T210) from 4h to 10h after release from dTB (Fig. 5A, lanes 5-8 and Table S1). Importantly, treatment of HBx-expressing cells with BI2536 reversed this reduction (Fig. 5A, lanes 9-12). Next, we determined the effect of the reduction in SUZ12 and ZNF198 proteins on global H3 modifications mediated by the PRC2 and LSD1/Co-REST/HDAC1 complexes, respectively. Concomitant with the reduction in SUZ12, we observed reduction in H3K27me3 levels in HBx-expressing cells. Conversely, the levels of activating modifications on H3K4 were increased. Treatment of HBx-expressing cells with BI2536 reversed these effects (Table S1 shows quantification of Fig. 5A).
Figure 5. HBx mediates global histone changes during cell cycle progression.
A. Immunoblots of endogenous proteins with indicated antibodies, employing WCE isolated at 0-10h after release from dTB. Tetracycline regulated HBx-expressing 4pX-1 cells, grown without (−) or with (+) HBx, were synchronized in G1/S as described (27). BI2536 (500nM) was added 2h prior to cell-harvesting. ( n=3) B. Chromatin immunoprecipitation assays (ChIP) with indicated antibodies employing 4pX-1 cells arrested in G2/M by nocodazole (100nM) addition for 12h, and released from nocodazole block for 4h. Expression of HBx was by tetracycline removal for 16h. C. Immunoblots of SUZ12 and ZNF198 employing WCE from 4pX-1 cells isolated at 4h after release from nocodazole block, without (−) or with (+) HBx expression for 16hr, without (−) or with (+) BI2536 addition for 4h, without (−) or with (+) knockdown of HOTAIR by siRNA (siHOTAIR) transfection. ( n=3) D. Immunoblots of SUZ12 and ZNF198 employing WCE from 4pX-1 cells isolated in a time course (0-6 hr) after release from nocodazole and addition of CHX (20μg/ml), with (+) HBx expression, with (+) or without (−) BI2536.
Since HBx, via Plk1-mediated down-regulation of SUZ12 and ZNF198, altered global histone modifications during cell cycle progression (Fig. 5A), we investigated whether these epigenetic changes are propagated to daughter cells. We synchronized 4pX-1 cells in G2/M by nocodazole, released cells from the block for 4h, allowing progression to G1 phase (Fig. S3B). Employing chromatin immunoprecipitation assays (ChIP), we examined H3 modifications of the EpCAM promoter, a target of the PRC2 and LSD1/Co-REST/HDAC1 complexes (15). ChIP assays show that HBx expression decreased the silencing H3K27me3 modification mediated by PRC2, while the activating histone modification H3K4me1, associated with loss of LSD1 function, was increased (Fig. 5B). This altered chromatin landscape of the EpCAM promoter in HBx-expressing cells after cell division is supported by immunoblots showing decreased SUZ12 and ZNF198 levels in nocodazole-treated cells released from the block for 4h (Fig. 5C). Moreover, the reduction in endogenous SUZ12 and ZNF198 levels was restored by treatment with Plk1 inhibitor BI2536 and siRNA knockdown of HOTAIR (Fig. 5C), supporting involvement of both Plk1 and HOTAIR in their downregulation. Lastly, protein levels of endogenous SUZ12 and ZNF198 were stabilized in CHX- and BI2536-treated cells (Fig. 5D), further supporting the role of Plk1 in SUZ12 and ZNF198 degradation.
Plk1 and HOTAIR are elevated in liver tumors of X/c-myc mice and chronic HBV-infected patients
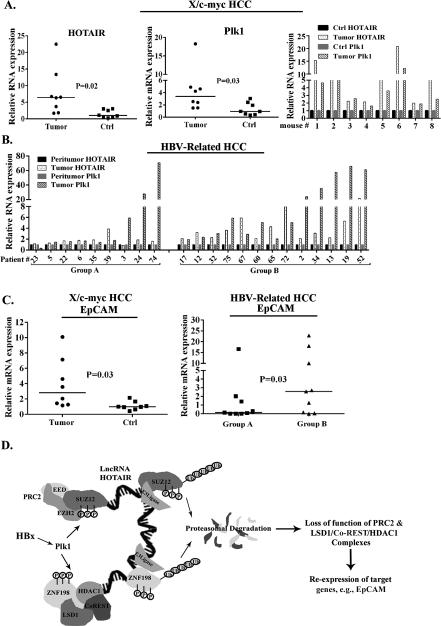
Liver tumors from X/c-myc bitransgenic mice (6) exhibit increased levels of Plk1 and reduced levels of SUZ12 and ZNF198 proteins (15). Based on the mechanism of Plk1-mediated degradation of SUZ12 and ZNF198 described in this study (Fig. 4), we quantified expression levels of Plk1 and HOTAIR RNAs in liver tumors from X/c-myc bitransgenic mice. We observed enhanced expression of Plk1 mRNA and HOTAIR in the same tumor, ranging from 2-fold to > than 10-fold (Fig. 6A, right panel). We also quantified Plk1 and HOTAIR RNA levels in liver tumors of chronically infected HBV patients (Fig. 6B). Group B patients exhibits at least 2-fold increased expression of both Plk1 and HOTAIR.
Figure 6. Plk1 and HOTAIR overexpression in liver tumors from X/c-myc mice and HBV-infected patients.
PCR quantification of Plk1 and HOTAIR RNAs using total RNA isolated from A. liver tumors of X/c-myc bitransgenic mice compared to control (Ctrl) RNA (normal mouse liver or peritumoral tissue), shown as box plots or histograms in liver tumors from individual mice. B. PCR quantification of Plk1 and HOTAIR RNAs using RNA isolated from HBV-related HCCs compared to peritumoral tissue. Quantitative PCR reactions were performed in identical triplicates (Plk1 and HOTAIR) using GAPDH as internal control. C. Quantification of EpCAM mRNA in liver tumors from X/c-myc mice (left panel) and HBV-mediated HCCs (right panel), expressed relative to Ctrl RNA. PCR quantification of RNAs from HBV-related HCC samples were performed in duplicates employing PCR arrays, and expressed relative to normal liver (average value from eight patients). p values are shown. D. Model illustrates the mechanism by which HBx-activated Plk1 by phosphorylation signals ubiquitination and proteasomal degradation of SUZ12 and ZNF198. HOTAIR accelerates ubiquitination and proteasomal degradation of SUZ12 and ZNF198, likely acting as scaffold for recruitment of RNA binding E3 ligases, as described by Yoon et al (41).
Since, liver tumors of X/c-myc bitransgenic mice display increased expression of EpCAM (15, and Fig. 6C left panel), we also quantified EpCAM expression in the HBV-related HCCs. Interestingly, increased EpCAM expression was observed in Group B HCCs, those displaying at least 2-fold increased expression of Plk1 and HOTAIR (Fig. 6C, right panel). Lastly, we determined the clinical significance of the expression of Plk1 and HOTAIR in HBV-related HCCs (Table 1). HOTAIR upregulation in Group B HCCs strongly correlates with the appearance of satelite nodules in the tumors (p=0.02), a well-recognized factor of aggressiveness of HCC; furthermore, Group B tumors are more prone to develop satellite nodules in comparison to those of Group A (p = 0.05).
Table 1.
Correlation of HOTAIR and Plk1 up-regulation between Tumor and Non-Tumor (ratio ≥ 2) to clinico-pathological features of 13 HBV-related HCCs (Group B). AFP = α-fetoprotein Spearman's correlation coefficient (P value) shown in parenthesis.
| Marker over-expression in Tumors | HOTAIR(+) | Plk1(+) | Group A vs. Group B |
|---|---|---|---|
| Presence of cirrhosis | −0.29 (.29) | −0.41 (.13) | 0.37 (.09) |
| Tumor size > 50 mm | 0.28 (.29) | 0.27 (.32) | −0.20 (.36) |
| Poor differentiation status (WHO classification) | 0.09 (.73) | −0.20 (.47) | 0.09 (.68) |
| AFP > 200 ng/mL | 0.001 (.99) | 0.27 (.32) | 0.05 (.81) |
| Microvascular invasion | 0.28 (.29) | 0.41 (.13) | −0.13 (.56) |
| Portal neoplastic embolization | −0.1 (.72) | 0.35 (.19) | 0.03 (.90) |
| Presence of satellite nodules | 0.57 (.02) | 0.06 (.80) | − 0.81 (.05) |
DISCUSSION
Liver tumors from X/c-myc bitransgenic mice and woodchucks chronically infected by WHV display an inverse relationship between Plk1 and chromatin regulating proteins SUZ12 and ZNF198 (15). In this study we provide evidence of a direct, causal link between Plk1 activation and reduced protein levels of SUZ12 and ZNF198. As shown in the model (Fig. 6D), Plk1 phosphorylates SUZ12 and ZNF198 at specific sites (Fig. 2), disrupts their association with the respective chromatin modifying complexes (Fig. 3) and signals proteasomal degradation of SUZ12 and ZNF198 by ubiquitination (Fig. 4). Interestingly, their ubiquitination is facilitated by HOTAIR, acting as ubiquitination scaffold (41). The combined action of active Plk1 and HOTAIR accelerates degradation of SUZ12 and ZNF198, decreasing their half-life by >15-fold, while the mutants of SUZ12 and ZNF198 that cannot be phosphorylated by Plk1 are stable (Fig. 4). The E3 ubiquitin ligases involved in this mechanism are presently unknown, although the RNA binding E3 ligases Dzip3 and Mex3b, shown to bind to HOTAIR (41), are likely candidates.
This novel mechanism that regulates SUZ12 and ZNF198 stability is significant for several reasons.
In eukaryotic cells transcription occurs in the context of chromatin, and often in response to extracellular signals. However, how signal transduction pathways communicate with chromatin is not well-understood. Such mechanisms could act directly to modify histones (42) or indirectly by regulating the activity of chromatin modifying and remodeling complexes (43). For example, EZH2, the methyltransferase of PRC2, is modified by phosphorylation by AKT and CDKs, thereby regulating its enzymatic activity (44-46) and substrate specificity (47). Recent studies have identified a series of molecular steps initiated by oncogenic Ras signaling that epigenetically silence tumor suppressor genes (48). However, in addition to silencing tumor suppressor genes, oncogenic signaling must epigenetically modify chromatin components to activate transcription of genes linked to proliferation and oncogenic transformation. In this study we identified such a mechanism, involving Plk1, overexpressed in many human cancers (28), regulating the stability of epigenetic regulators.
This mechanism is operational upon activation of Plk1 by HBx, shown in the model of Fig. 6D, occurring during cell cycle progression, in the G2 phase (Fig. 5). Downregulation of endogenous SUZ12 and ZNF198 coincides with Plk1 activation in HBx-expressing cells, reversed by inhibition of Plk1 with BI2536. The consequence of SUZ12 and ZNF198 downregulation is global changes in H3 modifications. Specifically, the EpCAM gene, silenced by PRC2 and ZNF198-LSD1/Co-REST/HDAC1 complexes (15), immediately after cell division exhibits H3 modifications associated with gene activation. We propose that with continued expression of HBx, as during chronic HBV infection, additional epigenetic alterations result in cellular reprogramming linked to oncogenic transformation. Indeed in SUZ12 knockdown cells, we have identified an active DNA demethylation mechanism that allows expression of EpCAM by HBx (Fan et al, pending). Importantly, EpCAM is expressed in hepatic cancer stem cells (49). Thus, down-regulation of SUZ12 beyond a threshold may be associated with acquisition of an oncogenic cellular program.
The involvement of HOTAIR in significantly decreasing the stability of SUZ12 and ZNF198 in the presence of activated Plk1 is another important discovery of this study (Fig. 4). The elevated expression of Plk1 and HOTAIR in liver tumors of X/c-myc bitransgenic mice as well as in a group of human HBV-related HCCs (Fig. 6) supports the relevance of this mechanism to HBV-mediated liver carcinogenesis. The enhanced expression of HOTAIR and Plk1 disrupts PRC2 and LSD1/Co-REST-HDAC1 complexes leading to re-expression of genes silenced by these complexes. Indeed, we detect enhanced expression of EpCAM both in liver tumors of X/c-myc mice as well as in Group B HCCs of HBV-infected patients (Fig. 6C), those dispaying elevated expression of both Plk1 and HOTAIR. The strong correlation between enhanced HOTAIR expression and the aggressive HCC phenotype supports the significance of this mechanism to HCC pathogenesis (Table 1), and the potential use of HOTAIR expression as a biomarker for aggressive HCC. Since Plk1 and HOTAIR are overexpressed in many human cancers (50), including a group of HBV-mediated HCCs (Fig. 6), we speculate this novel mechanism contributes to cellular reprograming observed during oncogenic transformation.
Supplementary Material
Acknowledgements
This work was supported by NIH grant DK044533 to OA. Shared Resources (flow cytometry and DNA sequencing) are supported by NIH grant P30CA023168 to Purdue Cancer Research Center. The authors thank Drs. X. Liu and J. Li for assistance and sharing of reagents for in vitro Plk1 kinase assays, M. Gorospe and H. Chang for providing the HOTAIR expression vector, E. Tran for critical review of this manuscript and P. Pascuzzi for bioinformatics analyses.
Footnotes
CONFLICT OF INTEREST: No conflict of interest to disclose.
Author Contribution:
H. Zhang : generated the majority of data
A. Diab, H. Fan, S. Mani: contributed in part to the genertation of data.
P. Merle: Analysis and interpretation of data.
R. Hullinger : figure illustration, manuscript review and editing.
O. Andrisani: study conception, design, supervision, funding and manuscript writing.
REFERENCES
- 1.Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981;2:1129–33. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 2.Zoulim F, Poynard T, Degos F, Slama A, El Hasnaoui A, Blin P, et al. A prospective study of the evolution of lamivudine resistance mutations in patients with chronic hepatitis B treated with lamivudine. Journal of viral hepatitis. 2006;13:278–88. doi: 10.1111/j.1365-2893.2005.00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. The New England journal of medicine. 2008;359:378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312–27. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hagen TM, Huang S, Curnutte J, Fowler P, Martinez V, Wehr CM, et al. Extensive oxidative DNA damage in hepatocytes of transgenic mice with chronic active hepatitis destined to develop hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1994;91:12808–12. doi: 10.1073/pnas.91.26.12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terradillos O, Billet O, Renard CA, Levy R, Molina T, Briand P, et al. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- 7.Madden CR, Finegold MJ, Slagle BL. Hepatitis B virus X protein acts as a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J Virol. 2001;75:3851–8. doi: 10.1128/JVI.75.8.3851-3858.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su Q, Schroder CH, Hofmann WJ, Otto G, Pichlmayr R, Bannasch P. Expression of hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatology. 1998;27:1109–20. doi: 10.1002/hep.510270428. [DOI] [PubMed] [Google Scholar]
- 9.Andrisani OM, Barnabas S. The transcriptional function of the hepatitis B virus X protein and its role in hepatocarcinogenesis (Review). Int J Oncol. 1999;15:373–9. doi: 10.3892/ijo.15.2.373. [DOI] [PubMed] [Google Scholar]
- 10.Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725–34. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–30. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Studach LL, Rakotomalala L, Wang WH, Hullinger RL, Cairo S, Buendia MA, et al. Polo-like kinase 1 inhibition suppresses hepatitis B virus X protein-induced transformation in an in vitro model of liver cancer progression. Hepatology. 2009;50:414–23. doi: 10.1002/hep.22996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang WH, Studach LL, Andrisani OM. Proteins ZNF198 and SUZ12 are down-regulated in hepatitis B virus (HBV) X protein-mediated hepatocyte transformation and in HBV replication. Hepatology. 2011;53:1137–47. doi: 10.1002/hep.24163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golsteyn RM, Schultz SJ, Bartek J, Ziemiecki A, Ried T, Nigg EA. Cell cycle analysis and chromosomal localization of human Plk1, a putative homologue of the mitotic kinases Drosophila polo and Saccharomyces cerevisiae Cdc5. J Cell Sci. 1994;107(Pt 6):1509–17. doi: 10.1242/jcs.107.6.1509. [DOI] [PubMed] [Google Scholar]
- 15.Studach LL, Menne S, Cairo S, Buendia MA, Hullinger RL, Lefrancois L, et al. Subset of Suz12/PRC2 target genes is activated during hepatitis B virus replication and liver carcinogenesis associated with HBV X protein. Hepatology. 2012;56:1240–51. doi: 10.1002/hep.25781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–36. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–79. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K, et al. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neri F, Krepelova A, Incarnato D, Maldotti M, Parlato C, Galvagni F, et al. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell. 2013;155:121–34. doi: 10.1016/j.cell.2013.08.056. [DOI] [PubMed] [Google Scholar]
- 21.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–53. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gocke CB, Yu H. ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers. PLoS One. 2008;3:e3255. doi: 10.1371/journal.pone.0003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richly H, Aloia L, Di Croce L. Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis. 2011;2:e204. doi: 10.1038/cddis.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moinzadeh P, Breuhahn K, Stutzer H, Schirmacher P. Chromosome alterations in human hepatocellular carcinomas correlate with aetiology and histological grade--results of an explorative CGH meta-analysis. Br J Cancer. 2005;92:935–41. doi: 10.1038/sj.bjc.6602448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao S, Nalabolu SR, Aster JC, Ma J, Abruzzo L, Jaffe ES, et al. FGFR1 is fused with a novel zinc-finger gene, ZNF198, in the t(8;13) leukaemia/lymphoma syndrome. Nat Genet. 1998;18:84–7. doi: 10.1038/ng0198-84. [DOI] [PubMed] [Google Scholar]
- 27.Studach L, Wang WH, Weber G, Tang J, Hullinger RL, Malbrue R, et al. Polo-like kinase 1 activated by the hepatitis B virus X protein attenuates both the DNA damage checkpoint and DNA repair resulting in partial polyploidy. J Biol Chem. 2010;285:30282–93. doi: 10.1074/jbc.M109.093963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo J, Liu X. Polo-like kinase 1, on the rise from cell cycle regulation to prostate cancer development. Protein Cell. 2012;3:182–97. doi: 10.1007/s13238-012-2020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mamely I, van Vugt MA, Smits VA, Semple JI, Lemmens B, Perrakis A, et al. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr Biol. 2006;16:1950–5. doi: 10.1016/j.cub.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 30.Cholewa BD, Liu X, Ahmad N. The role of polo-like kinase 1 in carcinogenesis: cause or consequence? Cancer Res. 2013;73:6848–55. doi: 10.1158/0008-5472.CAN-13-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu JC, Merlino G, Fausto N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc Natl Acad Sci U S A. 1994;91:674–8. doi: 10.1073/pnas.91.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarn C, Bilodeau ML, Hullinger RL, Andrisani OM. Differential immediate early gene expression in conditional hepatitis B virus pX-transforming versus nontransforming hepatocyte cell lines. J Biol Chem. 1999;274:2327–36. doi: 10.1074/jbc.274.4.2327. [DOI] [PubMed] [Google Scholar]
- 33.Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, et al. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother. 1997;41:1715–20. doi: 10.1128/aac.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunapuli P, Kasyapa CS, Chin SF, Caldas C, Cowell JK. ZNF198, a zinc finger protein rearranged in myeloproliferative disease, localizes to the PML nuclear bodies and interacts with SUMO-1 and PML. Exp Cell Res. 2006;312:3739–51. doi: 10.1016/j.yexcr.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Liu XS, Yang X, Song B, Wang Y, Liu X. Polo-like kinase 1 phosphorylation of p150Glued facilitates nuclear envelope breakdown during prophase. Proc Natl Acad Sci U S A. 2010;107:14633–8. doi: 10.1073/pnas.1006615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X, Li H, Zhou Z, Wang WH, Deng A, Andrisani O, et al. Plk1-mediated phosphorylation of Topors regulates p53 stability. J Biol Chem. 2009;284:18588–92. doi: 10.1074/jbc.C109.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101:12130–5. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, et al. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- 39.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–92. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto K, Sonoda M, Inokuchi J, Shirasawa S, Sasazuki T. Polycomb group suppressor of zeste 12 links heterochromatin protein 1alpha and enhancer of zeste 2. J Biol Chem. 2004;279:401–6. doi: 10.1074/jbc.M307344200. [DOI] [PubMed] [Google Scholar]
- 41.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- 43.Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol. 2013;14:211–24. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X, et al. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol. 2010;12:1108–14. doi: 10.1038/ncb2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–20. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–52. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wajapeyee N, Malonia SK, Palakurthy RK, Green MR. Oncogenic RAS directs silencing of tumor suppressor genes through ordered recruitment of transcriptional repressors. Genes Dev. 2013;27:2221–6. doi: 10.1101/gad.227413.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–24. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wan Y, Chang HY. HOTAIR: Flight of noncoding RNAs in cancer metastasis. Cell Cycle. 2010;9:3391–2. doi: 10.4161/cc.9.17.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.