Abstract
Purpose
BRAF-inhibition (BRAFi) therapy for advanced melanoma carries a high rate of secondary cutaneous squamous cell carcinoma (cSCC) and risk of other cancers. Ultraviolet (UV) radiation and α-genus human papillomavirus (HPV) are highly associated with SCC, but a novel role for β-genus HPV is suspected in BRAFi-cSCC. Cutaneous β-HPV may act in concert with host and environmental factors in BRAFi-cSCC.
Experimental Design
Primary BRAFi-cSCC tissue DNA isolated from patients receiving vemurafenib (Vem) or dabrafenib from two cancer centers was analyzed for the presence of cutaneous oncogenic viruses and host genetic mutations. Diagnostic specimens underwent consensus dermatopathology review. Clinical parameters for UV exposure and disease course were statistically analyzed in conjunction with histopathology.
Results
Twenty-nine patients contributed 69 BRAFi-cSCC lesions. BRAFi-cSCC had wart-like features (BRAFi-cSCC-WF) in 22% of specimens. During Vem therapy, BRAFi-cSCC-WF arose 11.6 weeks more rapidly than conventional-cSCC when controlled for gender and UV-exposure (p-value=0.03). Among all BRAFi-cSCC, β-genus HPV-17, HPV-38, HPV-111 were most frequently isolated and novel β-HPV genotypes were discovered (CTR, CRT-11, CRT-22). Sequencing revealed 63% of evaluated BRAFi-cSCCs harbored RAS mutations with PIK3CA, CKIT, ALK and EGFR mutations also detected.
Conclusions
We examined clinical, histopathologic, viral and genetic parameters in BRAFi-cSCC demonstrating rapid onset; wart-like histomorphology; β-HPV-17, HPV-38, and HPV-111 infection; UV damage; and novel ALK and CKIT mutations. Discovered β-HPV genotypes expand the spectrum of tumor-associated viruses. These findings enhance our understanding of factors cooperating with BRAF inhibition that accelerate keratinocyte oncogenesis as well as broaden the knowledge base of multifactorial mediators of cancer in general.
Introduction
Molecular inhibition of mutant BRAF protein in advanced melanoma with vemurafenib (Vem) or dabrafenib (Dab) (BRAFi) has improved patient survival but has also caused unanticipated adverse malignancies. BRAFi has been associated with cutaneous squamous cell carcinoma (cSCC) (1–7) with unusually aggressive histopathologic potential (8), and even recurrence of KRAS-mutant colonic adenocarcinoma and emergence of undiagnosed chronic myelomonocytic leukemia (9, 10). The frequency of cSCC (including cSCC and keratoacanthoma [KA]-type SCC) during BRAFi is reported in 7.0 – 26.7% of patients (mean 9.7%) (1–3, 11–18). In isolation, BRAFi-associated KA may complicate 5.8 – 14.3% of patients (mean 6.8%) (11, 13, 14, 16, 17). With the addition of MEK-inhibition, dabrafenib and trametinib (D+T) reduces the frequency of patients with cSCC/KA to 1.4 – 11.1% (D+T mean 2.2%) (12, 19, 20) while combination of Vem and MEK-inhibitor cobimetinib reduces cSCC to 2.8 – 9.3% (mean 5.0%) and KA to 0.8 – 1.6% (mean 1.0%) (17, 21).
The mechanism for cSCC development in otherwise healthy individuals is the result of ultraviolet (UV) radiation-induced transition mutations in host DNA (dipyrimidine or dipurine, e.g. A<–>G or C<–>T), which if they occur in tumor suppressor genes, such as TP53, can cause protein inactivation, loss of cell cycle control and cancer growth (22). UV radiation (particularly UVB) in conv.-cSCC induces mutations in HRAS (23). The limited studies on BRAFi-induced cSCC (BRAFi-cSCC) mechanisms have employed focused “hot-spot” genetic analyses to begin to understand the multiplicity of variables that may contribute to adverse BRAFi-cSCC (13, 24–26). The data from these efforts demonstrate in part that upstream RAS mutations act in concert with paradoxical activation of MAPK signaling caused by BRAFi in 30–60% of lesions (13, 24–26). The remaining lesions appear to lack RAS mutation, and several studies (4–7, 27) have reported a morphologic pattern of wart-like features (WF) in BRAFi-cSCC, suggesting an additional possible contribution of human papillomaviruses (HPV) to this secondary disease.
Squamous cell carcinoma of the cervix is the prototypical virus-mediated epithelial cancer (28, 29). Infection by the α-genus of HPV (particularly genotypes HPV-16, −18, −31, −33) is most often associated with this disease, while condylomata of genital-mucosal sites are associated with the α-HPVs HPV-6 and HPV-11. More recently, infection by the α-genus HPVs has been implicated as a cause of a subset of head and neck SCC (28) and of both cutaneous warts or verrucae and squamous cell carcinoma of genitomucosal surfaces and cutaneous sites in immunocompromised patient populations (29).
Human papillomavirus 16 (HPV-16) oncoproteins E6 and E7 drive carcinogenesis in genital mucosal sites. HPV-16 E6 and E7 oncoproteins inhibit cellular cell cycle regulatory proteins TP53 and pRB, respectively, leading to abnormal cell proliferation, lack of normal differentiation and prolonged survival. These features are all hallmarks of cancer.
Several studies attempting to link BRAFi-cSCC to α-HPV infection report negative results using immunohistochemical methods (5, 30, 31), based on detection of viral capsid protein L1 specific to α-HPVs (Dako K1H8 clone or ab2417, Abcam). A recent HPV DNA detection study of seven patients contributing 9 biopsies confirmed the lack of infection with α-HPVs (32).
Immunocompromised patients suffer an increased burden of cSCC, suggesting a pathophysiologic role for an infectious agent that may act with other host factors such as germline or somatic acquired genetic mutations. One promising candidate is β-HPV, which contribute to cSCC in solid organ transplant patients, and several immunodeficiency states including epidermodysplasia verruciformis (EV), WHIM syndrome (Warts, Hypogammaglobulinemia, Infections, and Myelokathexis), and chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) (see recent reviews in refs. 29, 33). The presence of wart-like histopathology in some BRAFi-cSCC, coupled with prior evidence for HPV-mediated carcinoma in healthy and immunosuppressed populations suggests that β-HPV contribute to the pathogenesis of BRAFi-cSCC.
Data support the linkage between BRAFi-SCC and non-α-HPV infection. In genital-mucosal SCC, p16 is a tumor suppressor overexpressed during carcinogenic HPV infection. Strong p16 immunoreactivity is a surrogate for HPV infection in these sites. Two studies found strong p16 expression in the majority of SCC and KA lesions arising during BRAFi, arguing for a tumorigenic viral role (14, 27).
Recently, the β-genus HPV-17 and Merkel cell polyomavirus were reported to coinfect one case of BRAFi-cSCC, again raising the possibility that β-HPV and other tumorigenic viruses participate in the development of BRAFi-cSCC (34). Further, in vitro evidence demonstrates cooperation between the paradoxical hyperactivation of the MAPK cascade with Vem and β-HPV infection (35).
Finally, while RAS mutations have been reported frequently in BRAFi-cSCC, identical mutations have been detected in benign epithelial skin lesions (36), indicating that other factors in addition to RAS may be necessary in the ontogeny of BRAFi-cSCC. The morphologic wart-like features we and others have observed in BRAFi-cSCC, combined with in vitro cooperation of Vem MAPK signaling with HPV infection, overexpression of p16 in BRAFi-cSCC and absence of α-HPV in BRAFi-cSCC, have served to establish the hypothesis that β-HPV infection may indeed be present in BRAFi-cSCC and serve a pathologic role. We sought to definitively examine and test for β-HPV and non-RAS genomic changes in primary clinical specimens from two independent cancer centers by employing central histomorphological review, robust viral analysis and next-generation sequencing to identify oncogenic changes in host DNA.
Methods
Case selection and patient protection
A retrospective analysis of previously procured diagnostic tissue samples was performed of patients treated with either study analogs of vemurafenib (Vem), or dabrafenib (Dab) with and without MEK-inhibition by trametinib (D+T) in clinical trials. Patients were seen by either a medical oncologist (JAS, DBJ, JRI) and/or dermatologist (BRM, WAL) in accordance with the clinical trial protocols (NCT00405587, NCT 00880321, NCT01006980, NCT01107418, NCT01072175, NCT01271803, NCT01037127) for metastatic or unresectable melanoma. Clinical signs and symptoms prompted biopsy of skin lesions for histopathologic evaluation and diagnosis by a board-certified dermatopathologist (JBR, JPZ, ASB). Twenty-nine patients had sufficient tissue from 69 biopsy specimens for morphologic examination and DNA extraction for viral genotype analysis as described below. Biopsies were excluded if they had insufficient residual tissue for nucleic acid extraction and histologic confirmation of lesional tissue in the post-sectioning H&E stained slide, or if clinical parameters were incomplete. Re-excisions of lesions previously considered in the study were excluded to eliminate repetition bias. This study was approved by the Vanderbilt University Medical Center Institutional Review board for patient safety and research integrity (#111786, #130461).
Clinical Parameters
The following patient-specific information was obtained for the study: gender, age, biopsy site, biopsy date, study medication, and date of initiation of therapy. Ultraviolet-light radiation from sun exposure was estimated by the biopsy site. In men, the head and neck and upper extremities were considered UV-exposed (UV +). In women, in addition to the criteria for men, the upper chest, back and lower legs were also considered “UV-exposed”. Biopsy sites that were not “UV-exposed” zones were considered “UV-protected” (UV –). Fitzpatrick skin-type was not utilized as a clinical criterion.
Pathology review
Histologic sections were prepared from formalin-fixed paraffin-embedded tissue and stained with hematoxylin and eosin under standard conditions. Lesional histomorphology was reviewed independently by four pathologists (DNC, JPZ, ASB, JBR) and descriptive and diagnostic consensus was reached. Histomorphology was determined to be conventional pattern of cutaneous squamous cell carcinoma (conv.-cSCC), keratoacanthoma-type cSCC (cSCC-KA), or cSCC with wart-like features (cSCC-WF). Conv. cSCC lesions are typified by keratinocyte proliferation in lobules with cytologic and nuclear atypia and invasion into the dermis. Cutaneous SCC-KAs were composed of a crateriform eosinophilic keratinaceous core with a pushing border of atypical keratinocytes with papillary dermal invasion. When either cSCC-KA or conv.-cSCC revealed wart-like features (WF) that included overlying hyperkeratosis, hyperparakeratosis, papillomatosis, irregular nuclear contours, glassy nuclear inclusions or koilocytic changes, the term cSCC-WF was applied.
DNA Extraction
Nucleic acid was extracted from formalin-fixed paraffin embedded tissue in the form of unstained slides or microtome-cut tissue curls and transferred to sterile microcentrifuge tubes. All sections were cut using a fresh knife-edge to prevent cross-contamination. Paraffin was removed via triplicate warmed xylene wash followed by centrifugation at 13,000 rpm for 5 minutes. Deparaffinized tissue was further washed three times with 100% ethanol followed by centrifugation at 13,000 rpm for 5 minutes. Tissues were dried at 37°C for 30 minutes, followed sequentially by proteinase digestion and DNA extraction using the QIAamp DNA Mini kit per manufacturer’s instructions (Qiagen, Germantown, MD).
DNA Quality determination and tumorigenic virus detection
The quality of the extracted DNA was assessed by β-globin reference gene PCR (37). HPV-DNA was amplified by PCR in the β-globin PCR-positive samples utilizing multiple nested primer systems designed for detection of broad ranges of HPV types (38–40). Isolation, cloning (TOPO TA cloning kit, Invitrogen Co. Carlsbad, CA) and sequencing of putative HPV-PCR products were carried out as described (34). HPV sequence data were aligned and identified to known HPV DNA sequences available through GenBank (National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD) using the BLAST program (41). A minimum of 5 clones was sequenced from the above TOPO-cloning procedure for HPV genotyping per specimen. Briefly, colony purified E. coli isolates were grown to an optimal density, and plasmids containing viral sequences were purified for sequencing. PCR was employed to detect MCPyV in the β-globin PCR-positive specimens. MCPyV small T gene-based primers were used (34). PCR products were resolved in 2.5% agarose gel and visualized with ethidium bromide for the expected 150bp band. MCPyV-PCR products were extracted from the agarose, TOPO-cloned (as described above for HPV) and sequenced. Viral DNA sequences were subjected to BLAST alignment.
Host Genetic Analysis
Multiplex PCR was performed on previously extracted genomic nucleic acids and next generation sequencing was performed using a custom amplicon panel (Ion Ampliseq Library 2.0, Ion OneTouch 200 Template v2, Ion PGM 200 Sequencing, Ion 318 Chip on Ion Torrent PGM using Torrent Suite 2.2 software). This assay interrogates regions of 35 known well-characterized cancer-related genes (Table S3). DNA quality was assured by qPCR for β-globin prior to library preparation. Sequence reads were analyzed in a CLIA-approved laboratory environment by determining target coverage, strand bias, read length, quality score and other parameters. Suitable reads were aligned to the reference human genome GrCH37 using CLCbio Genomics Workbench and CLCbio Genomics Server. Data was annotated using COSMIC and dbSNP databases. Allele frequency cut-off of ≥10% and/or ≥100x coverage with approximately equal strand bias were predefined inclusion criteria. Sanger sequencing was performed using standard methods with primers directed at corresponding oncogene mutations in PCR amplicons (Table S3) (Integrated DNA Technologies, Coralville, Iowa).
Statistical Analysis
A p-value less than or equal to 0.05 was defined as statistically significant prior to all analyses. The time from initiation of study medication (Vem, Dab, D+T) to biopsy of cSCC, termed hereafter as “latency,” was calculated by subtracting the initiation date from the biopsy date. To compare the latency of lesion onset between groups, we employed the Wilcoxon Rank Sum or Kruskal Wallis non-parametric tests using R (v.3.0.3 & 3.1.0, The R Foundation for Statistical Computing, Vienna, Austria). We studied the relationship between latency and morphology of Vem treated lesions while adjusting for UV exposure status and gender using a linear mixed-effects model with random intercept. The latencies of multiple lesions per patient were not considered as independent. Patient 6 (#6), on Dab therapy, was unusual with 7 lesions arising quickly. These displayed unique spindle-cell squamous carcinoma histomorphology and have been reported elsewhere (8). Another patient (#19) treated with D+T also had lesions arising quickly (Table S1). These two patients were omitted from multivariate model analyses as the number of lesions and rapidity of onset indicated these cases to be outliers.
Results
Demographics of the study
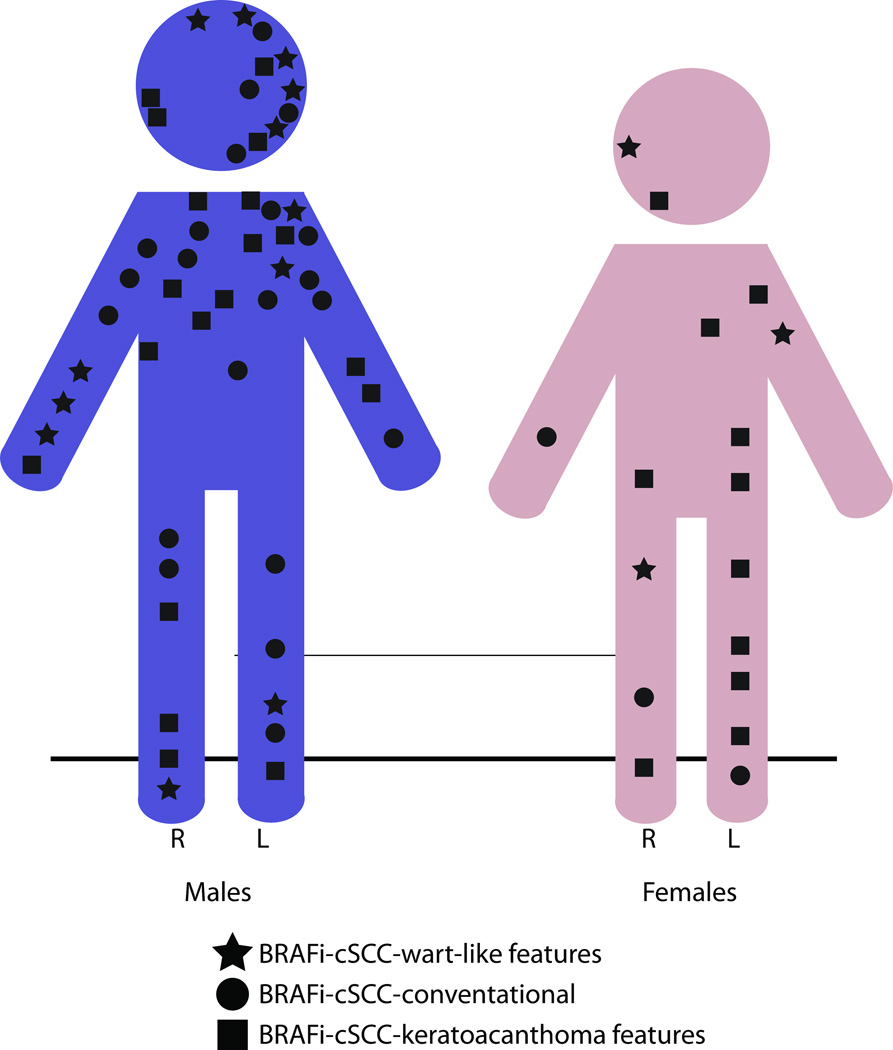
Histomorphological and viral analyses was performed on 69 cutaneous squamous cell carcinomas obtained during diagnostic procedures for 29 patients with BRAF V600 mutant-melanoma enrolled in clinical trial studies of vemurafenib (Vem, N = 22 patients, n = 53 lesions), dabrafenib (Dab, N= 4 patients, n = 10 lesions) or combination of dabrafenib with trametinib (MEK inhibitor) (D+T, N = 3 patients, n = 6 lesions) (Table 1). Twenty-one men and 8 women provided 52 and 17 skin samples for diagnosis, respectively (Table 1 and Fig 1). The overall mean patient age at biopsy was 63.2 years. Lesions in men were more often in areas of prior UV radiation exposure of the head, neck and upper extremities, with relative sparing of the trunk and abdomen (Fig. 1). A similar number of biopsies from UV-exposed and UV-protected sites, 37 (53.3%) and 32 (46.4%), respectively, were analyzed. The distribution of lesions in women included sun-exposed and sun-protected sites.
Table 1.
Patient and Specimen Parameters and Tumorigenic Virus Detection
| Patient | Sample | Age | Gender | Biopsy Site | UV-exposed | BRAF V600^ | Tx# | Latency (weeks) |
cSCC Morphology* |
HPV genotype^ | HPV genus | MCPyV# |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | P001 | 64 | M | Jaw line, L | Y | E | D | 11 | WF | 65, 80 | β,γ | NP |
| P003 | Forearm, R | Y | D | 11 | WF | 22 | β | N | ||||
| 2 | P005 | 58 | M | Lower leg, R | N | E | D | 50 | WF | ND | NP | |
| 3 | P006 | 73 | M | Shoulder, L | Y | E | V | 12 | KA | 36b | β | NP |
| 4 | P007 | 53 | F | Upper arm, L | Y | E | V | 4.0 | WF | ND | NP | |
| 5 | P008 | 63 | M | Eyelid, L | Y | E | V | 3.4 | WF | ND | NP | |
| P035 | Upper arm, L | Y | V | 11 | C | ND | N | |||||
| P036 | Upper arm, L | Y | V | 19 | KA | RTRX4/HPVX4b, CTR* |
β | N | ||||
| 6 | P010 | 63 | F | Flank, R | N | E | D | 4.6 | KA* | ND | NP | |
| P011 | Upper leg, L | N | D | 4.6 | KA* | ND | NP | |||||
| P012 | Lower abdomen, L | N | D | 4.6 | KA* | ND | NP | |||||
| P013 | Lower back, L | N | D | 4.6 | KA* | ND | NP | |||||
| P014 | Upper back, mid | N | D | 4.6 | KA* | ND | NP | |||||
| P015 | Scapular back, L | N | D | 4.6 | KA* | ND | NP | |||||
| 7 | P024 | 60 | F | Forearm, R | y | E | D+T | 52 | C | ND | Y | |
| P025 | Lower leg, R | N | D+T | 52 | C | 17 | β | N | ||||
| 8 | P040 | 63 | M | Upper forehead, L | Y | E | V | 12.0 | C | FA132 | β | N |
| P041 | Shoulder, R | Y | V | 12.0 | C | ND | Y | |||||
| 9 | P062 | 61 | M | Lateral neck, L | Y | E | V | 54 | C | 38, 17, FA14 | β | N |
| P067 | 62 | Chest, R | N | V | 77 | C | ND | N | ||||
| P069 | Upper leg, R | N | V | 62 | C | 17, 80 | β | N | ||||
| P070 | Chest, R | N | V | 64 | KA | 38, 76 | β | Y | ||||
| P071 | Lower leg, L | N | V | 64 | WF | 38, 105, 143, SIBX18 | β | N | ||||
| P073 | Lower leg, R | N | V | 65 | KA | 17, 24, 38, 124 | β | N | ||||
| 10 | P102 | 51 | M | Shoulder, L | Y | E | V | 28 | C | 17, 38 | β | N |
| 11 | P112 | 57 | M | Upper leg, L | Y | E | V | 16 | C | 17, 38, FA14 | β | N |
| P113 | Upper back, mid | N | V | 16 | C | 17, 22, 38, 111 | β | N | ||||
| 12 | P137 | 59 | F | Leg, L | Y | E | V | 19 | KA | 38, 111 | β | N |
| 13 | P144 | 70 | M | Back | N | E | D | 9.6 | C | 17, 151 | β | N |
| 14 | P151 | 64 | M | Lower leg, L | N | E | D+T | 28 | C | 7, 17, 38 | β,α | N |
| P153 | Upper arm, R | Y | D+T | 52 | C | 17, 22,38 | β | N | ||||
| 15 | P172 | 56 | M | Chest, L | N | E | V | 16 | WF | 17, 24, 38, 111 | β | N |
| P173 | Shoulder, L | Y | V | 16 | WF | 17, 38 | β | N | ||||
| 16 | P184 | 57 | F | Upper leg, R | N | E | V | 5.9 | WF | 38, FA7/FA162 | β | N |
| P187 | 58 | Ant. mandible, R | Y | V | 25 | KA | ND | Y | ||||
| 17 | P201 | 68 | M | Forearm, L | Y | E | V | 9.0 | C | SIBX20, vs42-1 | β | N |
| P202 | Lower leg, L | N | V | 9.0 | KA | ND | N | |||||
| P208 | Upper arm, R | Y | V | 21 | C | 110 | β | N | ||||
| P211 | Upper arm, L | Y | V | 24 | C | 17 | β | N | ||||
| P216 | Upper leg, R | N | V | 27 | C | 17, 38, 111 | β | Y | ||||
| P217 | Lower leg, R | N | V | 27 | KA | 111 | β | Y | ||||
| 18 | P220 | 83 | M | Face, temple, L | Y | E | V | 5.4 | WF | 15 | β | N |
| P235 | Upper back, R | N | V | 39 | KA | 23, 24 | β | N | ||||
| P237 | 84 | Face, cheek, R | Y | V | 46 | KA | SIBX20, vs42-1 | β | N | |||
| 19 | P238 | 70 | F | Lower leg, L | N | K | D+T | 4.3 | KA | 22, 38 | β | N |
| P239 | Lower leg, L | N | D+T | 4.3 | KA | 120 | β | Y | ||||
| 20 | P252 | 62 | M | Knee, L | N | E | V | 24 | C | 38, 111 | β | Y |
| P261 | Forehead, L | Y | V | 32 | KA | 38 | β | N | ||||
| 21 | P263 | 74 | M | Scalp, L | Y | E | V | 1.0 | WF | 24, 38, 111, DL231, FA14, VS20-4 |
β | N |
| P265 | Scalp, vertex | Y | V | 6.1 | WF | 38, 49, DL231, VS20-4, | β | N | ||||
| P269 | Forearm, R | Y | V | 24 | WF | 38, 111 | β | N | ||||
| P270 | Forearm, R | Y | V | 25 | WF | 38 | β | N | ||||
| 22 | P281 | 55 | M | Back, L | N | E | V | 13 | C | CRT-22*, VS42-1 SE68/KC49 |
β | N |
| P282 | Scalp, occiput | Y | V | 13 | C | 17, 38 | β | N | ||||
| P288 | Back, mid, R | N | V | 32 | KA | 38, 111 | β | N | ||||
| P290 | Upper back, L | N | V | 39 | C | 38 | β | N | ||||
| 23 | P333 | 55 | M | Hand, R | Y | E | V | 11 | KA | 38, 111 | β | N |
| P334 | Upper back, L | N | V | 11 | KA | 17, FA14 | β | N | ||||
| 24 | P335 | 60 | M | Lip, lower cut., L | Y | E | V | 11 | KA | 38 | β | N |
| P340 | Face, preauricular, R | Y | V | 5.6 | KA | 25 | β | N | ||||
| 25 | P353 | 42 | F | Lower leg, R | Y | D | V | 13 | C | ND | N | |
| P354 | Lower leg, L | Y | V | 13 | KA | 17, 76, 143 | β | Y | ||||
| 26 | P364 | 69 | F | Face, infraorbital, R | Y | E | V | 24 | WF | 8, 17, 80, 120 | β | N |
| 27 | P404 | 63 | M | Axilla, R | N | E | V | 16 | KA | 17, 38, 76 | β | N |
| 28 | P441 | 63 | M | Chest | N | E | V | 9.1 | KA | 20, 25, 145, CRT-11*, FA18 |
β | Y |
| P442 | Face, cheek | Y | V | 9.1 | C | 17, 124, FA14, FA15, vs42-1 |
β,γ | Y | ||||
| 29 | P467 | 69 | M | Upper arm, L | Y | R | V | 4.0 | KA | ND | Y | |
| P468 | Clavicle | Y | V | 7.3 | KA | 17, 38, 108 | β | N | ||||
| P469 | Lower leg | N | V | 12 | KA | 111, 124, FA14, SIBX18 | β | Y | ||||
Patient (P) identification sample number; male (M) or female (F) gender; left (L) or right (R) laterality; Ultraviolet (UV)-radiation exposed skin (Y); UV-radiation protected skin (N);
Primary melanoma tumor mutation: BRAF valine at amino-acid 600 (V600) mutated to glutamate (E), aspartate (D), lysine (K), or arginine (R);
Therapy (Tx): V = vemurafenib, D = dabrafenib, D+T = dabrafenib with trametinib.
Histomorphology of conventional (C) cutaneous squamous cell carcinoma (cSCC) with keratoacanthoma (KA) and invasive spindle-SCC (KA*) (ref 8) or with wart-like features (WF). Human papillomavirus (HPV) genotype identification, HPV genus of identified genotypes or not detected (ND); Merkel Cell Polyomavirus (MCPyV) identified: yes (Y), no (N), or not performed (NP).
Figure 1. Sun exposed sites most frequently harbor BRAFi-cSCC.
Distribution and histopathology of all adverse cutaneous squamous cell carcinoma (cSCC) lesions from male (blue, left) and female (rose, right) patients with advanced melanoma treated with BRAF-inhibition (BRAFi). Conventional-cSCC (circles), well-differentiated keratoacanthomatous–type SCC (squares) and conv.-cSCC or cSCC-KA with wart-like features (cSCC-WF, stars).
Primary melanoma genetics and therapy parameters
The majority of patients had metastatic or unresectable melanoma with mutations in the BRAF gene that resulted in valine (V) to glutamate (E) amino acid changes at position 600 (V600E, N = 26 patients). Three total patients (one with each mutation) had mutations that caused V600K (valine to lysine), V600D (valine to aspartate) and V600R (valine to arginine) contributing 2, 2 and 3 lesions, respectively (Table 1).
Histomorphology of BRAFi-cSCC
Features contributing to histomorphologic classification are reviewed in the Methods section. Thirty-six percent (36%, n=25) of lesions had conventional cSCC morphology, (BRAFi-conv.-cSCC), 42% (n=29) were BRAFi-cSCC with keratoacanthomatous features (BRAFi-cSCC-KA) and 22% (n=15) were BRAFi-cSCC with wart-like features (BRAFi-cSCC-WF). The morphologic distribution was similar to previously reported studies (1, 2, 18–20, 37, 38)(1, 2, 11, 13, 25, 26, 42).
Latency to development of lesions
For all patients, the median latency was 13 weeks with a maximum of 77 and minimum of 1 week (Table 1). The latency also depended on therapy administered. There were 53 lesions from patients on Vem, with 10 lesions from Dab-treated patients and 6 lesions from patients on D+T. The mean latency regardless of histopathology while on Vem (22 weeks) or Dab (10.9 weeks) was significantly shorter than D+T (32.1 weeks, p-value = 0.02) including patient #6 and #19.
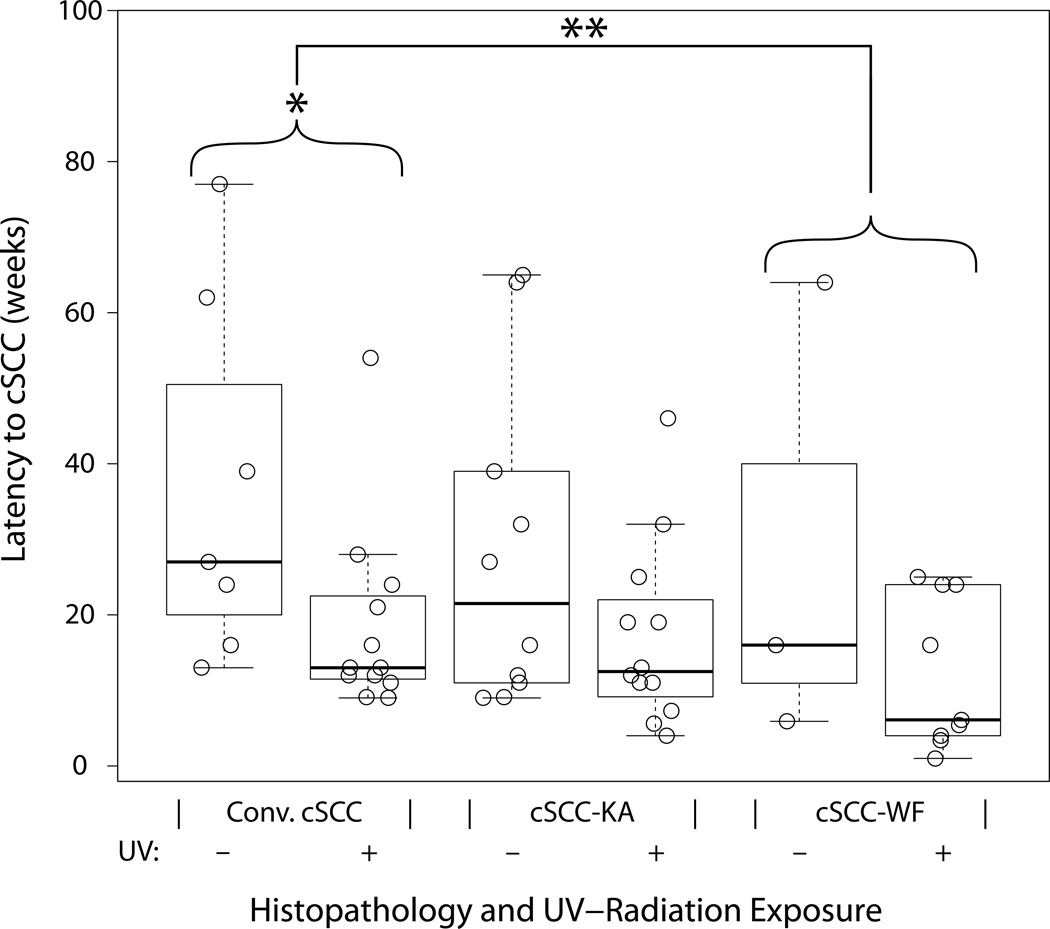
The median latency for patients on Vem therapy (N = 22 patients, n = 53 lesions) was shorter in UV-exposed sites (median 13 weeks, n = 33) compared to UV-protected regions (median 25.5 weeks, n = 20, p-value = 0.010), supporting the hypothesis that UV-DNA damage may accelerate BRAFi-cSCC. Further, the BRAFi-conv.-cSCC morphotype showed significantly more rapid onset from UV-exposed sites (Fig 3, median 13 weeks, n = 12) compared to UV-protected (median 27 weeks, n = 7, p-value = 0.016). Cutaneous BRAFi-SCC-KA did not have statistically significant UV-exposure latency differences: 21.5 weeks vs 12.5 weeks UV-protected vs UV-exposed (Fig. 3, n = 10 vs n=12, respectively).
Figure 3. BRAF-inhibition induced cutaneous squamous cell carcinoma latency by UV-radiation exposure and histopathology.
Time course of lesion development is multifactorial and statistically related to ultraviolet radiation exposure and is associated with characteristic histopathology. Box-whisker plot with median (black horizontal line), 25th and 75th percentile (box) and min and max (whiskers). All cSCC samples for vemurafenib treated patients are shown (circles) by histomorphologic subtype and presence (+) or absence (–) of ultraviolet (UV) radiation exposure. *Significant 14.0 week decrease in latency in conventional type cSCC with UV radiation exposure (p-value = 0.016). **Significant 11.6 week decrease in latency in cSCC-WF ver. conv.-cSCC when matched for gender and UV-exposure in a multivariate model (p-value = 0.030).
Latency to development of lesions by morphologic type
The morphologic subtype (BRAFi-conv.-cSCC, BRAFi-cSCC-KA or BRAFi-cSCC-WF) correlated with latency. Using a multivariate model we compared the mean latency difference between lesions with different morphology adjusted for UV-exposure and gender. The latency in BRAFi-cSCC-WF decreased significantly by 11.6 weeks (Fig. 3, p-value = 0.03) compared to BRAFi-conv.-cSCC for Vem treated patients’ lesions with the same UV-exposure and gender. Taken together, in the context of morphology regardless of UV exposure, Vem-treated patients tended to show longer median latency with cSCC-KA (13.0 weeks) and conv.-cSCC (18.5 weeks) compared to cSCC-WF (11 weeks); however, this difference did not meet statistical significance (p-value = 0.14). The D+T group showed no cSCC-WF and a majority of conv.-cSCC. The median latency for the D+T group is 52 weeks (n=4, after excluding patient #19).
Spectrum of HPV infection during BRAFi
Cutaneous β-genus HPVs were identified in the BRAFi-cSCC of all morphologic types, with the predominant subtypes being HPV-38 (n=28), HPV-17 (n=21), and HPV-111 (n=11) (Table 1, Fig S1, Table S1). Thirty-nine BRAFi-cSCC lesions were infected by more than one HPV genotype. Beta-HPVs previously associated with cSCC, HPV-5 (n=0) and HPV-8 (n=1), were infrequently found, and none of the subtypes associated with condylomata or SCC in genital-mucosal surfaces were identified (HPV-6, −11, −16, −18, etc.) (Table 1).
Seventeen samples were negative for virus detection in our study (Table 1) and were processed on the same day and same method as samples that were HPV or MCPyV positive. This reduces the likelihood of knife-blade contamination (43), supporting the reliability of the method to reduce false-positive testing.
Putative novel HPV genotypes identified
Three viral nucleic acid sequences from patients P005, P022, and P028, were unique, each with less than 90% homology to previously reported HPV genotypes in GenBank (NCBI) (Fig. S1, Table S1). These were designated as putative novel β-genus HPVs CTR, CRT-22, and CRT-11. Multiple alignment techniques demonstrate partial similarity of the novel HPV-CTR with previously described HPV-118, implying genus- or subgroup-level relatedness, while novel HPVs CRT-11 and CRT-22 demonstrate more distant relatedness to their closest known relatives, implying that they may represent novel HPV genotypes (Fig S2, Table S2) (40)(44). The phylogenetic relatedness of HPV isolates in this study are compared to more fully characterized α-, δ-, ε-, γ- and ξ-HPV viruses via available L1 genomic data (Fig S1).
Coincidence of HPV and MCPyV
Merkel cell polyomavirus (MCPyV) is frequently found in the skin and is associated with Merkel cell carcinoma, an aggressive cutaneous neuroendocrine carcinoma (28, 45). Thirteen BRAFi-cSCCs in our study were positive by genetic studies for MCPyV infection. Nine of these were coinfected with HPV (Table 1). Six of 9 were cSCC-KA type lesions, 3 were conv.-cSCC and none of the dual MCPyV/β-HPV infected lesions showed wart-like (cSCC-WF) morphology. Histomorphologic review confirmed that neuroendocrine carcinoma was not present in any of the MCPyV-positive cases of BRAFi-cSCC.
Cancer Gene Mutations and Polymorphisms in BRAFi-cSCC
Twenty-five (25) biopsies from 13 patients were available for next generation sequencing and analysis, excluding 3 biopsies with insufficient DNA quality or quanitity for further analysis. Nine lesions from 7 patients harbored mutations in major cancer genes (HRAS, KRAS, PIK3CA, CKIT, ALK, EGFR) with mean coverage of 214-fold. Fifteen (15) lesions harbored genetic mutations or single nucleotide polymorphisms (SNPs) in isolation or combination. Two (2) lesions had isolated HRAS Q61L (P003) and KRAS G12D (P144). In the remaining lesions, two (2) had mutations in CKIT (P005, P006), two (2) had mutations in PIK3CA (P008, P070), and one lesion each had mutations in ALK (P041) and EGFR (P071). All RAS mutations were confirmed by Sanger sequencing. Several single nucleotide polymorphisms (SNP) were also identified, most frequently PDGFRA V824 (8 of 25 lesion). Two of three PIK3CA mutations and one of two MET SNPs were in samples with sufficient DNA for Sanger confirmation (Table 2, italic font). PIK3CA mutations and RAS G12 codon mutations did not occur with the PDGFRA SNP V824. In the seven remaining lesions (7), we detected no mutations or polymorphisms (P007, P035, P036, P062, P069, P102, P153).
Table 2.
Host Genetic Mutations and Polymorphisms in Known Cancer-causing genes in BRAFi-cSCC.
| Specimen |
Parameters |
Oncoprotein Mutation |
SNPs in Oncoproteins |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tx-^ | Type* | UV# | HRAS | KRAS | PIK3CA | ALK | EGFR | CKIT | PDGFRA | CKIT | PTCH | Total | |
| P003 | Dab | WF | Y | Q61L | 1 | ||||||||
| P006 | Vem | KA | Y | G12T | M541L | 2 | |||||||
| P008 | Vem | WF | Y | Q61K | E542K | 2 | |||||||
| P040 | Vem | C | Y | Q61R | V824 | 2 | |||||||
| P041 | Vem | C | Y | F1174L | V824 | 2 | |||||||
| P144 | Dab | C | N | G12D | 1 | ||||||||
| P070 | Vem | KA | N |
E545K A1046T |
2 | ||||||||
| P071 | Vem | WF | N | V769M | V824 | 2 | |||||||
| P005 | Dab | WF | N | M541L | V824 | 2 | |||||||
| P024 | D+T | C | Y | V824 | 1 | ||||||||
| P025 | D+T | C | N | V824 | 1 | ||||||||
| P112 | Vem | C | Y | V824 | 1 | ||||||||
| P113 | Vem | C | N | V824 | 1 | ||||||||
| P137 | Vem | KA | Y | K546 | 1 | ||||||||
| P151 | D+T | C | N | T671 | 1 | ||||||||
Oncogenic amino acid changes from host genetic mutations and single nucleotide polymorphisms (SNPs) in oncogenes detected in BRAF-inhibition associated cutaneous squamous cell carcinomas during therapy for advanced melanoma. RAS mutations predominate with additional PIK3CA, ALK and EGFR mutations detected using Ampliseq (IonTorrent) with Sanger sequencing (italic) methods. Boxed pairs of samples from individual patients.
Tx: Treatment medication Vem – Vemurafenib; Dab-Dabrafenib; D+T- Dabrafenib with trametinib.
Type – morphologic subtype of BRAFi-cSCC: C-conventional-BRAFi-conv.-cSCC; KA – BRAFi-cSCC-KA; WF – BRAFi-cSCC-WF.
UV: UV-radiation exposure, yes – Y; no – N. DNA sequence changes: HRAS G12T (c.34GG>AC), Q61L (c.182A>T), Q61R(c.182A>G), Q61K (c.181C>A); KRAS G12D (c.35G>A); PIK3CA E542K (c.1624G>A), E545K (c.1633G>A), A1046T (c.3136G>A); EGFR V769M (c.2305G>A); ALK F1147L (c.3520T>C); CKIT M541L (c.1621A>C); PDGFRA V824 (c.2472C>T), CKIT K546 (c.1638A>G), PTCH T672 (c.2016G>A).
Discussion
BRAF inhibition therapy for melanoma improves patient survival, but is complicated by secondary cutaneous squamous cell carcinoma (BRAFi-cSCC) of unclear etiology and unknown biologic potential (8). These BRAFi-cSCCs are clinically relevant, as they can limit duration of therapy for some patients. BRAFi acts in concert with other factors to increase the frequency of cutaneous, hematologic and visceral cancer by paradoxical hyperactivation of the MAP kinase cascade. Although BRAFi-cSCC is a recognized complication of BRAFi, few clinical trials have employed central dermatopathology review of biopsy material. In this work we present the largest clinical and molecular study published to date of skin cancers arising related to BRAF-inhibition for melanoma. We have performed a detailed study of the secondary SCC latency during BRAFi, including 29 patients and 69 biopsy tissue samples from two cancer centers on one of three current drug regimens (Vem, Dab, or D+T) for metastatic or unresectable advanced melanoma.
The histomorphologic subtypes of BRAFi-cSCC include conventional cSCC (BRAFi-conv.-cSCC), BRAFi-cSCC with keratoacanthomatous features (BRAFi-cSCC-KA) and BRAFi-cSCC lesions with wart-like features (BRAFi-cSCC-WF). We identified approximately 20% of BRAFi-cSCC lesions as BRAFi-cSCC-WF similar to previous reports (5–7). The time from initiating therapy to diagnosis of cSCC (latency) was significantly associated with histopathology of BRAFi-cSCC-WF. The latency of BRAFi-cSCC-WF was 11.6 weeks shorter that BRAFi-conv.-cSCC (p-value = 0.03) when the two groups were compared while controlling for gender and UV-radiation exposure using a multivariate model (Fig. 3). A recent study found a similar latency of cSCC (10.5 weeks) (18) although morphologic subtyping of BRAFi-cSCC was not described and molecular analyses not performed. The observations that UV exposure did not significantly influence BRAFi-cSCC-KA or -WF may suggest that other factors play a more dominant role in these lesions compared to the UV-radiation pathogenesis via TP53 inactivation in conventional-type cSCC (22).
The latency to develop SCC was also influenced by therapy. The differential latency to initial cSCC onset by therapy type (Vem, Dab, D+T) and morphology (conv. > KA > wf) may have pathophysiologic consequences. We observed that lesions with wart-like features frequently have earlier onset than KA or conventional-type cSCC, extending previous work where the comparison did not reach statistical significance (14). The combination of D+T results in decreased incidence of cSCC (19); our study shows a trend towards prolongation of latency (52 weeks for D+T vs 16.3 weeks for Vem) which correlates with the effect previously proposed (19). The relatively small sample size of D+T and Dab-only groups in this study precludes definitive demonstration of significance. Recent studies indicate the BRAFi can inhibit apoptosis through off-target JNK pathway effects that may explain the incomplete elimination of adverse cSCC during D+T (46).
Because so many (>20%) of BRAFi-SCCs have wart-like features reminiscent of viral-driven processes, and because previous studies had failed to isolate α-HPVs from these samples, we took a broadly sensitive approach to identify other, more novel tumorigenic viruses, including non-α HPV and Merkel Cell Polyomavirus (MCPyV).
The human papillomaviruses have close homology within the L1 capsid gene that can be amplified by multiple primer sets: PGMY, GP5+/GP6+, FAP, EV-HPV (38–40, 47). In our study, several lesions with wart-like features were negative with one primer set and positive with another primer set, demonstrating the need for a redundant PCR approach not reported in prior studies. We found no oncogenic α-HPV genotypes, in agreement with a recent report (32), and the only α-HPV found was a non-oncogenic genotype (HPV-7) coinfected with two β-HPVs (HPV-38 and HPV-17).
Under normal conditions, the β-HPV genotypes commensally reside in the human skin, and associate with cSCC in immunocompromised patients (28). β-HPV-5, HPV-8 and other β-HPVs are also found in cSCC from immunocompromised renal transplant and epidermodysplasia verruciformis (EV) patients (28). While reports of HPV in cSCC from otherwise healthy patients also reveal γ-HPV genotypes (48), few patient lesions in our study demonstrated γ-HPV infection: 2 patients, #1 and #28, each had one BRAFi-cSCC infected by HPV-65 and FA15, respectively, and each BRAFi-cSCC with a γ-HPV infection was also β-HPV coinfected (Table 1).
The most frequently detected HPV genotypes in the present study (HPV-38, HPV-17 and HPV-111) are very closely related and as such subclassified together as β2 species that are phylogenetically distant from the α-genus HPV that infect genital/mucosal surfaces and cause cervical SCC (Fig S1). This may suggest that the cutaneous tropism and oncogenic activity of E6 and E7 under BRAFi paradoxical MAPK cascade activity may be restricted to a particular biologic subset of β-HPV types related to and including HPV-38, HPV-17, and HPV-111.
Beta-HPV has been associated with cSCC from healthy patients, with frequent serologic evidence and intralesional HPV nucleic acid detected; however, the frequency of detection with different methods is highly variable thus making study comparison difficult (49, 50). Of interest, β2-HPVs have previously been associated with cSCC from extensively sun-exposed sites (51). Examination of cSCC from otherwise healthy individuals identified HPV-38 and HPV-17 in 13.5% (12 of 89) of infected cSCCs using similar methods to our study (52). In our study, HPV-38 and HPV-17 composed a significantly higher rate of total infections: 40% (49 of 123, p-value = <0.0001 [2.7e-05], Fisher’s exact test [two-sided], Table 1). Further, studies have demonstrated transforming properties in vitro cell culture and in in vivo mouse models of HPV-38 E6 and E7 putative oncoproteins (53, 54) that have enhanced in vitro transforming activity in the presence of BRAFi (35).
Merkel cell polyomavirus (MCPyV) is a significant causative factor in the cutaneous neuroendocrine carcinoma known as Merkel cell carcinoma. In this study, MCPyV was found in 13 BRAF-cSCC lesions, often with coinfection of β-HPV (9 of 13 lesions). MCPyV infection and expression of the large T-antigen has been found to interrupt the cell cycle regulatory protein pRB (55). Coincidentally, the HPV E7 protein also inhibits pRB (28). The detection of MCPyV in these lesions, concurrent with HPV and in isolation, reinforces a prior observation of HPV-17 and MCPyV coinfection (34). MCPyV has also been detected in cSCC from immunocompetent individuals; however, the causative or commensal nature of MCPyV in this setting is unclear (56). Because of the frequent coinfection of MCPyV and HPV in this study, it is difficult to make a definitive statement regarding the causal nature of MCPyV infection in BRAFi-SCC.
Of the lesional biopsies that were available for NGS and had detectable mutations, all but 4 were infected by β-HPV. Canonical cancer-associated HRAS (Q61, G12) and KRAS (G12) amino acid mutations were confirmed in 5 of 8 lesions (62.5%, Table 2), similar to the frequency reported in other studies (13, 24–26). The present study adds two examples of HRAS mutation coincident with HPV infection (HRAS G12T with β-HPV36b – P006 and KRAS G12D β-HPV 17 and β-HPV 151 – P144). To our knowledge this has only been observed once before (57).
We also demonstrate the presence of relevent mutations in other cancer-associated proteins not previously reported in BRAFi-cSCC. EGFR, ALK, CKIT and PIK3CA mutations are most commonly associated with lung, breast, endometrial and colorectal carcinomas, gastrointestinal stromal tumors and lymphomas. Mutations in these oncoproteins are not frequently detected in cutaneous squamous cell carcinoma. PIK3CA mutations have been detected in two spontaneous cSCC and one KA lesion (24) and in two BRAFi-cSCC (26). One EGFR R108K mutation has been reported in a single BRAFi-cSCC (26). We report for the first time to our knowledge an ALK F1174L mutation, two tumors with CKIT M541L mutations, and an EGFR V769M mutation in BRAFi-cSCCs.
Prior mechanistic studies of conv.-cSCC have demonstrated a significant role for UV-B via increased rate of mutation in tumor suppressor TP53 (23). We also found in BRAFi-conv.-c-SCC that UV exposure significantly increased the rate of onset (decreased latency). By contrast, BRAFi-cSCC-WF does not show a significant effect of UV radiation on latency in the present study, indicating a possible independence from UV induction in this subtype of BRAFi-cSCC. Although the number of MCPyV-infected samples in this study were too low to draw specific conclusions, UV-B has been associated with induction of MCPyV small-T antigen expression (58). A larger sample size may reveal an interplay of UV-radiation, β-HPV and MCPyV coinfection during BRAFi-cSCC from primary patient cancer tissues.
An alternate hypothesis, which is intriguing but not possible to evaluate rigorously with our study, states that BRAFi-cSCC is the same lesion that has been sampled at different time points in a unique tumor progression, beginning with wart-like features and culminating with keratoacanthomatous or conventional cSCC morphology. Lesions biopsied early in this life history would morphologically have wart-like features and lesions biopsied later in development would have fewer wart-like features. Further study into the life history of BRAF-inhibition induced carcinogenesis using the cutaneous SCC as a model system may prove or disprove this hypothesis.
Study limitations: Although we included 69 biopsies from 29 patients, the study size was limiting for the non-vemurafenib (Vem) groups to preclude extensive analysis of dabrafenib (Dab) with and without the MEK-inhibitor trametinib (D+T). Additional biopsies were not identified for patients on the Vem and cobimetinib MEK-inhibition combination therapy. Further, the limitation of sample size prevents demonstration of statistical significance for several parameters suggestive but not definitive of relevance to BRAFi-cSCC carcinogenesis in our study, including coinfection of β-HPV and MCPyV with and without UV exposure and RAS gene mutation. The primary measure in the study of time from therapy initiation to lesion is limited by patient presentation. This bias would overestimate the true time to onset as the initiating genetic events leading to cancer precede epidermal overgrowth and thus precede discomfort, bleeding or other causes that draw then patients’ attention and therefore provider intervention. Ampliseq by Ion Torrent is a versatile platform for focused amplicon-based next generation sequencing. However the pipeline is not without infrequent fault. NGS and analysis in our experiments did report HRAS Q61L for P040 that on Sanger was confirmed as Q61R. Additionally, the annotation algorithm misidentified a G12-change in sample P006 that was confirmed G12T by Sanger. This correction was required because the CLCbio database contains duplicate entries, both single and double base mutations, for this codon that was misidentified in the algorithm. A possible double PIK3CA mutation resulting in A1046T (P070) was suggested by the NGS that was not confirmed by Sanger (Table 2). Also, CKIT, EGFR and ALK mutations were within Ampliseq study parameters, however there was insufficient biopsy DNA for further Sanger confirmation.
In conclusion, β-HPV infection is frequent in BRAFi-cSCC, and BRAFi-cSCC-WF is statistically associated with rapid onset (short latency) lesions. The large number of novel HPV genotypes identified in these lesions needs more complete genetic study to begin to understand putative E6/E7 oncoproteins in non-mucosal HPV infection, but indicates the possibility of subgroup-specific biologic factors in BRAFi oncogenesis. Further in-depth study into the host-genetic patterns of BRAFi-cSCC-WF and UV-damage host-genetic models in conjunction with HPV and MCPyV large and small T antigens may provide sufficient power for analysis. These novel studies will likely shed new insight to the pathogenesis of squamous cell carcinoma as well as multifactorial models for cancer in general.
Supplementary Material
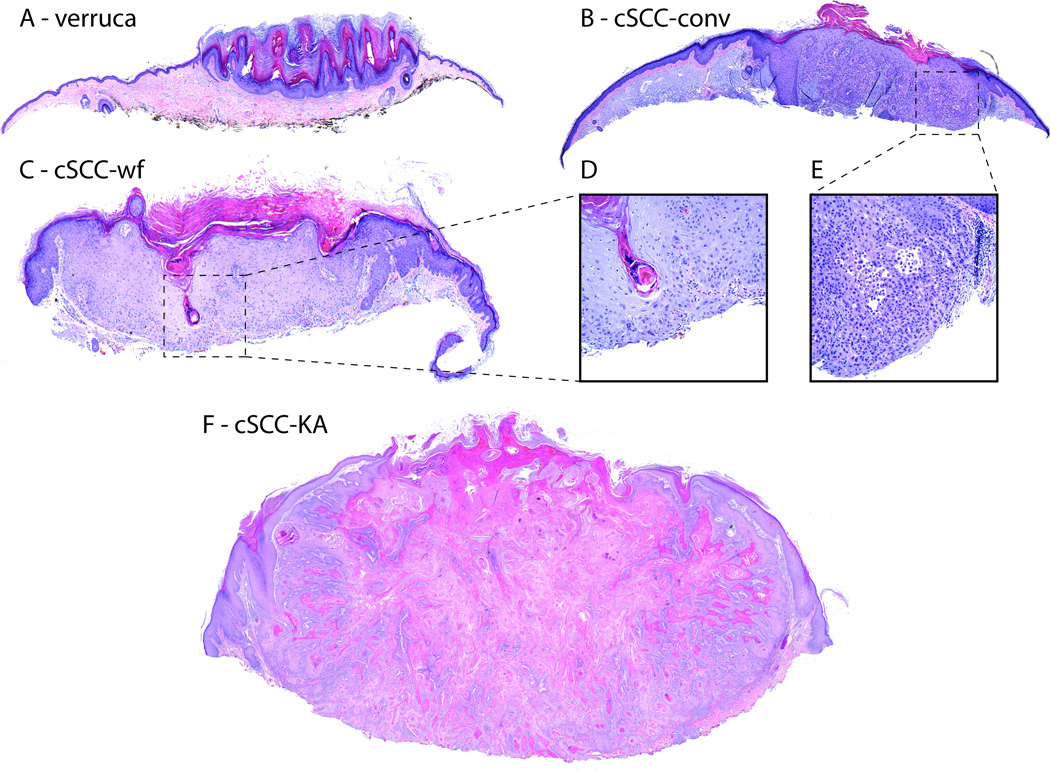
Figure 2. Adverse cutaneous neoplasia during BRAF-inhibition.
Histopathologic subtypes of squamoproliferative lesions include verruca (A) with papillomatosis, hyperkeratosis and hypogranulosis; actinic keratosis (not shown) with keratinocyte atypia without full thickness involvement or invasion; conventional (conv.) cutaneous squamous cell carcinoma (cSCC, B, atypical keratinocytes, inset E) with invasion of dermis; well differentiated keratoacanthomatous-type cSCC (cSCC-KA, F); and cSCC with wart-like features (cSCC-WF, C; atypical keratinocytes inset D). H&E stained sections, composite 4x orig. obj. mag. Insets (D, E) 20x orig. obj. mag.
Translational Relevance.
Targeted molecular therapy for advanced melanoma with BRAF inhibition (BRAFi) induces cSCC, frequently with morphologic resemblance to warts. Human papillomaviruses (HPV) are frequently detected in human cutaneous squamous cell carcinoma (cSCC). β-genus HPV, RAS mutation and UV-radiation exposure are associated with BRAFi-cSCC. Less frequently, Merkel Cell Polyomavirus (MCPyV) is found within BRAFi-cSCC with and without coinfection by β-HPV. The unique pathophysiologic interaction of β-HPV and MCPyV infection, UV-damage and host genetic predisposition and somatic mutation with BRAFi in cSCC is presently under investigation. A BRAFi-cSCC paradigm may spur vaccine and molecular intervention and better understanding of multifactorial oncogenesis mechanisms.
Acknowledgements
The authors thank Criziel Quinn, Susan Sefers, Dr. Kimberly Dahlman and Dr. Angela Rohwedder for DNA extraction and early PCR assay development assistance. We further wish to thank Lisa Pratt, William Powell, Molly Marinelli, Melissa Radford, MaryAnn Deathridge, Steven LaMontange, and Fran Blanpain for expert histology guidance and laboratory assistance.
Funding: This research was supported by the Clinical and Translational Research Enhancement Award from the Vanderbilt Department of Pathology, Microbiology and Immunology to D.N. Cohen and by NCI grant K24 CA097588-09 (J.A. Sosman), and K12 CA0906525 (D.B. Johnson), an ACS Melanoma Professorship in Translational Medicine SIMP-09-102-01-CCE (J.A. Sosman), and MRA Team Science Award 270010 (J.A. Sosman). Development experiments were performed in the Vanderbilt Innovative Translational Research Shared Resource supported by the Vanderbilt-Ingram Cancer Center and the TJ Martell Foundation (K Dahlman, CA68485).
Footnotes
Disclosures: D. Cohen, S. Lawson, A. Shaver, L. Du, H. Nguyen, Q. He, D. Johnson, W. Lumbang, J. Prescott, A. Boyd, J. Zwerner, J. Robbins, S. Tyring, P. Rady, Y. Shyr, and J. Chappell have no disclosures. J. Sosman has served as a consultant/advisory role to Bristol-Myers Squibb and Genentech. J. Infante has served as an uncompensated consultant to GlaxoSmithKline. B. Moody has received honoraria from Genentech. P. Chandra has served as a consultant/advisory role to and received honoraria from Pfizer.
References
- 1.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366(8):707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falchook GS, Long GV, Kurzrock R, Kim KB, Arkenau TH, Brown MP, et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: A phase 1 dose-escalation trial. Lancet. 2012;379(9829):1893–1901. doi: 10.1016/S0140-6736(12)60398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long GV, Trefzer U, Davies MA, Kefford RF, Ascierto PA, Chapman PB, et al. Dabrafenib in patients with Val600Glu or Val600Lys BRAF-mutant melanoma metastatic to the brain (BREAK-MB): A multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13(11):1087–1095. doi: 10.1016/S1470-2045(12)70431-X. [DOI] [PubMed] [Google Scholar]
- 4.Cohen DN, Chappell JD, Robbins JB. Squamous cell carcinoma in BRAF-inhibitor-treated melanoma patients.In abstracts presented at the 15th Joint Meeting of the International Society of Dermatopathology, March 14–15, San Diego, CA, USA. Amer J Dermatopath. 2012;34(5):e55–e72. doi: 10.1097/dad.0b013e31826018e7. [DOI] [PubMed] [Google Scholar]
- 5.Chu EY, Wanat KA, Miller CJ, Amaravadi RK, Fecher LA, Brose MS, et al. Diverse cutaneous side effects associated with BRAF inhibitor therapy: A clinicopathologic study. J Am Acad Dermatol. 2012;67(6):1265–1272. doi: 10.1016/j.jaad.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harvey NT, Millward M, Wood BA. Squamoproliferative lesions arising in the setting of BRAF inhibition. Am J Dermatopathol. 2012;34(8):822–826. doi: 10.1097/DAD.0b013e3182604873. [DOI] [PubMed] [Google Scholar]
- 7.Huang V, Hepper D, Anadkat M, Cornelius L. Cutaneous toxic effects associated with vemurafenib and inhibition of the BRAF pathway. Arch Dermatol. 2012;148(5):628–633. doi: 10.1001/archdermatol.2012.125. [DOI] [PubMed] [Google Scholar]
- 8.Cohen DN, Lumbang WA, Boyd AS, Sosman JA, Zwerner JP. Spindle cell squamous carcinoma during BRAF inhibitor therapy for advanced melanoma: An aggressive secondary neoplasm of undetermined biologic potential. JAMA Dermatol. 2014;150(5):575–577. doi: 10.1001/jamadermatol.2013.7784. [DOI] [PubMed] [Google Scholar]
- 9.Andrews MC, Behren A, Chionh F, Mariadason J, Vella LJ, Do H, et al. BRAF inhibitor-driven tumor proliferation in a KRAS-mutated colon carcinoma is not overcome by MEK1/2 inhibition. J Clin Oncol. 2013;31(35):e448–e451. doi: 10.1200/JCO.2013.50.4118. [DOI] [PubMed] [Google Scholar]
- 10.Callahan MK, Rampal R, Harding JJ, Klimek VM, Chung YR, Merghoub T, et al. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N Engl J Med. 2012;367(24):2316–2321. doi: 10.1056/NEJMoa1208958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattei PL, Alora-Palli MB, Kraft S, Lawrence DP, Flaherty KT, Kimball AB. Cutaneous effects of BRAF inhibitor therapy: A case series. Ann Oncol. 2013;24(2):530–537. doi: 10.1093/annonc/mds292. [DOI] [PubMed] [Google Scholar]
- 13.Lacouture ME, Duvic M, Hauschild A, Prieto VG, Robert C, Schadendorf D, et al. Analysis of dermatologic events in vemurafenib-treated patients with melanoma. Oncologist. 2013;18(3):314–322. doi: 10.1634/theoncologist.2012-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boussemart L, Routier E, Mateus C, Opletalova K, Sebille G, Kamsu-Kom N, et al. Prospective study of cutaneous side-effects associated with the BRAF inhibitor vemurafenib: A study of 42 patients. Ann Oncol. 2013;24(6):1691–1697. doi: 10.1093/annonc/mdt015. [DOI] [PubMed] [Google Scholar]
- 15.McArthur GA, Chapman PB, Robert C, Larkin J, Haanen JB, Dummer R, et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): Extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol. 2014;15(3):323–332. doi: 10.1016/S1470-2045(14)70012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larkin J, Del Vecchio M, Ascierto PA, Krajsova I, Schachter J, Neyns B, et al. Vemurafenib in patients with BRAF(V600) mutated metastatic melanoma: An open-label, multicentre, safety study. Lancet Oncol. 2014;15(4):436–444. doi: 10.1016/S1470-2045(14)70051-8. [DOI] [PubMed] [Google Scholar]
- 17.Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med. 2014;371(20):1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 18.Belum VR, Rosen AC, Jaimes N, Dranitsaris G, Pulitzer MP, Busam KJ, et al. Clinico-morphological features of BRAF inhibition-induced proliferative skin lesions in cancer patients. Cancer. 2015;121(1):60–68. doi: 10.1002/cncr.28980. [DOI] [PubMed] [Google Scholar]
- 19.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367(18):1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014;371(20):1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 21.Ribas A, Gonzalez R, Pavlick A, Hamid O, Gajewski TF, Daud A, et al. Combination of vemurafenib and cobimetinib in patients with advanced BRAF(V600)-mutated melanoma: A phase 1b study. Lancet Oncol. 2014;15(9):954–965. doi: 10.1016/S1470-2045(14)70301-8. [DOI] [PubMed] [Google Scholar]
- 22.Ziegler A, Jonason A, Simon J, Leffell D, Brash DE. Tumor suppressor gene mutations and photocarcinogenesis. Photochem Photobiol. 1996;63(4):432–435. doi: 10.1111/j.1751-1097.1996.tb03064.x. [DOI] [PubMed] [Google Scholar]
- 23.Popp S, Waltering S, Herbst C, Moll I, Boukamp P. UV-B-type mutations and chromosomal imbalances indicate common pathways for the development of merkel and skin squamous cell carcinomas. Int J Cancer. 2002;99(3):352–360. doi: 10.1002/ijc.10321. [DOI] [PubMed] [Google Scholar]
- 24.Oberholzer PA, Kee D, Dziunycz P, Sucker A, Kamsukom N, Jones R, et al. RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. J Clin Oncol. 2012;30(3):316–321. doi: 10.1200/JCO.2011.36.7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med. 2012;366(3):207–215. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anforth R, Tembe V, Blumetti T, Fernandez-Penas P. Mutational analysis of cutaneous squamous cell carcinomas and verrucal keratosis in patients taking BRAF inhibitors. Pigment Cell Melanoma Res. 2012;25(5):569–572. doi: 10.1111/j.1755-148X.2012.01031.x. [DOI] [PubMed] [Google Scholar]
- 27.Cohen DN, Chappell JD, Robbins JB. Human papillomavirus mechanisms in squamous cell carcinomagenesis in BRAF-inhibitor treated melanoma patients (#110)In Carcinogenesis, Growth factors, and Cancer genetics, abstracts of the Society for Investigative Dermatology 72nd Annual Meeting, May 9–12, Raleigh, NC, USA. J Invest Derm. 2012;132:S19–S30. [Google Scholar]
- 28.White MK, Pagano JS, Khalili K. Viruses and human cancers: A long road of discovery of molecular paradigms. Clin Microbiol Rev. 2014;27(3):463–481. doi: 10.1128/CMR.00124-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sri JC, Dubina MI, Kao GF, Rady PL, Tyring SK, Gaspari AA. Generalized verrucosis: A review of the associated diseases, evaluation, and treatments. J Am Acad Dermatol. 2012;66(2):292–311. doi: 10.1016/j.jaad.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Anforth RM, Blumetti TC, Kefford RF, Sharma R, Scolyer RA, Kossard S, et al. Cutaneous manifestations of dabrafenib (GSK2118436): A selective inhibitor of mutant BRAF in patients with metastatic melanoma. Br J Dermatol. 2012;167(5):1153–1160. doi: 10.1111/j.1365-2133.2012.11155.x. [DOI] [PubMed] [Google Scholar]
- 31.Ko CJ, McNiff JM, Iftner A, Iftner T, Choi JN. Vemurafenib (PLX-4032)-induced keratoses: Verrucous but not verrucae. J Am Acad Dermatol. 2013;69(2):e95–e96. doi: 10.1016/j.jaad.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 32.Dika E, Patrizi A, Venturoli S, Fanti PA, Barbieri D, Strammiello R, et al. Human papillomavirus evaluation of vemurafenib induced skin epithelial tumors: A case series. Br J Dermatol. 2014 doi: 10.1111/bjd.13275. [DOI] [PubMed] [Google Scholar]
- 33.Quint KD, Genders RE, de Koning MN, Borgogna C, Gariglio M, Bouwes Bavinck JN, et al. Human beta-papillomavirus infection and keratinocyte carcinomas. J Pathol. 2014;235(2):342–354. doi: 10.1002/path.4425. [DOI] [PubMed] [Google Scholar]
- 34.Falchook GS, Rady P, Hymes S, Nguyen HP, Tyring SK, Prieto VG, et al. Merkel cell polyomavirus and HPV-17 associated with cutaneous squamous cell carcinoma arising in a patient with melanoma treated with the BRAF inhibitor dabrafenib. JAMA Dermatol. 2013;149(3):322–326. doi: 10.1001/jamadermatol.2013.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holderfield M, Lorenzana E, Weisburd B, Lomovasky L, Boussemart L, Lacroix L, et al. Vemurafenib cooperates with HPV to promote initiation of cutaneous tumors. Cancer Res. 2014;74(8):2238–2245. doi: 10.1158/0008-5472.CAN-13-1065-T. [DOI] [PubMed] [Google Scholar]
- 36.Hassel JC, Groesser L, Herschberger E, Weichert W, Hafner C. RAS mutations in benign epithelial tumors associated with BRAF inhibitor treatment of melanoma. J Invest Dermatol. 2015;135(2):636–639. doi: 10.1038/jid.2014.360. [DOI] [PubMed] [Google Scholar]
- 37.Resnick RM, Cornelissen MT, Wright DK, Eichinger GH, Fox HS, terSchegget J, et al. Detection and typing of human papillomavirus in archival cervical cancer specimens by DNA amplification with consensus primers. J Natl Cancer Inst. 1990;82(18):1477–1484. doi: 10.1093/jnci/82.18.1477. [DOI] [PubMed] [Google Scholar]
- 38.FuesselHaws AL, He Q, Rady PL, Zhang L, Grady J, Hughes TK, et al. Nested PCR with the PGMY09/11 and GP5(+)/6(+) primer sets improves detection of HPV DNA in cervical samples. J Virol Methods. 2004;122(1):87–93. doi: 10.1016/j.jviromet.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Harwood CA, Spink PJ, Surentheran T, Leigh IM, de Villiers EM, McGregor JM, et al. Degenerate and nested PCR: A highly sensitive and specific method for detection of human papillomavirus infection in cutaneous warts. J Clin Microbiol. 1999;37(11):3545–3555. doi: 10.1128/jcm.37.11.3545-3555.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forslund O, Ly H, Reid C, Higgins G. A broad spectrum of human papillomavirus types is present in the skin of australian patients with non-melanoma skin cancers and solar keratosis. Br J Dermatol. 2003;149(1):64–73. doi: 10.1046/j.1365-2133.2003.05376.x. [DOI] [PubMed] [Google Scholar]
- 41.de Oliveira WR, He Q, Rady PL, Hughes TK, Neto CF, Rivitti EA, et al. HPV typing in Brazilian patients with epidermodysplasia verruciformis: High prevalence of EV-HPV 25. J Cutan Med Surg. 2004;8(2):110–115. doi: 10.1007/s10227-003-0100-6. [DOI] [PubMed] [Google Scholar]
- 42.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyd AS, Annarella M, Rapini RP, Adler-Storthz K, Duvic M. False-positive polymerase chain reaction results for human papillomavirus in lichen planus potential laboratory pitfalls of this procedure. J Am Acad Dermatol. 1996;35(1):42–46. doi: 10.1016/S0190-9622(96)90494-6. [DOI] [PubMed] [Google Scholar]
- 44.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324(1):17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 45.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human merkel cell carcinoma. Science. 2008;319(5866):1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vin H, Ojeda SS, Ching G, Leung ML, Chitsazzadeh V, Dwyer DW, et al. BRAF inhibitors suppress apoptosis through off-target inhibition of JNK signaling. Elife. 2013;2:e00969. doi: 10.7554/eLife.00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3’ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995;76(Pt 4):1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 48.Ekstrom J, Bzhalava D, Svenback D, Forslund O, Dillner J. High throughput sequencing reveals diversity of human papillomaviruses in cutaneous lesions. Int J Cancer. 2011;129(11):2643–2650. doi: 10.1002/ijc.26204. [DOI] [PubMed] [Google Scholar]
- 49.Asgari MM, Kiviat NB, Critchlow CW, Stern JE, Argenyi ZB, Raugi GJ, et al. Detection of human papillomavirus DNA in cutaneous squamous cell carcinoma among immunocompetent individuals. J Invest Dermatol. 2008;128(6):1409–1417. doi: 10.1038/sj.jid.5701227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iannacone MR, Wang W, Stockwell HG, O’Rourke K, Giuliano AR, Sondak VK, et al. Sunlight exposure and cutaneous human papillomavirus seroreactivity in basal cell and squamous cell carcinomas of the skin. J Infect Dis. 2012;206(3):399–406. doi: 10.1093/infdis/jis374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forslund O, Iftner T, Andersson K, Lindelof B, Hradil E, Nordin P, et al. Cutaneous human papillomaviruses found in sun-exposed skin: Beta-papillomavirus species 2 predominates in squamous cell carcinoma. J Infect Dis. 2007;196(6):876–883. doi: 10.1086/521031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plasmeijer EI, Neale RE, Buettner PG, de Koning MN, Ter Schegget J, Quint WG, et al. Betapapillomavirus infection profiles in tissue sets from cutaneous squamous cell-carcinoma patients. Int J Cancer. 2010;126(11):2614–2621. doi: 10.1002/ijc.24991. [DOI] [PubMed] [Google Scholar]
- 53.Caldeira S, Zehbe I, Accardi R, Malanchi I, Dong W, Giarre M, et al. The E6 and E7 proteins of the cutaneous human papillomavirus type 38 display transforming properties. J Virol. 2003;77(3):2195–2206. doi: 10.1128/JVI.77.3.2195-2206.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Viarisio D, Mueller-Decker K, Kloz U, Aengeneyndt B, Kopp-Schneider A, Grone HJ, et al. E6 and E7 from beta HPV38 cooperate with ultraviolet light in the development of actinic keratosis-like lesions and squamous cell carcinoma in mice. PLoS Pathog. 2011;7(7):e1002125. doi: 10.1371/journal.ppat.1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, et al. T antigen mutations are a human tumor-specific signature for merkel cell polyomavirus. Proc Natl Acad Sci U S A. 2008;105(42):16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dworkin AM, Tseng SY, Allain DC, Iwenofu OH, Peters SB, Toland AE. Merkel cell polyomavirus in cutaneous squamous cell carcinoma of immunocompetent individuals. J Invest Dermatol. 2009;129(12):2868–2874. doi: 10.1038/jid.2009.183. [DOI] [PubMed] [Google Scholar]
- 57.Zaravinos A, Kanellou P, Spandidos DA. Viral DNA detection and RAS mutations in actinic keratosis and nonmelanoma skin cancers. Br J Dermatol. 2010;162(2):325–331. doi: 10.1111/j.1365-2133.2009.09480.x. [DOI] [PubMed] [Google Scholar]
- 58.Mogha A, Fautrel A, Mouchet N, Guo N, Corre S, Adamski H, et al. Merkel cell polyomavirus small T antigen mRNA level is increased following in vivo UV-radiation. PLoS One. 2010;5(7):e11423. doi: 10.1371/journal.pone.0011423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.