Abstract
The growth factor PDGF controls the development of glioblastoma (GBM) but its contribution to the function of GBM stem-like cells (GSC) has been little studied. Here we report that the transcription factor FoxM1 promotes PDGFA-STAT3 signaling to drive GSC self-renewal and tumorigenicity. In GBM we found a positive correlation between expression of FoxM1 and PDGF-A. In GSC and mouse neural stem cells, FoxM1 bound to the PDGF-A promoter to upregulate PDGF-A expression, acting to maintain the stem-like qualities of GSC in part through this mechanism. Analysis of the human cancer genomic database TCGA revealed that GBM express higher levels of STAT3, a PDGF-A effector signaling molecule, as compared with normal brain. FoxM1 regulated STAT3 transcription through interactions with the β-catenin/TCF4 complex. FoxM1 deficiency inhibited PDGF-A and STAT3 expression in neural stem cells and GSC, abolishing their stem-like and tumorigenic properties. Further mechanistic investigations defined a FoxM1-PDGFA-STAT3 feed-forward pathway that was sufficient to confer stem-like properties to glioma cells. Collectively, our findings showed how FoxM1 activates expression of PDGF-A and STAT3 in a pathway required to maintain the self-renewal and tumorigenicity of glioma stem-like cells.
Keywords: FoxM1, glioblastoma stem-like cells, self-renewal, PDGF-A, STAT3, tumorigenicity
Introduction
Glioblastoma is the most lethal primary malignant brain tumor in adults. Recent evidence from human and animal studies suggests that cancer stem cells are the cellular origin of glioma. GBM stem-like cells (GSCs) can drive brain tumorigenesis through differentiation into highly proliferative tumor cells and transdifferentiation into endothelial cells and vascular pericytes in animal models (1–3). GSCs are resistant to chemotherapy and radiation, and therefore they give rise to tumor recurrence by sustaining long-term tumor growth (4,5).
Several studies have demonstrated that gliomagenesis involves aberrant signaling through PDGF pathway (6–14). Activation of the PDGF/PDGFR pathway may be triggered by binding between PDGFR (PDGFRa and PDGFRb) with PDGF ligands (PDGF-A, B, C, and D). However, high expression of PDGF-A was found in 70% of gliomas, and PDGF-A expression directly correlates with grade with higher expression in glioblastoma multiforme (GBM) (8,15). Moreover, inhibition of PDGF-A was able to inhibit growth of GBM cell lines in vivo, indicating an important role of PDGF-A in mediating the growth of glioma cells (16,17). Furthermore, Jackson et al showed that the PDGF-A pathway may link adult NSCs to glioma, because infusion of PDGF-AA (PDGF-A homodimer) into the lateral ventricles led to formation of atypical hyperplasias while withdrawal of PDGF-AA resulted in regression of hyperplasias and increased differentiation of NSCs (18). Thus, the study provides compelling evidence that PDGF-A signaling regulates NSC self-renewal and increased PDGF signaling may be important for glioma initiation (18). However, whether GSCs express PDGF-A is unknown. Also, the mechanism driving PDGF-A expression in glioma has not been elucidated.
The signal transducer and activator of transcription factor 3 (STAT3) is a critical signaling node in tumor that can be activated by a panel of cytokines and growth factors, such as EGF, PDGF, and IL-6, as well as by oncogenic proteins, such as Src and Ras (19–21). STAT3 is activated at high frequency in human tumors including GBM (22,23). Recently, STAT3 activity was found to be required for maintenance of the stem-like characteristics of GSCs (24). Moreover, IL-6, bone marrow X-linked nonreceptor tyrosine kinase, and EZH2 activate STAT3 signaling to maintain self-renewal and tumorigenic potential of GSCs (25–27), confirming the importance of STAT3 signaling in GSCs. Inhibition of STAT3 expression in GBM cells and GSCs results in downregulated STAT3 phosphorylation (24,28), indicating that the level of STAT3 is important to its phosphorylation. Moreover, STAT3 may drive downstream targets gene expression in the absence of phosphorylation (29,30). However, the molecular mechanisms underlying STAT3’s own gene expression in glioma are poorly defined.
The transcription factor FoxM1 is also substantially elevated in a majority of human tumors and contributes to oncogenesis in different organs, including brain (31,32). Independent profiling studies showed that the expression of FoxM1 in high-grade anaplastic astrocytomas and glioblastomas is significantly higher than that in low-grade astrocytomas (33,34). Overexpression of FoxM1 in GBM specimens was further confirmed by a study which analyzed a set of data from The Cancer Genome Atlas (TCGA) (35). Our previous studies showed that FoxM1 promotes the uncontrolled proliferation, invasion, and angiogenesis of GBM cells (36,37).
In the present study, we examined the mechanisms underlying PDGF-A expression in glioma cells and GSCs. We show that FoxM1 plays critical roles in both expression and activation of PDGF-A. We also show a unique role of FoxM1 in STAT3 expression and activation. Collectively, our findings demonstrate that FoxM1 is required for the self-renewal and tumorigenicity of GSCs and that FoxM1-induced expression of PDGF-A and STAT3 and activation of STAT3 is a key mechanism for FoxM1-induced GSC self-renewal and tumorigenicity.
Materials and Methods
Cell lines and culture conditions
Human glioma Hs683 and SW1783 cells and 293T cells were obtained from the American Type Culture Collection and cultured in DMEM with 10% FBS. The cell lines were tested and authenticated by DNA typing at the MD Anderson Cancer Center Characterized Cell Line Core. GBM stem cell lines were isolated from fresh surgical specimens of human GBM and cultured as GBM neurospheres in neurosphere medium (NM): DMEM/F12 medium supplemented with B27 supplement (Invitrogen) and bFGF and EGF (20 ng/ml each) as described previously (38). The study was approved by the Institutional Review Board of MD Anderson Cancer Center. Only early-passage cell lines were used for the study.
Immunohistochemical staining
Human tissue sections of GBM were de-paraffinized and then stained with a primary antibody against human FoxM1, pSTAT3 or PDGF-A. To quantify FoxM1 and PDGF-A expression, staining of each protein was scored as 0–8 according to the percentage of positive cells and staining intensity, as we previously described (39). Two separate individuals who were blinded to the slides examined and scored each sample. An average value of the two scores was reported.
Intracranial tumor assay
GSC and glioma cells were injected intracranially into nude mice as described previously (31). Animals showing general or local symptoms were euthanized; the remaining animals were euthanized 90 days after tumor cell injection. Tumor formation was determined by histologic analysis of hematoxylin-eosin-stained brain sections. Tumor volumes were measured by using length (a) and width (b) in H&E-stained sections and were calculated using the equation: V = ab2/2 (40). All mouse experiments were approved by the Institutional Animal Care and Use Committee of MD Anderson Cancer Center.
Western blot analysis
Standard Western blotting was done with antibody against FoxM1, β-catenin, or β-actin (Santa Cruz Biotechnology), PDGF-A (Orbigen), SSEA-1 or Nestin (BD Transduction Laboratories), SOX2 or CD133 (Abcam), GFAP (DAKO), STAT3, pSTAT3 (Y705), Tuj-1 or OCT4 (Cell Signaling).
Neurosphere formation assays
The sphere formation assay was performed by plating dissociated single cells at a density of 1 cell/μl in 96-well plates, and counting the number of spheres that formed after 7 days of culture in neurosphere medium (38).
Promoter reporters and dual luciferase assays
Cells were transfected with PDGF-A promoter reporter plasmids (41) or TOPflash (38) or STAT3 promoter reporter. Transfection efficiency was normalized by co-transfection with a pB-actin-RL reporter containing a Renilla luciferase gene under the control of a human β-actin promoter. The activities of firefly luciferase and Renilla luciferase were quantified using the dual-luciferase reporter assay system (Promega).
Statistical analysis
Correlations between positive staining for FoxM1 and positive staining for PDGF-A or pSTAT3 in the GBM specimens were assessed using Pearson’s correlation. The significance of the in vitro results was determined with Student’s t-test (two-tailed). The significance of in vivo survival study was determined by the Log-rank test. p < 0.05 was considered to be significant.
Results
FoxM1 regulates PDGF-A in GSCs and NSCs
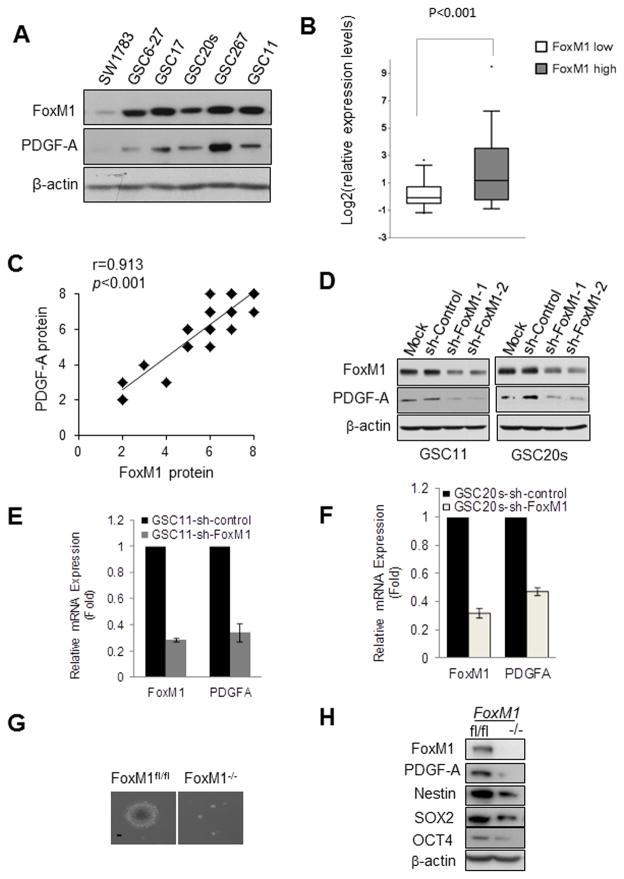
To examine the role of PDGF-A in GSCs, we first detected PDGF-A expression in SW1783 glioma cells and in GSC11, GSC20s, GSC17, GSC6-27, and GSC267 GBM stem cells. PDGF-A expression was much higher in GSCs than in SW1783 cells (Fig. 1A), suggesting that higher PDGF-A expression may be associated with GSC phenotype. Next, we examined the expression levels of FoxM1, a transcriptional factor associated with GSCs, in SW1783 cells and GSCs. The levels of FoxM1 might be associated with the levels of PDGF-A in the cells (Fig. 1A). To determine whether the finding has clinical relevance, we evaluated the expression of PDGF-A and FoxM1 in the 154 GBM tumors with RNA sequencing data available from TCGA (42). When the GBM tumors were characterized into FoxM1high and FoxM1low subpopulation, higher PDGF-A RNA expression was enriched in the FoxM1high subgroup (Fig. 1B, P<0.001). Moreover, we examined the protein levels of FoxM1 and PDGF-A in serial sections of 50 human primary GBM samples by immunohistochemical analyses. The levels of PDGF-A expression significantly correlated with the levels of FoxM1 expression in these samples (Fig. 1C).
Figure 1. FoxM1 regulates PDGF-A in GSCs and NSCs.
(A) Western blotting of FoxM1 and PDGF-A proteins in SW1783 cells and GSCs. (B) Box plots of relative expression of PDGF-A in FoxM1low or FoxM1high GBMs from a TCGA data sets. Boxes show median 25th and 75th percentiles, while whiskers represent the 5th and the 95th percentiles. Outliers are shown as individual points. t-test was used to test statistical significance. (C) FoxM1 expression correlated with elevated expression of PDGF-A in GBM patients. The correlation was significant as determined by Pearson’s correlation test. Note that some of the symbols on the graphs represent more than one specimen (i.e., there were overlapping scores). (D) Western blotting of FoxM1 and PDGF-A proteins in sh-control-, sh-FoxM1-1-, or sh-FoxM1-2-GSC11 and GSC20s cells. (E,F) FoxM1 and PDGF-A mRNA levels detected by real-time PCR in sh-control- or sh-FoxM1-GSC11 and GSC20s cells. (G) Photographs of neurosphere in immortalized FoxM1fl/fl and FoxM1−/− NSCs. Bar, 10 μm. (H) Expression of indicated proteins in immortalized FoxM1fl/fl and FoxM1−/− NSCs determined by Western blotting.
To determine whether FoxM1 is an upstream regulator of PDGF-A, we examined PDGF-A expression in stable FoxM1-knockdown GSCs. Knocking down FoxM1 significantly downregulated PDGF-A protein and mRNA levels as compared with sh-control (Fig. 1D–F and Fig. S1A&B). To ascertain the effect of FoxM1 on PDGF-A expression, we examined PDGF-A expression in FoxM1-null NSCs. Deletion of FoxM1 in FoxM1fl/fl NSCs resulted in diminished sphere formation ability (Fig. 1G) and diminished expression of PDGF-A (Fig. 1H and Fig. S1C), as well as downregulated expression of stem cell markers Nestin, SOX2, and OCT4 (Fig. 1H). Taken together, these results indicated that FoxM1 plays an important role in regulating PDGF-A expression.
FoxM1 directly interacts with the PDGF-A promoter and regulates its expression
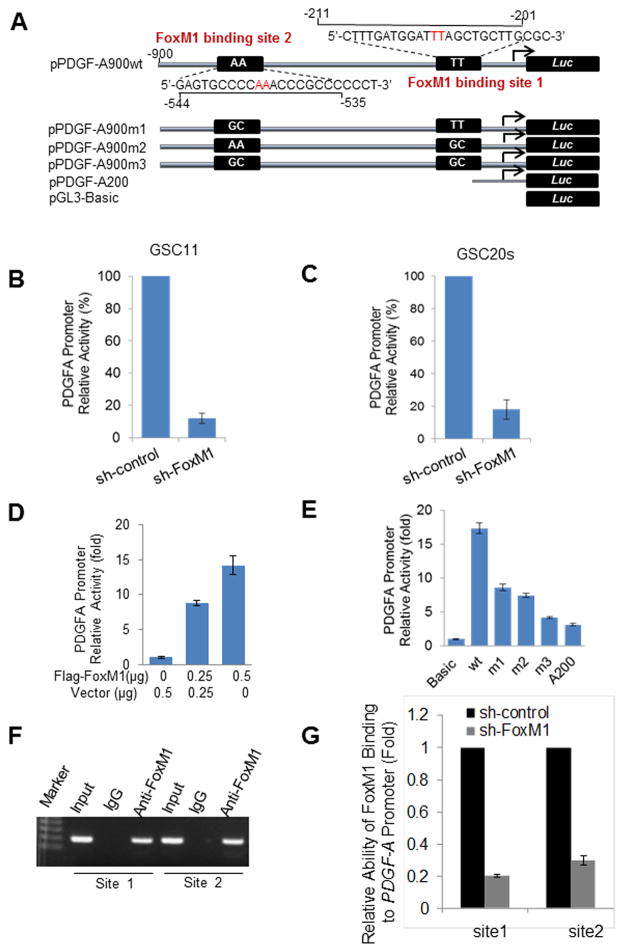
To test whether FoxM1 directly regulates PDGF-A expression, we first analyzed the sequence of the PDGF-A promoter by using the FoxM1 consensus binding sequences and identified two putative FoxM1-binding sites (Fig. 2A). Moreover, knocking down FoxM1 in GSC11 and GSC20s cells significantly decreased PDGF-A promoter activity in the cells (Fig. 2B, C), while overexpression of FoxM1 in 293T cells increased PDGF-A promoter activity (Fig. 2D). Next, we generated various mutants of PDGF-A promoter (Fig. 2A). The mutants containing mutations in binding site 1, binding site 2, or both sites and the deletion mutant A200 exhibited lower promoter activity than wild-type (wt) PDGF-A promoter in 293T cells (Fig. 2E). Also, disruption of one or both of the FoxM1 binding sites significantly attenuated PDGF-A promoter activity in GSC11 and GSC20s cells (Fig. S2A&B), indicating that the FoxM1-binding sites were critical for PDGF-A promoter activation.
Figure 2. FoxM1 directly interacts with the PDGF-A promoter and regulates its expression.
(A) Schematic illustration of the intact and various mutated PDGF-A promoters that were fused with a luciferase reporter gene. (B,C) PDGF-A promoter luciferase reporter was transfected into GSC11 cells and GSC20s cells expressing sh-control or sh-FoxM1. Values are mean ± SD for triplicate samples. (D) 293T cells were cotransfected with the PDGF-A luciferase reporter and an increasing amount of Flag-FoxM1 or vector. A Renilla luciferase reporter was used as a control to standardize the transfection efficiency. Values are mean ± SD for triplicate samples from a representative experiment. (E) The relative luciferase activity of wild-type (A900WT; wt) or the various mutant PDGF-A promoters described in (A) was determined in 293T cells transfected with 0.5 μg of Flag-FoxM1 plasmid. pGL3-Basic was used as a control. Values are mean ± SD for triplicate samples from a representative experiment. (F) ChIP assays on two FoxM1-binding sites of PDGF-A promoter were performed in GSC11 cells. (G) ChIP assays were performed in GSC11-sh-control or -sh-FoxM1 cells. The immunoprecipitated DNA was analyzed for the relative ability of FoxM1 binding to PDGF-A promoter site 1 and site 2 by real-time PCR. Values are mean ± SD for triplicate samples.
To provide direct evidence that FoxM1 binds to the endogenous PDGF-A promoter during transcription in vivo, we performed ChIP assays using GSC11 cells. Both of the FoxM1-binding regions of the PDGF-A promoter bound specifically to endogenous FoxM1 protein in vivo (Fig. 2F), and FoxM1 knockdown strikingly inhibited the FoxM1 binding to both regions (Fig. 2G & Fig. S2C). Taken together, these results clearly indicate that FoxM1 upregulates PDGF-A expression through direct binding to the PDGF-A promoter.
FoxM1 maintains stemness of GSCs partially via PDGF-A
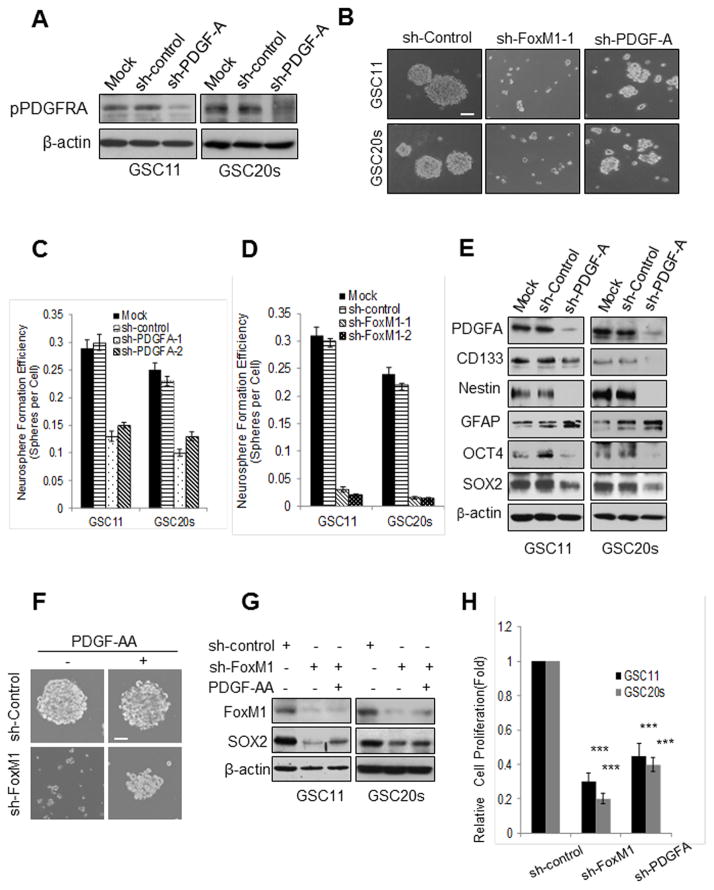
We next examined whether the FoxM1-PDGF-A axis regulates the stemness of GSCs. PDGF-A knockdown substantially decreased PDGFRA phosphorylation (Fig. 3A) and resulted in reduced size and number of spheres (Fig. 3B,C), indicating that knockdown of PDGF-A inhibited self-renewal of GSCs. PDGF-A knockdown also suppressed the expression of stem cell markers CD133, Nestin, SOX2, and OCT4 but upregulated the expression of differentiation marker GFAP (Fig. 3E), indicating that knockdown of PDGF-A inhibited the stemness of GSCs. FoxM1 knockdown also reduced the size and number of spheres (Fig. 3B,D and Fig. S3A,B) and suppressed the expression of stem cell markers (Fig. S3C) but upregulated the expression of GFAP (Fig. S3C). However, FoxM1 knockdown exhibited much stronger inhibitory effects on GSC self-renewal than did PDGF-A knockdown, as determined by the size and number of spheres in each group (Fig. 3B, D).
Figure 3. FoxM1 maintains the stemness of GSCs partially via PDGF-A.
(A) Western blotting of PDGFRA phosphorylation levels in GSC11 and GSC20s cells expressing sh-control or sh-PDGF-A. (B) Photographs of neurosphere of GSC11 and GSC20s cells expressing control, FoxM1, or PDGF-A shRNA. Bar, 20 μm. (C,D) Neurosphere formation efficiency of the cells in (B). Values are mean ± SD for triplicate samples. (E) Western blotting of stem cell and differentiation markers in GSC11 and GSC20s cells expressing sh-control or sh-PDGF-A. (F) Photographs of neurosphere formation of GSC11-sh-control and GSC11-sh- FoxM1 cells treated with or without PDGF-AA (50 ng/ml) for 10 days. Bar, 10 μm. (G) SOX2 expression detected by Western blotting in GSC11-sh-FoxM1 and GSC20s-sh-FoxM1 cells treated with or without PDGF-AA (50 ng/ml) for 72 hr. (H) Relative cell proliferation of GSC11 and GSC20s cells expressing control, FoxM1, or PDGF-A shRNA in 72 hr was determined by in vitro cell proliferation assay.
To determine the role of PDGF-A in FoxM1-mediated stemness of GSCs, we tested whether exogenous PDGF-A rescued the inhibitory effects of FoxM1 knockdown on the stemness of GSCs. Exogenous PDGF-AA (50 ng/ml) only partially rescued the effect of downregulation of FoxM1 on the self-renewal of GSC11 and GSC20s cells (Fig. 3F,G, Fig. S3D,E) or the effect of FoxM1 depletion on the self-renewal of NSCs (Fig. S3G). Exogenous PDGF-AA also only partially reversed the effect of FoxM1 knockdown on the expression of SOX2 (Fig. 3G) and Nestin (Fig. S3F). These findings indicated that FoxM1 maintains the stemness of GSCs partially through PDGF-A.
Inhibition of FoxM1 decreased cell proliferation and increased chemosensitivity of GSCs to TMZ
Since cell proliferation is ultimately required, although not sufficient, for the self-renewal of GSCs, we examined the effects of FoxM1 or PDGF-A on GSCproliferation. FoxM1 or PDGF-A knockdown significantly decreased cell proliferation of GSC11 and GSC20s (Fig. 3H). Also, a small fraction of apoptotic cells was observed in FoxM1 knockdown cells and to a less extent in PDGF-A knockdown cells (Fig. S4A). Furthermore, GSCs have been postulated to have intrinsic resistance to chemotherapy including temozolomide (TMZ), a standard chemotherapy for newly diagnosed GBM patients. Because the above finding indicated that FoxM1 is important to the stemness of GSC, thus, we determined whether FoxM1 inhibition leads to an increase in sensitivity to TMZ. FoxM1 knockdown significantly decreased cell viability of GSC11, GSC20s and GSC267 cells after TMZ treatment as compared with sh-control (Fig. S4B–D). PDGF-A knockdown also significantly decreased cell viability after TMZ treatment, but to a less extent than FoxM1 knockdown (Fig. S4B–D). Together, these results indicate that FoxM1 inhibition decreased cell proliferation and increases the chemosensitivity of GSCs to TMZ.
FoxM1 regulates both STAT3 expression and STAT3 activation
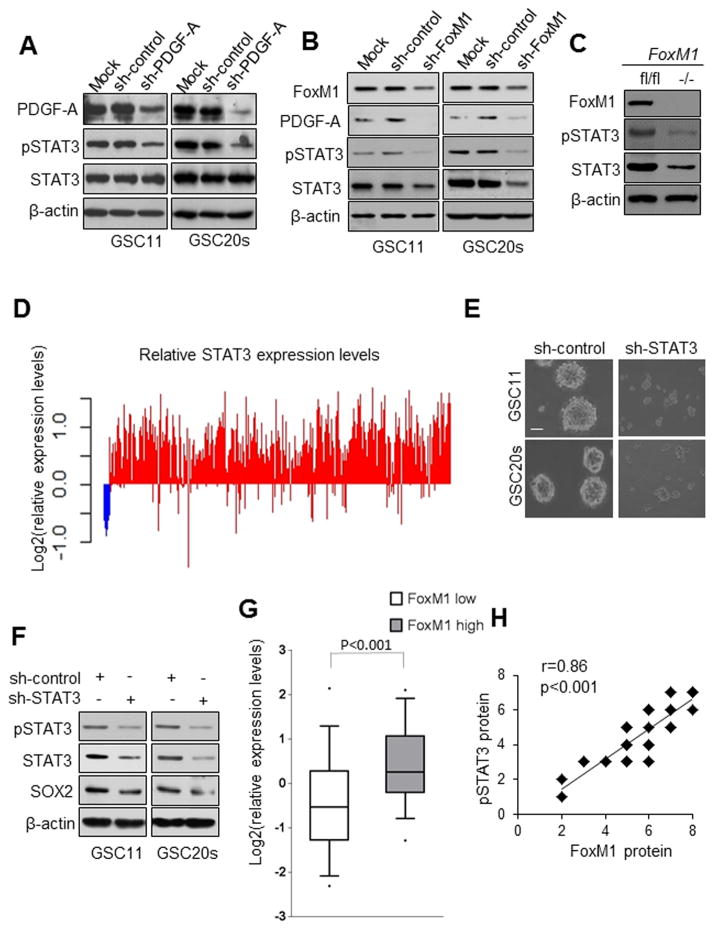
We next examined the mechanism by which the FoxM1-PDGF-A axis regulates self-renewal of GSCs. Since STAT3 is a critical node in PDGF signaling, we investigated whether STAT3 is a downstream component of FoxM1 signaling in GSCs. Knocking down PDGF-A or FoxM1 reduced STAT3 phosphorylation in both GSC11 and GSC20s cells (Fig. 4A,B). However, surprisingly, knocking down FoxM1 resulted in not only decreased STAT3 phosphorylation but also decreased STAT3 expression (Fig. 4B). In contrast to FoxM1 knockdown, PDGF-A knockdown decreased STAT3 phosphorylation but did not change STAT3 expression (Fig. 4A). Moreover, deletion of FoxM1 in FoxM1fl/fl NSCs resulted in a strikingly lower level of STAT3 expression compared with the level in FoxM1 wild-type NSCs (Fig. 4C), indicating that STAT3 is a downstream target of FoxM1. These results suggest that FoxM1 plays dual roles in STAT3 signaling, i.e., FoxM1 regulates both STAT3 expression and STAT3 activation.
Figure 4. FoxM1 regulates both STAT3 expression and STAT3 activation.
(A) Activated STAT3 (pSTAT3) and total STAT3 proteins detected in GSCs expressing sh-control or sh-PDGF-A. (B) Cellular levels of PDGF-A, pSTAT3, and total STAT3 proteins detected by Western blotting in GSCs expressing sh-control or sh-FoxM1. (C) pSTAT3 and total STAT3 protein detected in FoxM1fl/fl and FoxM1−/− NSCs. (D) Assessment of Stat3 gene expression from TCGA was conducted on GBMs tumors (n=594, red) and nontumor controls (n=10, blue). The fold changes were relative to the median log2 value. (E) Photographs of neurosphere of GSC11 cells expressing sh-control or sh-STAT3. Bar, 10μm. (F) Western blotting of SOX2 in GSC11 and GSC20s cells expressing sh-control or sh-STAT3. (G) Box plots of relative expression of STAT3 in FoxM1low or FoxM1high GBMs from a TCGA data set. Boxes show median 25th and 75th percentiles, while whiskers represent the 5th and the 95th percentiles. Outliers are shown as individual points. t-test was used to test statistical significance. (H) FoxM1 expression correlated with elevated expression of pSTAT3 in 50 GBM patient samples. The correlation was significant as determined by Pearson’s correlation test.
STAT3 expression is elevated in GBM specimen and correlated with FoxM1 expression
To determine the importance of STAT3 in GBM, we have assessed the gene expression of STAT3 from TCGA on 594 cases of GBM tumors and 10 cases of normal brain tissues; and found that most GBM tumors expressed higher STAT3 than normal tissue (Fig. 4D). Moreover, STAT3 knockdown inhibited the self-renewal of both GSC11 and GSC20s cells (Fig. 4E) and downregulated the levels of pSTAT3 and SOX2 (Fig. 4F), indicating that STAT3 is required for maintaining the stemness of GSCs.
The above results provide a compelling rationale for studying the mechanisms for the regulation of STAT3 expression in tumor cells. We evaluated the association of STAT3 and FoxM1 expression in the GBM tumors from TCGA dataset which were characterized into FoxM1high and FoxM1low subpopulation as described in Figure 1B, and found that higher STAT3 expression was enriched in the FoxM1high subgroup (Fig. 4G). Consistently, the level of activated STAT3 detected by nuclear staining of phosphorylated STAT3 protein, also correlated with the level of FoxM1 expression in the 50 GBM specimens described in Figure 1C (Fig. 4H).
FoxM1 increases STAT3 expression by enhancing β-catenin/TCF4 binding to the STAT3 gene promoter
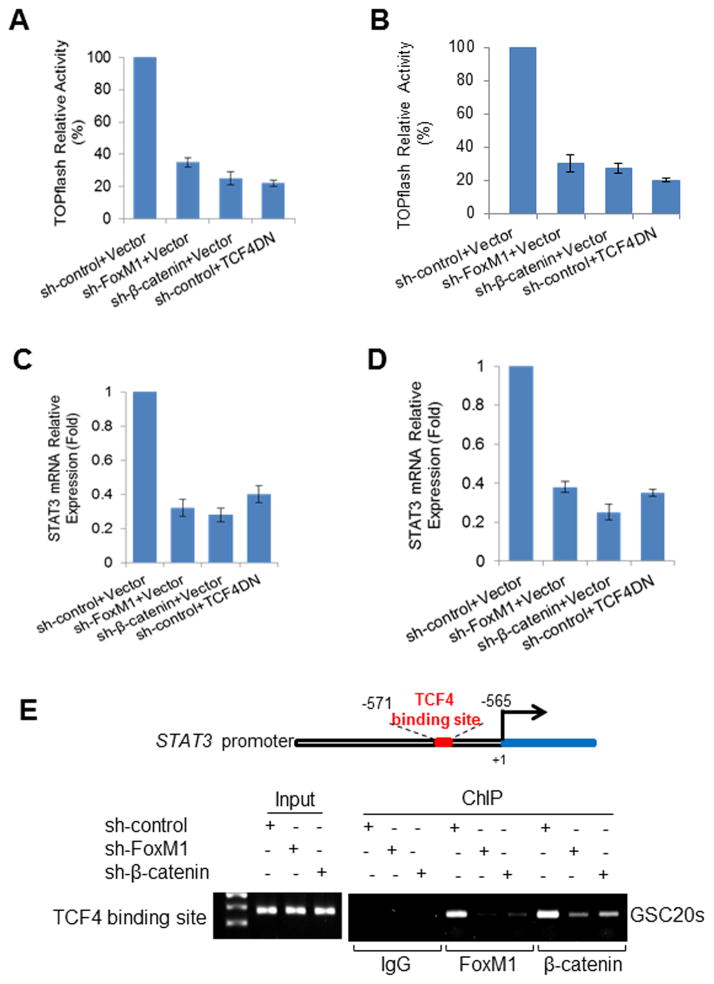
We showed that FoxM1 can bind to β-catenin to enhance expression of its target genes (38) and a previous study reported that β-catenin may affect STAT3 transcription (43). Thus, we tested whether FoxM1 regulates STAT3 expression via β-catenin. Knocking down β-catenin or inhibiting TCF4, the key transcriptional factor of β-catenin pathway, by TCF4-DN (dominate-negative TCF4) significantly decreased the transcriptional activity of β-catenin in GSCs, as indicated by TOPflash activity (Fig. 5A, 5B). Knocking down FoxM1 also significantly decreased the transcriptional activity of β-catenin in GSCs (Fig. 5A, 5B). Consistent with this finding, knocking down FoxM1 or β-catenin or inhibiting TCF4 in GSCs significantly decreased STAT3 mRNA expression (Fig. 5C, 5D), suggesting that FoxM1 regulates STAT3 expression via the β-catenin/TCF4 transcriptional complex. Moreover, by using primers flanking the TCF4-binding site (−571 to −565 bp) in the STAT3 promoter, we found that both β-catenin and FoxM1 bound to the STAT3 promoter (Fig. 5E). Furthermore, β-catenin knockdown inhibited FoxM1 binding to the STAT3 promoter, and FoxM1 knockdown inhibited β-catenin binding to the STAT3 promoter (Fig. 5E), indicating that β-catenin and FoxM1 are dependent on each other for binding to the promoter. These results demonstrated that FoxM1 enhances STAT3 expression by promoting β-catenin/TCF4 binding to the STAT3 promoter.
Figure 5. FoxM1 increases STAT3 expression by enhancing β-catenin/TCF4 binding to the STAT3 gene promoter.
(A, B) GSC cells expressing different combinations of shRNAs for control, FoxM1, or β-catenin and expression vector for control or TCF4DN were subjected to TOP-Flash luciferase assay. (C, D) The above cells were subjected to real-time PCR to detect STAT3 mRNA expression levels. Values are mean ± SD for triplicate samples. (E) ChIP assays on TCF4/LEF-1 binding site of the STAT3 promoter were performed in GSC20s cells expressing shRNAs for control, FoxM1, or β-catenin.
FoxM1 regulates growth factor- and cytokine-induced STAT3 activation
To determine the mechanism by which FoxM1 maintains the stemness of GSCs, we investigated whether FoxM1 affects STAT3 activation by growth factors and cytokines in GSCs. Exogenous PDGF-AA induced phosphorylation of STAT3 in both sh-control and sh-FoxM1 cells (Fig. 6A). However, the pSTAT3 level increased less in sh-FoxM1 than in sh-control cells, indicating that FoxM1 knockdown inhibited STAT3 activation induced by PDGF-AA because of lower expression of total STAT3. Next, we investigated whether FoxM1 affects STAT3 activation by EGF or IL-6 in GSCs, since STAT3 is a common node in multiple stem cell maintenance pathways, including EGF and IL-6, both of which promote self-renewal and tumorigenic potential of GSCs by activating STAT3 (27). Both EGF and IL-6 immediately activated STAT3, but the GSCs with FoxM1 knockdown were less responsive to EGF or IL-6 treatment than were the normal control cells (Fig. 6A). These results indicated that FoxM1 regulates growth factor- and cytokine-induced STAT3 activation.
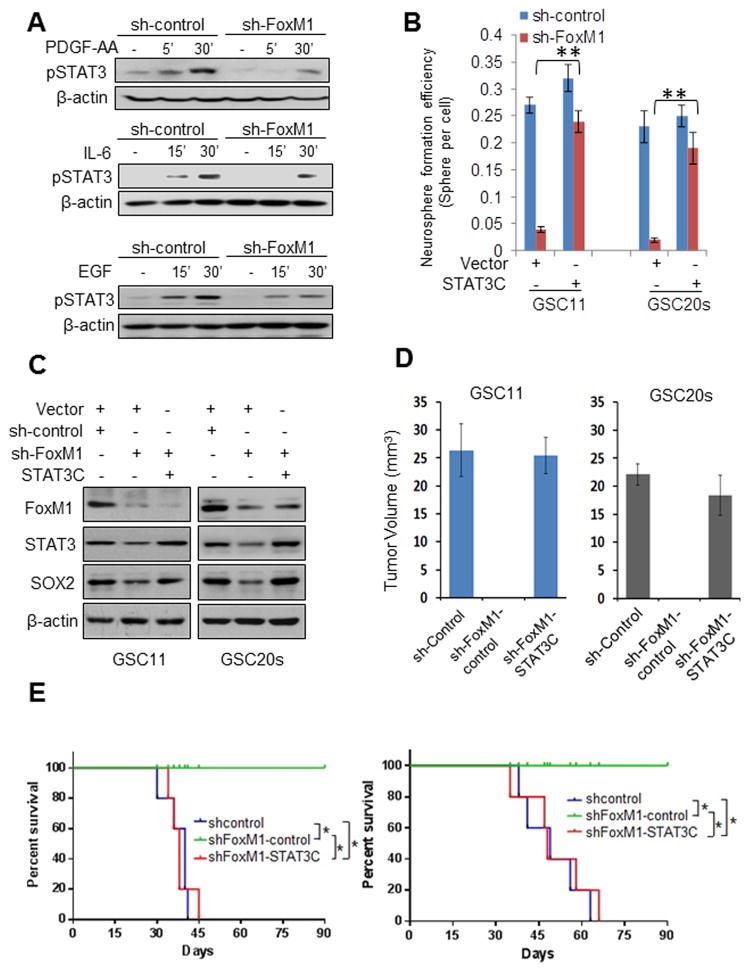
Figure 6. FoxM1 regulates STAT3 activation, and STAT3C reinstates self-renewal and tumor growth of GSCs with depleted FoxM1.
(A) GSC11-sh-control or GSC11-sh-FoxM1 cells were deprived of growth factors overnight and washed with fresh medium and then treated with PDGF-AA, EGF, or IL-6 for different times. pSTAT3 levels were detected by Western blotting in the cells. (B) Neurosphere formation efficiency of in GSC11 and GSC20s cells expressing sh-control or sh-FoxM1 and with STAT3C or a vector control. Values are mean ± SD for triplicate samples. (C) SOX2 and STAT3 expression levels were detected by Western blotting in GSC11 and GSC20s cells expressing sh-control or sh-FoxM1 and with STAT3C or a vector control. (D) GSC11 and GSC20s cells (5 × 105 cells/mouse) expressing the indicated shRNAs and plasmids were implanted intracranially in nude mice. After 40 days, the mice were sacrificed. Tumor volumes were measured by using length (a) and width (b) in H&E-stained sections and were calculated using the equation: V = ab2/2. Data represent results of 5 mice per group of two independent experiments. Values are mean ± SD. (E) The tumor cells in (A) were implanted intracranially in nude mice (n = 5). Mice were killed when they were moribund or 90 days after implantation. Survival of mice was evaluated by Kaplan-Meier analysis. *P <0.001. The result is from one representative experiment of two.
Constitutively active STAT3 rescues the inhibitory effects of FoxM1 knockdown on the stemness and tumorignecity of GSCs
To ascertain FoxM1 regulates the stemness of GSCs by affecting STAT3, we tested whether adding back activated STAT3 could reverse the effects of FoxM1 inhibition by using GSC cell lines that stably expressed STAT3C (constitutively active STAT3). In sphere formation assays, STAT3C largely rescued the consequences of FoxM1 knockdown: the sphere formation efficiency in sh-FoxM1-STAT3C GSCs was up to 90% than that in sh-control-vector GSCs (Fig. 6B, S5A). Moreover, SOX2 expression levels were restored in sh-FoxM1-STAT3C GSCs compared to sh-control-vector GSCs (Fig. 6C). These results indicated that activated STAT3 is important for FoxM1’s impact on the stemness of GSCs.
Next, we examined whether ectopic expression of STAT3C rescued tumor growth inhibition caused by FoxM1 knockdown. All mice injected with GSC11 and GSC20s cells expressing sh-control developed GBM but the mice injected with sh-FoxM1 GSCs did not, as indicated by tumor volumes (Fig. 6D). Forced expression of STAT3C in sh-FoxM1 cells largely abolished the tumor growth inhibition (Fig. 6D) and the decreased survival of mice by FoxM1 downregulation (Fig. 6E). Furthermore, Sox2 and Ki67, a cellular marker for proliferation, were highly expressed in the tumor tissues of sh-control group and sh-FoxM1+STAT3C group (Fig. S5B). These data demonstrated that STAT3 is critical for FoxM1’s promotion of the stem cell phenotype and tumorigenic potential of GSCs.
FoxM1 promotes glioma cells reprogramming to GSC-like cells via PDGF-A and STAT3
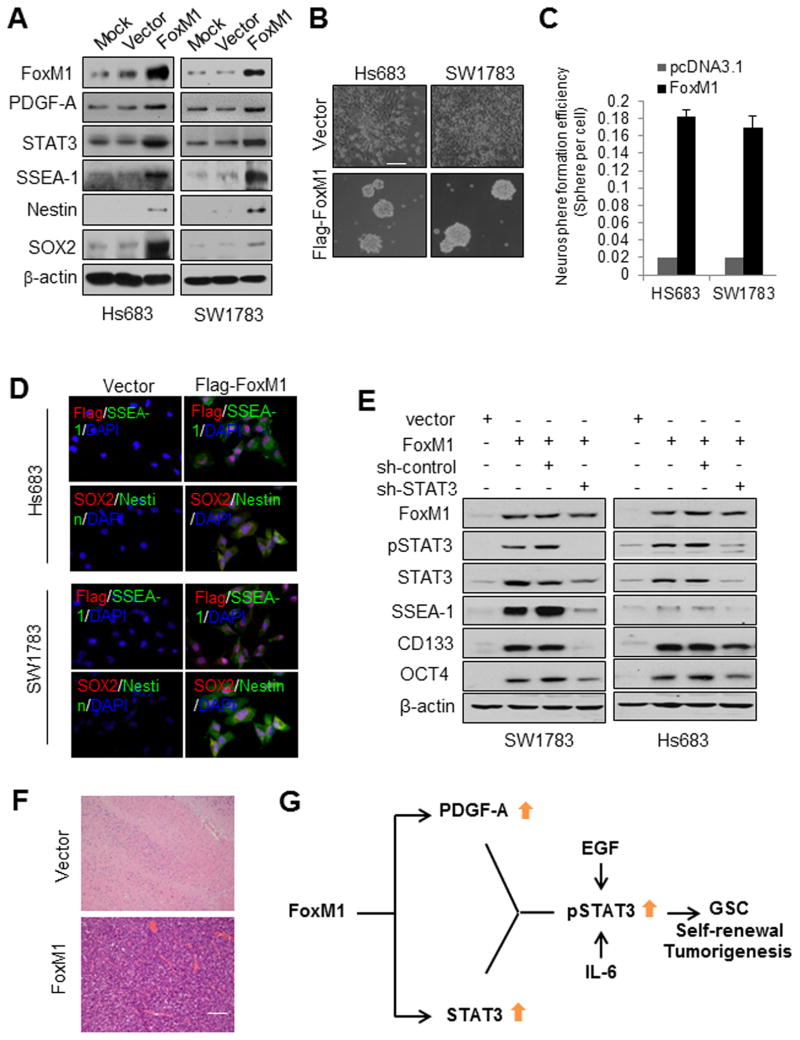
To further ascertain whether PDGF-A and STAT3 mediates the tumorigenic effect of FoxM1, we examined the tumorigenicity of non-GSC glioma cells overexpressing FoxM1 but deficient in STAT3. SW1783 and Hs683 glioma cells have low levels of the FoxM1 protein (31). Overexpression of FoxM1 in SW1783 and Hs683 cells caused them to exhibit GSC characteristics: SW1783-FoxM1 and Hs683-FoxM1 cells were able to form neurospheres and expressed stem cell markers SSEA-1, SOX2, and Nestin (Fig. 7A–D, Fig. S6). Furthermore, overexpression of FoxM1 in SW1783 and Hs683 cells significantly increased PDGF-A and STAT3 expression levels and activity of STAT3 (Fig. 7A, Fig. S7A). In contrast, knockdown of STAT3 in SW1783-FoxM1 and Hs683-FoxM1 cells abolished elevated expression of stem cell markers SSEA-1, SOX2, CD133, OCT4 and Nestin (Fig. 7E). Taken together, these findings indicated that FoxM1 overexpression is responsible for reprogramming non-stem glioma cells to GSC-like cells and that FoxM1’s impact on stemness of glioma cells depends on STAT3.
Figure 7. FoxM1 reprograms Hs683 and SW1783 glioma cells to GSC-like cells and initiates tumor formation.
(A) Expression of the indicated proteins was determined by Western blotting in Hs683 and SW1783 cells stably expressing Flag-FoxM1 or control vector. (B) Photographs of neurosphere of the cells in (A) cultured in neurosphere medium for 10 days. (C) Neurosphere formation efficiency of the cells in (B). Values are mean ± SD for triplicate samples. (D) Cells expressing Flag-FoxM1 or control vector were dissociated from spheres and plated on coverslips coated with poly-l-ornithine and fibronectin, and the indicated markers were examined via immunofluorescence. Cells were processed for triple immunofluorescence staining for Flag-FoxM1 (red), SSEA-1 (green), and nuclei (DAPI, blue) or SOX2 (red), Nestin (green), and nuclei (DAPI, blue). (E) Western blotting of stem cell markers in Hs683 and SW1783 cells expressing different combinations of expression vectors for control and FoxM1 and shRNAs for control and STAT3. (F) Sections of tumors produced by Hs683-FoxM1 cells or brain tissues from mice injected with Hs683-vector cells were HE-stained. Bar is 200μm. (G) The working model: a FoxM1-driven feed-forward STAT3-activation signaling loop is required for maintenance of self-renewal and tumorigenicity of GSCs.
Furthermore, SW1783 and Hs683 cells did not form brain tumors in nude mice, but SW1783-FoxM1 and Hs683-FoxM1 cells did form brain tumors, which possessed features of human GBM, including necrosis and invasion (Fig. 7F, Fig. S7B). The tumors also expressed SSEA-1 and Nestin (Fig. S7C&D). These results indicate that FoxM1 promotes glioma cells reprogramming to GSC-like cells and initiates tumor formation via PDGF-A and STAT3.
Discussion
Here, we demonstrate that FoxM1 is required for the expression of PDGF-A and STAT3 in GSCs. Constitutively active STAT3 rescued the effects of FoxM1 knockdown on the stemness of GSCs, indicating that FoxM1 signals through STAT3 in GSCs. Overexpression of FoxM1 reprogrammed glioma cells to GSC-like cells and enhanced their PDGF-A expression and STAT3 activation, whereas inhibition of STAT3 in GSCs diminished the FoxM1-induced stemness. Therefore, FoxM1-induced expression of PDGF-A and STAT3 represents a novel and critical mechanism for controlling GSC self-renewal and tumorigenesis (Fig. 7G).
FoxM1 plays dual roles in PDGF-A signaling
In this study, we found that FoxM1 plays dual roles in PDGF-A signaling to maintain GSC growth. FoxM1 regulates both PDGF-A expression and activation of STAT3. Overexpression of PDGF-A signaling components has been observed in GBMs, possibly associated with tumor initiation and malignant progression. For example, exogenous PDGF-AA or PDGF-A long isoform (PDGF-AL) can, to different extents, induce a glioma-like mass (18,44). These findings imply that autocrine or paracrine PDGF is a key signal for glioma initiation and progression. We found that PDGF-A signaling driven by FoxM1 was required for self-renewal of GSCs and tumorigenesis, which further confirms the role of PDGF-A in glioma initiation and progression.
PDGF-A promoter structure and function have previously been investigated (45). Several transcription factors, such as SP-1, Egr-1, and Pura, regulate PDGF-A expression in vitro via promoter binding pattern (45). The current study is the first, however, to show that FoxM1 regulates the transcription of PDGF-A. Our study also elucidated a mechanism for overexpression of PDGF-A in glioma for the first time.
FoxM1 establishes a link between PDGF signaling and Wnt signaling associated with GSC biology
A growing body of evidence suggests that PDGF signaling is required for NSC differentiation and oligodendrocyte precursor cell proliferation (46–48). Hyperactivated PDGF signaling is a common event in gliomagenesis and has been implicated in tumor initiation. On the other hand, our recent studies suggested that the Wnt-FoxM1/β-catenin pathway is critical for GSC maintenance and tumorigenicity (38). Thus, our results from the current study not only support the importance of PDGF signaling in GSC biology but also uncover a key node in FoxM1-mediated crosstalk between the two key forms of stem cell signaling: PDGF signaling and Wnt/β-catenin signaling.
FoxM1 and STAT3 are part of the key signaling node in GSC self-renewal
Recent accumulating evidence indicates that FoxM1 is required for maintaining stemness and self-renewal of stem cells, including neuronal precursors and GSCs (49) (50) (38). For example, FoxM1 stimulates Bmi1 expression to promote self-renewal of neural stem/progenitor cells (50). Also, FoxM1 signaling is essential for proliferation, self-renewal, and tumorigenesis of GSCs by regulating β-catenin activation (38).
STAT3 regulates diverse cellular processes, including cell growth, differentiation, and apoptosis, and is frequently activated during tumorigenesis. Moreover, STAT3 is required for maintenance of the stem-like characteristics of GSCs (24). Whereas the above findings suggest that both FoxM1 and STAT3 are critical for NSC and GSC growth, our results in the current study establish a direct link between FoxM1 and STAT3 and reveal an example for transcription regulation network in maintaining renewal of GSCs. This finding further suggests that the FoxM1 and STAT3 transcription regulation network is a potential target for novel therapeutic strategies against malignant glioma.
Supplementary Material
Acknowledgments
We thank Drs. Tucker Collins and Ryozo Nagai for PDGF-A promoter (wild-type and mutants). We thank Ms. Stephanie Deming in MD Anderson’s Department of Scientific Publications for editing the manuscript. This work was supported in part by NCI grants R01CA157933, R01CA182684, R21CA152623, and P50CA127001, and the National Science Foundation of China (No. 81372718 to AG).
References
- 1.Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468(7325):824–8. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 2.Wang R, Chadalavada K, Wilshire J, Kowalik U, Hovinga KE, Geber A, et al. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468(7325):829–33. doi: 10.1038/nature09624. [DOI] [PubMed] [Google Scholar]
- 3.Cheng L, Huang Z, Zhou W, Wu Q, Donnola S, Liu JK, et al. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153(1):139–52. doi: 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–6. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nister M, Heldin CH, Westermark B. Clonal variation in the production of a platelet-derived growth factor-like protein and expression of corresponding receptors in a human malignant glioma. Cancer research. 1986;46(1):332–40. [PubMed] [Google Scholar]
- 7.Lokker NA, Sullivan CM, Hollenbach SJ, Israel MA, Giese NA. Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells: evidence that the novel PDGF-C and PDGF-D ligands may play a role in the development of brain tumors. Cancer research. 2002;62(13):3729–35. [PubMed] [Google Scholar]
- 8.Hermanson M, Funa K, Hartman M, Claesson-Welsh L, Heldin CH, Westermark B, et al. Platelet-derived growth factor and its receptors in human glioma tissue: expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer research. 1992;52(11):3213–9. [PubMed] [Google Scholar]
- 9.Di Rocco F, Carroll RS, Zhang J, Black PM. Platelet-derived growth factor and its receptor expression in human oligodendrogliomas. Neurosurgery. 1998;42(2):341–6. doi: 10.1097/00006123-199802000-00080. [DOI] [PubMed] [Google Scholar]
- 10.Martinho O, Longatto-Filho A, Lambros MB, Martins A, Pinheiro C, Silva A, et al. Expression, mutation and copy number analysis of platelet-derived growth factor receptor A (PDGFRA) and its ligand PDGFA in gliomas. British journal of cancer. 2009;101(6):973–82. doi: 10.1038/sj.bjc.6605225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guha A, Dashner K, Black PM, Wagner JA, Stiles CD. Expression of PDGF and PDGF receptors in human astrocytoma operation specimens supports the existence of an autocrine loop. International journal of cancer Journal international du cancer. 1995;60(2):168–73. doi: 10.1002/ijc.2910600206. [DOI] [PubMed] [Google Scholar]
- 12.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA: a cancer journal for clinicians. 2010;60(3):166–93. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y, Kim E, Wu Q, Guryanova O, Hitomi M, Lathia JD, et al. Platelet-derived growth factor receptors differentially inform intertumoral and intratumoral heterogeneity. Genes & development. 2012;26(11):1247–62. doi: 10.1101/gad.193565.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng H, Hu B, Liu KW, Li Y, Lu X, Cheng T, et al. Activation of Rac1 by Src-dependent phosphorylation of Dock180(Y1811) mediates PDGFRalpha-stimulated glioma tumorigenesis in mice and humans. The Journal of clinical investigation. 2011;121(12):4670–84. doi: 10.1172/JCI58559. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Westermark B, Heldin CH, Nister M. Platelet-derived growth factor in human glioma. Glia. 1995;15(3):257–63. doi: 10.1002/glia.440150307. [DOI] [PubMed] [Google Scholar]
- 16.Kaetzel DM, Reid JDt, Pedigo N, Zimmer SG, Boghaert ER. A dominant-negative mutant of the platelet-derived growth factor A-chain increases survival of hamsters implanted intracerebrally with the highly invasive CxT24-neo3 glioblastoma cell. Journal of neuro-oncology. 1998;39(1):33–46. doi: 10.1023/a:1005905217361. [DOI] [PubMed] [Google Scholar]
- 17.Shamah SM, Stiles CD, Guha A. Dominant-negative mutants of platelet-derived growth factor revert the transformed phenotype of human astrocytoma cells. Molecular and cellular biology. 1993;13(12):7203–12. doi: 10.1128/mcb.13.12.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, et al. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51(2):187–99. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 19.Zhong Z, Wen Z, Darnell JE., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264(5155):95–8. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 20.Wang YZ, Wharton W, Garcia R, Kraker A, Jove R, Pledger WJ. Activation of Stat3 preassembled with platelet-derived growth factor beta receptors requires Src kinase activity. Oncogene. 2000;19(17):2075–85. doi: 10.1038/sj.onc.1203548. [DOI] [PubMed] [Google Scholar]
- 21.Garcia R, Yu CL, Hudnall A, Catlett R, Nelson KL, Smithgall T, et al. Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research. 1997;8(12):1267–76. [PubMed] [Google Scholar]
- 22.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nature reviews Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luwor RB, Stylli SS, Kaye AH. The role of Stat3 in glioblastoma multiforme. Journal of clinical neuroscience : official journal of the Neurosurgical Society of Australasia. 2013;20(7):907–11. doi: 10.1016/j.jocn.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem cells. 2009;27(10):2383–92. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer cell. 2013;23(6):839–52. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guryanova OA, Wu Q, Cheng L, Lathia JD, Huang Z, Yang J, et al. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer cell. 2011;19(4):498–511. doi: 10.1016/j.ccr.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Lathia JD, Wu Q, Wang J, Li Z, Heddleston JM, et al. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem cells. 2009;27(10):2393–404. doi: 10.1002/stem.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abou-Ghazal M, Yang DS, Qiao W, Reina-Ortiz C, Wei J, Kong LY, et al. The incidence, correlation with tumor-infiltrating inflammation, and prognosis of phosphorylated STAT3 expression in human gliomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(24):8228–35. doi: 10.1158/1078-0432.CCR-08-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Stark GR. Roles of unphosphorylated STATs in signaling. Cell research. 2008;18(4):443–51. doi: 10.1038/cr.2008.41. [DOI] [PubMed] [Google Scholar]
- 30.Gao SP, Bromberg JF. Touched and moved by STAT3. Science’s STKE : signal transduction knowledge environment. 2006;2006(343):pe30. doi: 10.1126/stke.3432006pe30. [DOI] [PubMed] [Google Scholar]
- 31.Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer research. 2006;66(7):3593–602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- 32.Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochimica et biophysica acta. 2012;1819(1):28–37. doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Lin B, Madan A, Yoon JG, Fang X, Yan X, Kim TK, et al. Massively parallel signature sequencing and bioinformatics analysis identifies up-regulation of TGFBI and SOX4 in human glioblastoma. PloS one. 2010;5(4):e10210. doi: 10.1371/journal.pone.0010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Boom J, Wolter M, Kuick R, Misek DE, Youkilis AS, Wechsler DS, et al. Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. The American journal of pathology. 2003;163(3):1033–43. doi: 10.1016/S0002-9440(10)63463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hodgson JG, Yeh RF, Ray A, Wang NJ, Smirnov I, Yu M, et al. Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro-oncology. 2009;11(5):477–87. doi: 10.1215/15228517-2008-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Zhang N, Dai B, Liu M, Sawaya R, Xie K, et al. FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer research. 2008;68(21):8733–42. doi: 10.1158/0008-5472.CAN-08-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R, et al. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26(42):6212–9. doi: 10.1038/sj.onc.1210443. [DOI] [PubMed] [Google Scholar]
- 38.Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, et al. FoxM1 promotes beta-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer cell. 2011;20(4):427–42. doi: 10.1016/j.ccr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu M, et al. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-beta-dependent cancer metastasis. The Journal of clinical investigation. 2014;124(2):564–79. doi: 10.1172/JCI71104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue J, Chen Y, Wu Y, Wang Z, Zhou A, Zhang S, et al. Tumour suppressor TRIM33 targets nuclear beta-catenin degradation. Nature communications. 2015;6:6156. doi: 10.1038/ncomms7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aizawa K, Suzuki T, Kada N, Ishihara A, Kawai-Kowase K, Matsumura T, et al. Regulation of platelet-derived growth factor-A chain by Kruppel-like factor 5: new pathway of cooperative activation with nuclear factor-kappaB. J Biol Chem. 2004;279(1):70–6. doi: 10.1074/jbc.M306621200. [DOI] [PubMed] [Google Scholar]
- 42.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hao J, Li TG, Qi X, Zhao DF, Zhao GQ. WNT/beta-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Developmental biology. 2006;290(1):81–91. doi: 10.1016/j.ydbio.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 44.Nazarenko I, Hedren A, Sjodin H, Orrego A, Andrae J, Afink GB, et al. Brain Abnormalities and Glioma-Like Lesions in Mice Overexpressing the Long Isoform of PDGF-A in Astrocytic Cells. PloS one. 2011;6(4) doi: 10.1371/journal.pone.0018303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khachigian LM, Williams AJ, Collins T. Interplay of Sp1 and Egr-1 in the Proximal Platelet-Derived Growth-Factor-a-Chain Promoter in Cultured Vascular Endothelial-Cells. J Biol Chem. 1995;270(46):27679–86. doi: 10.1074/jbc.270.46.27679. [DOI] [PubMed] [Google Scholar]
- 46.Raff MC, Lillien LE, Richardson WD, Burne JF, Noble MD. Platelet-Derived Growth-Factor from Astrocytes Drives the Clock That Times Oligodendrocyte Development in Culture. Nature. 1988;333(6173):562–65. doi: 10.1038/333562a0. [DOI] [PubMed] [Google Scholar]
- 47.Fruttiger M, Karlsson L, Hall AC, Abramsson A, Calver AR, Bostrom H, et al. Defective oligodendrocyte development and severe hypomyelination in PDGF-A knockout mice. Development. 1999;126(3):457–67. doi: 10.1242/dev.126.3.457. [DOI] [PubMed] [Google Scholar]
- 48.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Gene Dev. 2008;22(10):1276–312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuller U, Zhao Q, Godinho SA, Heine VM, Medema RH, Pellman D, et al. Forkhead transcription factor FoxM1 regulates mitotic entry and prevents spindle defects in cerebellar granule neuron precursors. Molecular and cellular biology. 2007;27(23):8259–70. doi: 10.1128/MCB.00707-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang ZB, Park HJ, Carr JR, Chen YJ, Zheng Y, Li J, et al. FoxM1 in Tumorigenicity of the Neuroblastoma Cells and Renewal of the Neural Progenitors. Cancer research. 2011;71(12):4292–302. doi: 10.1158/0008-5472.CAN-10-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.