Abstract
Background
African American (AA) women are diagnosed with more advanced breast cancers and have worse survival than white women, but a comprehensive understanding of the basis for this disparity remains unclear. Analysis of DNA methylation, an epigenetic mechanism that can regulate gene expression, could help to explain racial differences in breast tumor clinical biology and outcomes.
Methods
DNA methylation was evaluated at 1287 CpGs in the promoters of cancer-related genes in 517 breast tumors of AA (n=216) or non-AA (n=301) cases in the Carolina Breast Cancer Study.
Results
Multivariable linear regression analysis of all tumors, controlling for age, menopausal status, stage, intrinsic subtype, and multiple comparisons (FDR), identified 7 CpG probes that showed significant (adjusted p<0.05) differential methylation between AAs and non-AAs. Stratified analyses detected an additional 4 CpG probes differing by race within hormone receptor-negative (HR−) tumors. Genes differentially methylated by race included DSC2, KCNK4, GSTM1, AXL, DNAJC15, HBII-52, TUSC3 and TES; the methylation state of several of these genes may be associated with worse survival in AAs. TCGA breast tumor data confirmed the differential methylation by race and negative correlations with expression for most of these genes. Several loci also showed racial differences in methylation in peripheral blood leukocytes (PBLs) from CBCS cases, indicating that these variations were not necessarily tumor-specific.
Conclusions
Racial differences in the methylation of cancer-related genes are detectable in both tumors and PBLs from breast cancer cases.
Impact
Epigenetic variation could contribute to differences in breast tumor development and outcomes between AAs and non-AAs.
Keywords: methylation, breast cancer, racial variation, epigenetics
INTRODUCTION
Breast cancer, the most common cancer among women in the United States (1), is a heterogeneous disease with multiple clinical, histopathologic and molecular subtypes exhibiting different therapeutic responses and prognoses (2-4). Racial differences exist in its presentation and outcome, with African American (AA) women, especially younger women, being diagnosed with more advanced cancers (5) and having worse survival than white women even after controlling for known prognostic factors or treatment (5-7). Prior work using protein markers to classify breast tumor intrinsic subtypes in the Carolina Breast Cancer Study (CBCS) found a higher prevalence of basal-like breast tumors among AA women compared with white women (3); however, AA women also have worse outcomes across all intrinsic subtypes (8).
In an effort to better understand the molecular factors contributing to breast cancer development, outcomes and racial disparities, we evaluated DNA promoter methylation profiles in invasive breast tumors from the CBCS. An epigenetic modification to DNA that does not alter the nucleotide sequence (9), DNA methylation usually occurs as the addition of a methyl group to a cytosine within a CpG dinucleotide. Aberrant hypermethylation of CpG islands in tumor suppressor genes can result in their silencing in cancer, while hypomethylation can lead to increased oncogene expression (9, 10). Such methylation changes and their effects on gene expression have the potential to influence breast tumor phenotypes and clinical outcomes (10). Several prior studies using targeted methylation approaches for a few genes have suggested that racial variation exists in methylation between AAs and non-AAs (or Caucasians) (11-14), and a recent whole-genome study using the Illumina 450K platform supports these earlier reports (15).
In this study, use of a cancer-focused promoter methylation array to assess methylation in a large population-based series of breast tumors from AA and non-AA breast cancer cases in the CBCS revealed differential tumor methylation of several genes, and most of these were also differentially methylated in white blood cells, thereby enabling a more informed interpretation of which epigenetic differences might potentially contribute to breast cancer development, influence breast tumor chemosensitivity or the disparity in outcomes.
MATERIALS AND METHODS
Carolina Breast Cancer Study (CBCS) cases and specimens
The CBCS is a population-based, case-control study of incident invasive breast cancer in North Carolina. Details of the study design have been described (16). Randomized recruitment was used to oversample younger and African American cases to ensure that they comprised roughly half the study sample. Race/ethnicity was self-reported. Among non-AA cases, 97% self-reported as Caucasian (n=291) and the other 3% included 4 cases with Hispanic ethnicity, 3 American Indians, 6 Asian/Native Islanders, and 1 other; therefore, we refer to this group as non-AA.
Blood samples were obtained for DNA extraction from peripheral blood leukocytes (PBLs). Ancestry informative markers (AIMs) to estimate the proportion of African versus European ancestry were previously evaluated from PBL DNA in the CBCS (17), AIMs genotypes were derived from 144 single nucleotide polymorphisms (SNPs), and an AIMs score ranging from 0 to 1.0 was generated for each case representing lower to higher African ancestry.
Formalin-fixed paraffin-embedded (FFPE) breast tumors were sectioned and mounted on slides, histopathologically reviewed, and tumor areas macrodissected for DNA extraction (18). Clinical data were obtained from medical records or histopathologic review of tumor tissue. Breast tumor intrinsic subtypes were identified using estrogen receptor (ER), progesterone receptor (PR), HER2, CK5, CK6, and EGFR protein markers (3). Hormone receptor (HR) positivity was considered to be ER+ and/or PR+, while tumors designated HR negative were both ER− and PR−.
Methylation analysis
DNA lysates prepared from breast tumors were sodium bisulfite treated using the EZ DNA Methylation Gold kit (Zymo Research). Methylation profiling was accomplished using the Illumina GoldenGate Cancer Panel I array (Illumina, San Diego, CA) on 517 breast tumors and 69 PBLs from cases, 61 of which were matched to individual cases. Methylation data were preprocessed using GenomeStudio Methylation software (Illumina). Methylation is represented by β, an estimate of the fraction of methylated DNA, and ranges from 0 (unmethylated) to 1.0 (fully methylated). The array interrogated 1,505 CpG sites located in the upstream regulatory region (promoter or exon 1) of 807 genes. Of the 1,505 CpG probes, a total of 1287 CpGs (Table S1) were included in the final dataset after filtering (18), including probes previously identified as overlapping an underlying sequence variant (19). Compared with 163 cases not evaluated due to inadequate quantity or quality of DNA (of 680 total with tumors in CBCS phase 1), the 517 breast cancer cases evaluated were younger (p=0.03) but did not differ in other characteristics. Array data were deposited to Gene Expression Omnibus under accession number GSE51557.
Statistical analyses
Statistical analyses were carried out using the R package (http://www.r-project.org/) or SAS 9.3. CpGs differentially methylated between breast tumors of AAs and non-AAs were identified via generalized linear models (GLM), modeling the methylation value with logit link function and including race (AA vs non-AA) as a predictor, while also adjusting for age, menopausal status, stage, and intrinsic subtype (20). P-values were adjusted for multiple comparisons via the Benjamini-Hochberg method for controlling the false discovery rate (FDR). A GLM model to compare methylation by race was performed first for all breast tumors, and then was repeated with stratification of breast tumors on hormone receptor (HR) status. Box and whisker plots were constructed to display the distribution of methylation β values for each group, and included the mean, median, and interquartile range of methylation values, and the group minimum and maximum values. Univariate analysis of methylation by race in peripheral blood leukocytes (PBLs) was determined by the Student’s t-test. Correlation of methylation in paired tumors and PBLs from cases was determined by the Spearman rank correlation coefficient. Kaplan-Meier plots and log-rank p-values were used to illustrate disease-specific survival according to methylation level (high, intermediate, low) for select probes within the AA or non-AA case groups. For each probe, cutpoints were based on the tertiles in the combined AA/non-AA dataset.
Validation studies
Independent validation of racial differences in breast tumor methylation and correlations of methylation with gene expression were conducted using breast tumor data from The Cancer Genome Atlas (TCGA) (21). Methylation analysis in TCGA was performed using the Illumina Infinium 450K methylation array, while gene expression data was generated using RNA sequencing. Only 371 of the 1,505 CpG probes interrogated on the Illumina GoldenGate methylation platform exactly match those on the 450K methylation array; however, prior analysis of 450K probes showed that methylation levels were concordant for matched probes between distinct methylation platforms (22). Therefore, we compared methylation at probes from the 450K array in TCGA that interrogated direct match CpG sites when available, or those closest within 200 base pairs upstream or downstream of the GoldenGate probes of interest between AA and white patients. We also tested associations between methylation and gene expression for these probes. Two separate subsets of TCGA data were used for these analyses. For comparisons of breast tumor methylation, beta values for each probe of interest from the 450K array were compared between 42 AA women and 291 white women in the TCGA dataset. T-tests were used to identify probes that differed by race at p<0.05. TCGA data from 581 breast cancer patients were used to examine relationships between methylation and gene expression. Pearson correlation coefficients were calculated in all 581 TCGA breast tumors, or within hormone receptor-positive or hormone receptor-negative subsets, based on RNAseq (Illumina) log2 RSEM gene normalized expression values with methylation beta values for 450K CpG probes, with significance set at p<0.05. Independent validation of racial differences in methylation was also performed using an Illumina 450K methylation dataset in Gene Expression Omnibus on lymphoblastoid cell lines from healthy female African American (n=80) and Caucasian American subjects (n=49) (23) (accession #GSE36369). Moreover, technical validation was conducted to compare gene methylation from the GoldenGate array with that from quantitative methylation-specific polymerase chain reaction (Q-MSP) (24).
RESULTS
Characteristics of breast cancer cases
Characteristics of AA and non-AA cases and their tumors evaluated for DNA methylation are detailed in Table 1. Breast cancer cases were mostly early stage (>86% stages 1 or 2). AA cases were older, more frequently postmenopausal, and more likely to have hormone receptor (HR)-negative, high grade, or basal-like breast tumors compared with non-AAs, as previously reported (3). PBL samples derived from a subset of AA and non-AA cases did not differ in age (p=0.44) or menopausal status (p=0.12).
Table 1.
Demographic, tumor and clinical characteristics of the African American and non-African American breast cancer cases evaluated for DNA methylation
| Characteristic | African No. |
American % |
Non-African No. |
American % |
p-valuea | ||
|---|---|---|---|---|---|---|---|
| Cases with tumors | |||||||
| Total N | 216 | 301 | |||||
| Age | |||||||
| 50+ years | 97 | 44.9 | 102 | 33.9 | 0.01 | ||
| <50 years | 119 | 55.1 | 199 | 66.1 | |||
| Menopausal Status | |||||||
| Postmenopausal | 113 | 52.3 | 129 | 42.9 | 0.03 | ||
| Premenopausal | 103 | 47.7 | 172 | 57.1 | |||
| Stageb | |||||||
| I | 69 | 34.5 | 109 | 38.8 | 0.64 | ||
| II | 104 | 52.0 | 141 | 50.2 | |||
| III | 22 | 11.0 | 23 | 8.2 | |||
| IV | 5 | 2.5 | 8 | 2.8 | |||
| Primary Tumor Size | |||||||
| ≤2 cm | 98 | 47.1 | 152 | 52.6 | 0.23 | ||
| >2 cm | 110 | 52.9 | 137 | 47.4 | |||
| Lymph Node Status | |||||||
| Negative | 115 | 55.6 | 176 | 60.5 | 0.27 | ||
| Positive | 92 | 44.4 | 115 | 39.5 | |||
| Hormone Receptor Expression | |||||||
| ER+/PR+ | 88 | 42.5 | 162 | 55.3 | 0.01 | ||
| ER+/PR− | 20 | 9.7 | 28 | 9.5 | |||
| ER−/PR+ | 15 | 7.2 | 24 | 8.2 | |||
| ER−/PR− | 84 | 40.6 | 79 | 27.0 | |||
| Intrinsic Subtypec | |||||||
| Luminal A | 81 | 47.9 | 131 | 53.7 | 0.04 | ||
| Luminal B | 22 | 13.0 | 43 | 17.6 | |||
| HER2+/HR− | 13 | 7.7 | 13 | 5.3 | |||
| Basal-like | 46 | 27.2 | 40 | 16.4 | |||
| Unclassified | 7 | 4.1 | 17 | 7.0 | |||
| Cases with PBLs | |||||||
| Total N | 29 | 40 | |||||
| Age | |||||||
| 50+ years | 9 | 31.0 | 16 | 40.0 | 0.44 | ||
| <50 years | 20 | 69.0 | 24 | 60.0 | |||
| Menopausal Status | |||||||
| Postmenopausal | 9 | 31.0 | 20 | 50.0 | 0.12 | ||
| Premenopausal | 20 | 69.0 | 20 | 50.0 | |||
Chi-Square p-values.
According to the AJCC breast tumor staging guidelines.
Intrinsic subtypes determined by a panel of immunohistochemical markers included luminal A (ER+ and/or PR+, HER2−), luminal B (ER+ and/or PR+, HER2+), basal-like (ER−, PR−, HER2−, cytokeratins CK5+ and/or CK6+, or epidermal growth factor receptor+), HER2+ (ER−, PR−, HER2+), and unclassified (all markers negative).
Racial differences in breast tumor methylation
Comparison of β values for 1287 CpG probes in 517 breast tumors using GLM while controlling for age, menopausal status, stage, intrinsic subtype, and multiple comparisons (FDR) identified a total of 24 CpG probes that differed significantly at q<0.05 between AA (n=216) and non-AA (n=301) cases (Table 2). The majority of these probes (n=17), however, were considered technically ambiguous or unreliable because they were previously reported to overlap a site of a potential single nucleotide polymorphism (SNP) or repeat (19), or based on review of updated probe target sequence information in NCBI databases (e.g., Ensembl, dbSNP, Blast) were subsequently found to overlap underlying sequence variants or show ancestral population differences of >10% (as detailed in Table S1). In total, 7 probes showing differential methylation by race were retained for further analysis, including four that had no known underlying variant (DSC2_E90_F, KCNK4_E3_F, GSTM1_P266_F, HBII-52_E142_F) and three that overlapped a single SNP but that was unlikely to impact probe performance due to its location toward the end of the target sequence and the lack of evidence for ancestral differences in allele frequencies (AXL_P223_R, DNAJC15_E26_R, and TES_P182_F) (Table 2). While the GSTM1 gene is polymorphic for a deletion variant, this deletion occurs within the coding sequence and does not involve the probe target region in the promoter (25). Importantly, racial variation in methylation for these 7 loci was unlikely to be appreciably related to the known racial differences in breast tumor subtype distribution or differences in methylation patterns between the major intrinsic subtypes because we adjusted for IHC-based subtype, stage, age, and menopausal status in the GLM. Racial variation in breast tumor methylation was evident for these top CpG probes whether race was self-reported or defined by ancestry informative markers (Figure S1).
Table 2.
CpG probes differentially methylated by race in multivariable GLM analysis of all breast tumors
| Gene/Probe | CpG ID | Gene description | Mean βa AA (n=216) |
Mean βa Non- AA (n=301) |
Delta β |
Mean β Normal Breast (n=9) |
Coefb | Age-only adjusted q-valuec |
Fully- adjusted p-valued |
Fully- adjusted q-valuee |
Variant | Chr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No overlapping variantf | ||||||||||||
| DSC2_E90_F | cg08156793 | desmocollin 2 | 0.3564 | 0.2666 | 0.0898 | 0.1547 | 0.3777 | 6.48E-07 | 1.80E-06 | 3.31E-04 | None | 18 |
| KCNK4_E3_F | cg01352108 | potassium channel, subfamily K, member 4 | 0.4318 | 0.3502 | 0.0816 | 0.2877 | 0.2644 | 4.07E-06 | 1.27E-04 | 9.60E-03 | None | 11 |
| GSTM1_P266_Fg | cg19763514 | glutathione S-transferase mu 1 | 0.5757 | 0.6269 | −0.0512 | 0.4544 | −0.2681 | 3.27E-02 | 3.23E-04 | 2.08E-02 | None | 1 |
| HBII-52_E142_F | cg24301180 | SNORD115-1/small nucleolar RNA, C/D box 115-1 |
0.4026 | 0.5402 | −0.1376 | 0.5768 | −0.5283 | 4.97E-12 | 3.30E-10 | 1.06E-07 | None | 15 |
| Overlaps variant but minimal impact expectedh | ||||||||||||
| AXL_P223_R | cg09524393 | AXL receptor tyrosine kinase | 0.3311 | 0.2489 | 0.0822 | 0.1055 | 0.3015 | 6.15E-06 | 5.32E-04 | 3.11E-02 | SNP | 19 |
| DNAJC15_E26_R | cg10157207 | DnaJ (Hsp40) homolog, subfamily C, member 15 |
0.1399 | 0.1938 | −0.0539 | 0.1303 | −0.3789 | 2.30E-05 | 1.57E-05 | 1.68E-03 | SNP | 13 |
| TES_P182_F | cg00626984 | testis derived transcript (3 LIM domains) | 0.2223 | 0.2553 | −0.0330 | 0.1462 | −0.2989 | 8.61E-02 | 6.26E-04 | 3.50E-02 | SNP | 7 |
| Overlaps variant and/or ancestral allelic differencei | ||||||||||||
| CCL3_P543_R | cg05481196 | chemokine (C-C motif) ligand 3 | 0.8906 | 0.8598 | 0.0308 | 0.9081 | 0.3098 | 9.80E-03 | 3.76E-04 | 2.30E-02 | SNP | 17 |
| MAP2K6_E297_F | cg23296889 | mitogen-activated protein kinase kinase 6 | 0.1651 | 0.1160 | 0.0491 | 0.1085 | 0.3967 | 2.30E-05 | 2.24E-03 | 2.24E-03 | SNP | 17 |
| DNAJC15_P65_F | cg05035143 | DnaJ (Hsp40) homolog, subfamily C, member 15 |
0.8318 | 0.8682 | −0.0364 | 0.8381 | −0.2449 | 2.49E-04 | 9.20E-04 | 4.94E-02 | SNP | 13 |
| ID1_P659_R | cg09569033 | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
0.1850 | 0.1336 | 0.0514 | 0.1040 | 0.4381 | 5.33E-04 | 1.68E-03 | 1.68E-03 | SNP | 20 |
| PADI4_P1158_R | cg19159961 | peptidyl arginine deiminase, type IV | 0.4753 | 0.3589 | 0.1164 | 0.4671 | 0.3442 | 7.89E-11 | 9.24E-06 | 1.32E-03 | SNP | 1 |
| NTRK1_E74_F | cg18744444 | neurotrophic tyrosine kinase, receptor, type 1 |
0.7743 | 0.6043 | 0.1700 | 0.7435 | 0.7792 | 2.08E-05 | 1.52E-05 | 1.68E-03 | SNP | 1 |
| NNAT_P544_R | cg10288563 | neuronatin | 0.8504 | 0.8836 | −0.0332 | 0.8716 | −0.4324 | 3.24E-02 | 1.82E-05 | 1.80E-03 | SNP | 20 |
| WT1_P853_F | cg08219028 | Wilms tumor 1 | 0.1816 | 0.2852 | −0.1036 | 0.1518 | −0.5808 | 6.06E-05 | 2.96E-05 | 2.54E-03 | SNP | 11 |
| HLA-DQA2_P282_R | cg09782137 | major histocompatibility complex, class II, DQ alpha 2 |
0.7065 | 0.7625 | −0.0560 | 0.7280 | −0.3296 | 4.73E-03 | 9.38E-05 | 7.55E-03 | SNP | 6 |
| ABCB4_P892_F | cg02810586 | ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
0.6734 | 0.8491 | −0.1757 | 0.9052 | −0.9207 | 1.14E-15 | 5.33E-13 | 2.29E-10 | R, SNP |
7 |
| MSH3_P13_R | cg14636131 | mutS homolog 3 (E. coli) | 0.3998 | 0.6643 | −0.2645 | 0.5660 | −1.1511 | 1.75E-30 | 1.32E-27 | 1.70E-24 | Indel | 5 |
| MSH3_E3_F | cg06210628 | mutS homolog 3 (E. coli) | 0.5338 | 0.8121 | −0.2783 | 0.7250 | −1.4138 | 2.89E-27 | 5.40E-22 | 5.40E-22 | Indel | 5 |
| NOTCH1_P1198_F | cg26924342 | Notch 1 | 0.1761 | 0.1342 | 0.0419 | 0.1536 | 0.3496 | 1.32E-05 | 1.05E-06 | 2.71E-04 | R | 9 |
| ERCC1_P440_R | cg13282827 | excision repair cross-complementing rodent repair deficiency, complementation group 1 |
0.1702 | 0.1237 | 0.0465 | 0.0817 | 0.3462 | 7.14E-07 | 1.80E-06 | 3.31E-04 | R | 19 |
| PTPRF_E178_R | cg09322748 | protein tyrosine phosphatase, receptor type, F |
0.1429 | 0.1889 | −0.0460 | 0.1073 | −0.3715 | 1.71E-04 | 6.22E-06 | 1.00E-03 | R | 1 |
| ELL_P693_F | cg09597048 | elongation factor RNA polymerase II | 0.3922 | 0.3117 | 0.0805 | 0.2612 | 0.2937 | 2.34E-05 | 1.95E-04 | 1.39E-02 | R | 19 |
| GML_E144_F | cg21475536 | glycosylphosphatidylinositol anchored molecule like |
0.7043 | 0.7929 | −0.0886 | 0.7469 | −0.4365 | 3.63E-04 | 2.84E-04 | 1.92E-02 | indel | 8 |
Multivariable GLM was conducted using 1287 CpG probes from the Illumina Cancer Panel I array in 517 breast tumors.
Mean of β values, which ranged from 0 (completely unmethylated) to 1 (fully methylated).
Coefficients for comparison of AA cases to non-AA cases; positive coefficients indicate higher methylation in AA cases compared with non-AA cases, while negative values indicate higher methylation in non-AA cases than AA cases.
Adjusted for age and multiple comparisons (FDR).
Adjusted for age, menopausal status, stage, intrinsic subtype.
Adjusted for age, menopausal status, stage, intrinsic subtype, and multiple comparisons.
No known variant overlaps CpG probe based on review of Ensembl, dbSNP, or as annotated in Byun et al (19).
GSTM1del variant occurring in the coding sequence shows allele frequency difference between AAs and whites of approximately 15% (38).
Probe overlaps one SNP, but impact may be negligible due to location at end of probe, rarity of the minor allele, and/or no appreciable population difference (>10% in African vs European) reported; therefore, probes were retained.
Probes reported to overlap a SNP or repeat (R) according to Byun et al (19), or based on updated information in Ensembl and dbSNP that are likely to disrupt probe binding.
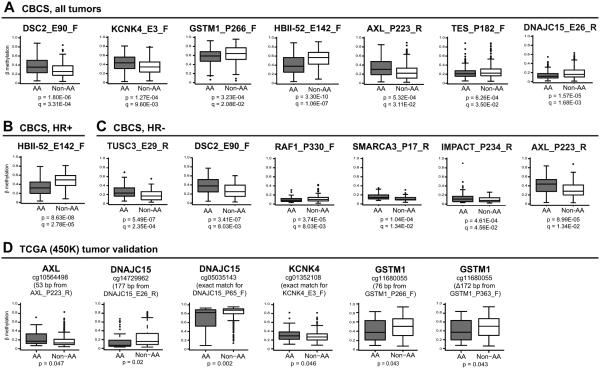
Figure 1. Box and whisker plots showing CpG probes exhibiting racial differences in breast tumor methylation.
CpG probes shown differed significantly (at q<0.05) in tumor methylation between AA and non-AA cases in GLM after controlling for age, menopausal status, stage, intrinsic subtype and multiple comparisons (FDR). Each box plot shows the median β-value (dark bar within box), the inter-quartile range (outer boundaries of box), and the whiskers extend to the group minimum and maximum β values. Probes shown either do not overlap a sequence variant or overlap a single SNP that is likely to have negligible impact on probe performance. Probes show significant racial differences in methylation in (A) all CBCS breast tumors (n=517), (B) HR+ positive breast tumors (n=337), and (C) HR− negative breast tumors (n=163). Shown are adjusted p-values and q-values from GLM. (D) Validation of racial variation in methylation for candidate genes in TCGA breast tumors. Data is shown for 450K CpG probes that exactly matched or were closest to (within 200 base pairs) probes on the GoldenGate array. P-values were determined by the T-test.
Stratified GLM was subsequently performed to determine whether the racial differences initially observed in CpG methylation were evident within hormone receptor (HR)-defined subtypes. As shown in Table S2, 4 additional probes that did not previously meet the threshold of q<0.05 showed significant racial differences among cases with HR−negative tumors, including TUSC3_E29_R, RAF1_P330_F, SMARCA3_P17_R, and IMPACT_P234_R. Notably, more CpG loci showed racial variation within HR− tumors than HR+ tumors. Only HBII-52_E142_F showed significant differential methylation within HR+ tumors of AAs versus non-AAs. Box and whisker plots summarizing the distribution of methylation β values for probes significantly varying by race (at q<0.05), overall or within HR-defined tumor subsets, are shown in Figure 1A-C. Technical validation comparing methylation obtained from the GoldenGate array with Q-MSP confirmed the comparability of the methylation measurements obtained by these two methods (Figure S2).
Figure 2. Methylation in matched pairs of breast tumors and PBLs from AA and non-AA cases in the CBCS.
(A) Box and whisker plots showing the distribution of methylation β values in 61 matched pairs of breast tumors and PBLs from AA and non-AA cases. (B) Bar charts showing individual methylation β values of matched tumor (black) and PBL (gray) pairs by race (26 AA, 35 non-AA) and age group (40 <50 years, 21 50+ years).
Although there were too few genes differentially methylated by racial group to perform formal gene ontology or pathway analyses, several of the genes showing epigenetic racial differences have roles in DNA repair, transcription, or mediate other DNA interactions (GSTM1, DNAJC15, SMARCA3/HLTF), are involved in cell adhesion (DSC2, TES), or are kinases important in signal transduction (AXL, RAF1).
Because AAs experience more adverse breast cancer outcomes than non-AAs, we determined whether the top candidate CpG markers might be differentially associated with disease-specific survival among AAs versus non-AAs. Kaplan-Meier plots in Figure S3 indicate that a low or intermediate (versus high) level of methylation at several probes (HBII-52_E142_F, TES_P182_F, DNAJC15_P65_F, DNAJC15_E26_R, DSC2_E90_F) was marginally associated with worse survival in AAs but not in non-AAs.
Validation in TCGA breast tumors
Breast tumor methylation and gene expression data from The Cancer Genome Atlas (TCGA) (21) were used to validate both our methylation findings and to test for correlations with expression for genes differentially methylated by race in CBCS. Comparison of 450K methylation beta values for probes that either directly matched or were located within 200 base pairs of GoldenGate probes in 42 AA and 291 white TCGA patients confirmed our findings of racial differences in methylation for AXL, GSTM1, KCNK4 and DNAJC15 (Table 3 and Figure 1D). Methylation was also significantly inversely correlated with gene expression for the majority of our candidate genes (Table S3).
Table 3.
TCGA validation of differential breast tumor methylation in genes showing racial variation in CBCS
| 450K Probe |
Chr | Gene | GoldenGate Probe |
Distance b/w GG & TCGA probes (bp)a |
Mean β AA |
Mean β Non-AA |
p-valueb |
|---|---|---|---|---|---|---|---|
| cg10564498 | 19 | AXL | AXL_P223_R | 53 | 0.223 | 0.182 | 0.05 |
| cg03247049 | 19 | AXL | AXL_E61_F | 14 | 0.086 | 0.063 | 0.22 |
| cg12722469 | 19 | AXL | AXL_E61_F | 169 | 0.189 | 0.147 | 0.12 |
| cg05035143 | 13 | DNAJC15 | DNAJC15_P65_F | 0 | 0.086 | 0.818 | 0.002 |
| cg12504148 | 13 | DNAJC15 | DNAJC15_P65_F | 34 | 0.610 | 0.690 | 0.02 |
| cg14729962 | 13 | DNAJC15 | DNAJC15_E26_R | 177 | 0.151 | 0.222 | 0.02 |
| cg00566759 | 18 | DSC2 | DSC2_E90_F | 58 | 0.065 | 0.055 | 0.34 |
| cg13870990 | 18 | DSC2 | DSC2_E90_F | 121 | 0.207 | 0.207 | 0.99 |
| cg00196671 | 18 | DSC2 | DSC2_E90_F | −58 | 0.062 | 0.060 | 0.87 |
| cg11680055 | 1 | GSTM1 | GSTM1_P266_F | 76 | 0.410 | 0.490 | 0.04 |
| cg24275769 | 18 | IMPACT | IMPACT_P234_R | −2 | 0.137 | 0.108 | 0.18 |
| cg13981356 | 18 | IMPACT | IMPACT_P234_R | −78 | 0.205 | 0.196 | 0.69 |
| cg03400437 | 18 | IMPACT | IMPACT_P234_R | 7 | 0.059 | 0.036 | 0.21 |
| cg22757447 | 18 | IMPACT | IMPACT_P234_R | −132 | 0.170 | 0.148 | 0.45 |
| cg01352108 | 11 | KCNK4 | KCNK4_E3_F | 0 | 0.329 | 0.281 | 0.05 |
| cg09396196 | 11 | KCNK4 | KCNK4_E3_F | 176 | 0.104 | 0.046 | 0.06 |
| cg06129498 | 11 | KCNK4 | KCNK4_E3_F | 59 | 0.169 | 0.142 | 0.09 |
| cg14673256 | 11 | KCNK4 | KCNK4_E3_F | 179 | 0.105 | 0.046 | 0.04 |
| cg02830576 | 3 | RAF1 | RAF1_P330_F | 0 | 0.255 | 0.246 | 0.40 |
| cg18568714 | 3 | RAF1 | RAF1_P330_F | −2 | 0.072 | 0.061 | 0.27 |
| cg13703021 | 3 | RAF1 | RAF1_P330_F | 157 | 0.146 | 0.125 | 0.21 |
| cg11297934 | 3 | RAF1 | RAF1_P330_F | −139 | 0.020 | 0.018 | 0.12 |
| cg24506533 | 3 | RAF1 | RAF1_P330_F | −122 | 0.022 | 0.022 | 0.93 |
| cg23032965 | 3 | RAF1 | RAF1_P330_F | −173 | 0.023 | 0.022 | 0.49 |
| cg15438497 | 3 | HLTF | SMARCA3_P17_R | 46 | 0.103 | 0.081 | 0.20 |
| cg24621354 | 7 | TES | TES_P182_F | −17 | 0.077 | 0.094 | 0.15 |
| cg20879085 | 7 | TES | TES_P182_F | 42 | 0.061 | 0.063 | 0.93 |
| cg19743881 | 7 | TES | TES_P182_F | 39 | 0.092 | 0.094 | 0.92 |
| cg16379337 | 7 | TES | TES_P182_F | 51 | 0.084 | 0.070 | 0.60 |
| cg00254079 | 7 | TES | TES_P182_F | −23 | 0.035 | 0.049 | 0.16 |
| cg03127174 | 8 | TUSC3 | TUSC3_E29_R | −28 | 0.079 | 0.083 | 0.83 |
| cg18145877 | 8 | TUSC3 | TUSC3_E29_R | −30 | 0.057 | 0.061 | 0.88 |
| cg03032098 | 8 | TUSC3 | TUSC3_E29_R | 140 | 0.112 | 0.094 | 0.46 |
Infinium 450K probes targeting CpG loci within 200 bp of the GoldenGate probes were assessed for racial differences in methylation in breast tumors from 42 African Americans and 291 whites in TCGA; negative value indicates the 450K probe was 5’ of the GoldenGate probe.
P-values determined by the T-test. There were no 450K probes for HBII-52 (alias SNORD115-1) within 200 bp of the GoldenGate probe.
c This 450K probe interrogates a CpG site (cg12504148) that is 55 bp upstream of the CpG targeted by the DNAJC15_E26_R GoldenGate probe.
Racial differences in methylation in PBLs or matched tumor/PBL pairs
Methylation patterns are considered to be tissue-specific, yet, if methylation at certain loci differs by race rather than tissue type, we expect that these differences might also be detectable in other normal cells, even if derived from a different tissue. To determine whether the racial differences in CpG methylation were restricted to tumors or occurred more broadly, we compared methylation in PBLs from 29 AAs and 40 non-AA cases matched on age and menopausal status. As shown in Table 4, the majority of CpG markers that were differentially methylated by race in tumors also differed significantly in PBLs, and the directionality of the difference in PBLs reflected that of the tumors. For example, similar to the pattern in tumors, DSC2_E90_F, GSTM1_P266_F, KCNK4_E3_F and AXL_P223_R all showed higher methylation in PBLs of AAs than in non-AAs even though the absolute levels of methylation varied somewhat between tumor and PBL. Differential methylation by race was also observed for CpG sites in AXL, GSTM1, DSC2, and DNAJC15 in an independent series of lymphoblastoid cell lines from 80 female AAs compared with 49 female Caucasian Americans described by Heyn et al (23) (Table S4).
Table 4.
Differential methylation in PBLs of AA versus non-AA cases in CBCS
| CpG probe | AA Mean β (n=29) |
Non-AA Mean β (n=40) |
Delta β |
Unadjusted p-value |
Adjusted p-valuea |
Overlapping variant |
|---|---|---|---|---|---|---|
| DSC2_E90_F | 0.4721 | 0.2904 | 0.1817 | <0.0001 | <0.0001 | None |
| KCNK4_E3_F | 0.5203 | 0.4144 | 0.1059 | <0.0001 | <0.0001 | None |
| GSTM1_P266_F | 0.3252 | 0.4637 | −0.1385 | 0.004 | 0.01 | None |
| TUSC3_E29_R | 0.1552 | 0.1040 | 0.0512 | 0.001 | 0.001 | None |
| SMARCA3_P17_R | 0.1079 | 0.0898 | 0.0181 | 0.003 | 0.002 | None |
| IMPACT_P234_R | 0.0972 | 0.0727 | 0.0245 | 0.001 | 0.001 | None |
| RAF1_P330_F | 0.0901 | 0.1105 | −0.0204 | 0.43 | 0.19 | None |
| HBII_52_E142_F | 0.5930 | 0.7702 | −0.1772 | 0.0001 | <0.0001 | None |
| AXL_P223_R | 0.6699 | 0.5306 | 0.1393 | <0.0001 | <0.0001 | SNPb |
| DNAJC15_E26_R | 0.1308 | 0.1582 | −0.0274 | 0.0003 | <0.0001 | SNPb |
| DNAJC15_P65_F | 0.9427 | 0.9565 | −0.0138 | 0.0003 | <0.0001 | SNP |
| TES_P182_F | 0.1642 | 0.1439 | 0.0203 | 0.10 | 0.05 | SNPb |
Comparison of transformed beta values in AAs versus non-AAs were performed using the T-test.
Adjusted for age and menopausal status.
Probe overlaps one SNP, but impact may be negligible due to SNP location at end of probe, rarity of the minor allele, and/or no appreciable difference in population (African vs European) frequencies reported.
We also compared methylation between matched pairs of breast tumors and PBLs within individual AA and non-AA cases (n=61) for several probes exhibiting the largest methylation differences by race. Methylation in tumor/PBL pairs was significantly correlated (with p<0.05) for the majority of probes (not shown). The box and whisker plots in Figure 2A show that among the 61 cases for whom tumor and PBL samples are matched, the methylation differences observed in tumors are also evident within PBLs. The bar chart in Figure 2B provides a comparison of individual β values in matched tumors and PBLs from breast cancer cases, further highlighting the differential methylation detected between AAs and non-AAs.
DISCUSSION
In this study, using a methylation array targeted to the promoters of genes important in cancer, we identified several genes differing in tumor DNA methylation between AAs and non-AAs even after controlling for potential confounders, including intrinsic subtype distributions which are known to vary by race (3). Previous studies, including the CBCS, have shown that breast tumor methylation varies between intrinsic subtypes (10, 18, 21, 26) or hormone receptor status (27), and by clinicopathologic characteristics such as tumor size and grade (18). Importantly, most of the CpG loci differing by race in CBCS breast tumors were also similarly differentially methylated in normal white blood cells of cases, although the absolute levels of methylation were not necessarily the same. A recent study that used the whole-genome Illumina 450K methylation array to assess racial differences in breast tumor methylation excluded probes that overlapped SNPs or that were otherwise ambiguous (15); however, we opted to retain such probes but to note their status. The impact of a probe that overlaps a target sequence potentially containing a SNP is difficult to predict, as it would depend on the SNP frequency in a given population as well as the probe sequence and precise location of the underlying SNP. Because many probes on the Illumina methylation arrays often overlap multiple CpG sites, the effect of an additional SNP-based single nucleotide mismatch on probe binding and methylation detection may be minimal. Consistent with this, we have previously observed in CBCS that some probes overlapping a SNP performed similarly in their detection of methylation (i.e., generated similar β values) to nearby probes in the same gene that do not (22). Genotyping for SNPs located within gene promoters was not performed in the CBCS and thus we could not rule out that racial variation in methylation might be related to the effects of underlying SNPs for certain probes; however, for most probes that overlapped a SNP, the variant alleles were relatively rare (minor allele frequency <10%) but in some cases differed by ancestry. Thus, to focus on CpG probes that would most reliably detect methylation differences (rather than underlying SNPs), we reviewed all probe target sequences against NCBI databases, assessed reported racial differences in allelic frequencies if available, and then removed those for which the underlying feature was likely to impede a reliable measurement of methylation. In the end, a total of 11 CpG probes were retained that were likely to provide a reasonably accurate assessment of racial variation in methylation.
Several prior studies using targeted methylation approaches to examine a small number of candidate genes known to be involved in breast cancer reported differences in breast tumor methylation between AAs and those of European ancestry. Consistent with our findings of more methylation differences by race within HR− tumors, Mehrotra et al (14) found significantly higher methylation in AAs than in Caucasian woman for four genes within a five-gene marker panel (HIN-1, TWIST1, CCND1, RASSF1A); these differences were only evident within ER-negative tumors and among women diagnosed before age 50 years. Similarly, Wang et al (13) examined a nine-marker panel (p16, RASSF1A, RARβ2, ESR1, CDH13, HIN1, SFRP1 and LINE1) and reported differential methylation for CDH13 between ER-negative tumors from younger AAs and European American (EA) women. Using the Illumina 450K whole-genome methylation array, the same array platform employed in TCGA, Ambrosone et al (15) also reported that many more loci were differentially methylated in AAs compared with EAs in ER-negative than in ER-positive breast cancers. The genes exhibiting differential promoter methylation by race in the CBCS are unique, and not among the candidate markers examined previously or the top loci reported from the 450K analysis in the Ambrosone study (15). Importantly, it should be noted that our analyses controlled for differences in intrinsic subtype, stage and age, whereas these prior studies did not. Therefore, it is unclear whether some loci reported previously as varying by race or ancestry may actually reflect differences in tumor intrinsic subtype distributions between the racial groups, such as within ER-negative tumors that are comprised of the basal-like, claudin-low and HER2+/ER− subtypes. Intriguingly, several genes showing racial variation in tumor methylation (HBII-52, TES, DNAJC15, DSC2) may be associated with differences in survival among AAs, while no such differences were observed among non-AAs. Further work is needed to confirm these results and to assess the possibility that underlying copy number or other changes contributed to these findings.
In the CBCS, we also observed racial differences in methylation in normal PBL samples from cases, with the overall patterns of tumor and PBL methylation for these probes being correlated, although the absolute levels of methylation in tumor versus PBL differed for some loci. Differences in the quality of DNA derived from FFPE tumors versus PBLs could potentially contribute to such methylation differences; however, PBL samples did not consistently show higher or lower methylation levels than in tumors. Although variation in PBL methylation profiles primarily reflect the admixture of white blood cell populations (28, 29), we still observed epigenetic differences by race. The finding of racial variation in methylation in both tumor and PBLs suggests underlying ancestral differences in the epigenetic state of some genes, with about half of differentially methylated loci being potentially explained by known sequence variations that could disrupt probe binding and methylation measurements. This is consistent with several prior studies noting that such genetic variants contributed to a substantial portion of racial differences in DNA methylation (23, 30-32). Racial differences in DNA methylation patterns in normal tissues have also been detected in PBLs from women in a multi-ethnic New York City Birth Cohort (33), in umbilical cord blood from newborns (34), and in breast tissue from reduction mammoplasty (11), raising the question of whether such normal epigenetic variation contributes to cancer risk. In a study of normal human prostate tissue and prostate cancer, several genes exhibited differential methylation between African Americans and EAs (12).
The genes displaying racial variation in tumor methylation in the CBCS included several involved in transcription or interactions with DNA (DNAJC15, SMARCA3 or HLTF), signal transduction (AXL, RAF1), carcinogen detoxification (GSTM1), cell adhesion (DSC2), or are known tumor suppressors (TUSC3, TES). Desmocollin 2 (DSC2), a membrane glycoprotein involved in cell-cell adhesion and maintenance of normal epithelial architecture is an independent prognostic marker for esophageal (35) and pancreatic cancers (36), and high DSC2 expression may be a marker of basal-like breast tumors (37). The carcinogen detoxifying enzyme, GSTM1, exhibits an intragenic deletion variant resulting in loss of enzyme activity (25), elevated cancer risk, and shows racial variation between blacks and whites (38); however, whether the variant is linked with promoter methylation state is unclear. Testin (TES) and TUSC3 are tumor suppressor genes that are silenced in part via promoter methylation (39); loss of expression of TES is an independent poor prognostic marker in breast cancer (40), while silencing of TUSC3 may be a poor prognostic factor in ovarian cancer (41). AXL is a receptor tyrosine kinase and an epithelial-to-mesenchymal transition (EMT)-induced regulator of breast cancer metastasis and patient survival (42-45). AXL expression is regulated in part by Sp1 transcription factor binding and methylation (42, 44, 46). In contrast to most other genes showing epigenetic racial variation in the CBCS, AXL methylation was not inversely correlated with mRNA expression in the TCGA tumor set, most likely because the 450K AXL CpG probes analyzed in relation to gene expression appear to be outside of these critical Sp1 binding sites. Intriguingly, AXL was reported to be aberrantly hypermethylated in association with prenatal tobacco smoke exposure (47), suggesting that epigenetic racial variation may be associated with lifestyle or environmental exposures and could potentially contribute to cancer risk.
The strengths of this study include the large, well-characterized population-based series of mostly early-stage breast cancer cases, providing the power to detect even modest differences in methylation. Epigenetic racial differences for several of the genes in CBCS were independently confirmed in TCGA breast tumors although the number of AAs in this dataset was limited. Moreover, the availability of demographic, clinical and subtype information allowed us to control for factors that are known to vary by patient or tumor subsets. However, our study was limited by use of the targeted cancer-focused array rather than a whole-genome methylation array.
The implications of the racial differences in methylation discovered in this study for breast cancer development, progression or outcomes in AA or non-AA women are unclear, and further work is needed to confirm these findings. It is of interest that the epigenetic differences were most evident within HR-negative tumors despite controlling for intrinsic subtype; however, it is possible that these racial variations signify different proportions of HR-negative subtypes in AA and non-AA women. Interestingly, most of the genes exhibiting differential tumor methylation by race in the CBCS (DSC2, AXL, DNAJC15, TES, TUSC3) are among the genes defining subclasses of triple-negative breast cancer as reported by Lehmann et al (48). Whether the prevalence of triple negative subtypes, which were not defined within CBCS, vary between AAs and non-AAs is presently unknown. The results of this study highlight the possibility that methylation patterns of breast tumors, particularly the more aggressive HR− tumors, may differ by ancestry, and that the racial differences may not be a tumor-specific phenomenon, suggesting that such variations could also contribute to cancer risk. Clearly, more research is needed to validate these findings and determine whether they contribute to the known racial disparity in breast cancer survival.
Supplementary Material
Acknowledgements
We are grateful to Dr. Andy Olshan for critical review of this manuscript and to the participants and study staff of the Carolina Breast Cancer Study for their continuing support of the study. This work is dedicated to the memory of our colleague, the late Dr. Robert Millikan, principal investigator of the CBCS.
Financial support: This research was supported by grants to KC from Susan G. Komen Foundation (grant #KG081397) and from the University Cancer Research Fund of the Lineberger Comprehensive Cancer Center at the University of North Carolina at Chapel Hill. The Carolina Breast Cancer Study was also funded by the University Cancer Research Fund of North Carolina and the National Cancer Institute Specialized Program of Research Excellence (SPORE) in Breast Cancer (NIH/NCI P50-CA58223).
Footnotes
Disclosure of Potential Conflict of Interest: There are no conflicts of interest.
REFERENCES
- 1.DeSantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA Cancer J Clin. 2011;61:409–18. doi: 10.3322/caac.20134. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumors. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 4.Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–34. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 5.Smigal C, Jemal A, Ward E, Cokkinides V, Smith R, Howe H, et al. Trends in breast cancer by race and ethnicity: update 2006. CA: Cancer J Physicians. 2006;56:168–183. doi: 10.3322/canjclin.56.3.168. [DOI] [PubMed] [Google Scholar]
- 6.Maskarinec G, Sen C, Koga K, Conroy SM. Ethnic differences in breast cancer survival: status and determinants. Womens Health (Lond Engl) 2011;7:677–87. doi: 10.2217/whe.11.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shavers VL, Brown ML. Racial and ethnic disparities in the receipt of cancer treatment. J Natl Cancer Inst. 2002;94:334–57. doi: 10.1093/jnci/94.5.334. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien KM, Cole SR, Tse CK, Perou CM, Carey LA, Foulkes WD, et al. Intrinsic breast tumor subtypes, race, and long-term survival in the Carolina Breast Cancer Study. Clin Cancer Res. 2010;16:6100–10. doi: 10.1158/1078-0432.CCR-10-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herman JG. Epigenetic changes in cancer and preneoplasia. Cold Spring Harb Symp Quant Biol. 2005;70:329–33. doi: 10.1101/sqb.2005.70.036. [DOI] [PubMed] [Google Scholar]
- 10.Kamalakaran S, Varadan V, Giercksky Russnes HE, Levy D, Kendall J, Janevski A, et al. DNA methylation patterns in luminal breast cancers differ from non-luminal subtypes and can identify relapse risk independent of other clinical variables. Mol Oncol. 2011;5:77–92. doi: 10.1016/j.molonc.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumitrescu RG, Marian C, Krishnan SS, Spear SL, Kallakury BVS, Perry DJ, et al. Familial and racial determinants of tumour suppressor genes promoter hypermethylation in breast tissues from healthy women. J Cell Mol Med. 2010;14:1468–1475. doi: 10.1111/j.1582-4934.2009.00924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwabi-Addo B, Wang S, Chung W, Jelinek J, Patierno SR, Wang B, et al. Identification of differentially methylated genes in normal prostate tissues from African American and Caucasian men. Clinical Cancer Research. 2010;16:3539–3547. doi: 10.1158/1078-0432.CCR-09-3342. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Dorsey TH, Terunuma A, Kittles RA, Ambs S, Kwabi-Addo B. Relationship between tumor DNA methylation status and patient characteristics in African-American and European-American women with breast cancer. PLoS One. 2012;7:e37928. doi: 10.1371/journal.pone.0037928. doi: 10.1371/journal.pone.0037928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehrotra J, Ganpat MM, Kanaan Y, Fackler MJ, McVeigh M, Lahti-Domenici J, et al. Estrogen receptor/ progesterone receptor-negative breast cancers of young African-American women have a higher frequency of methylation of multiple genes than those of Caucasian women. Clin Cancer Res. 2004;10:2052–2057. doi: 10.1158/1078-0432.ccr-03-0514. [DOI] [PubMed] [Google Scholar]
- 15.Ambrosone CB, Young AC, Sucheston LE, Wang D, Yan L, Liu S, et al. Genome-wide methylation patterns provide insight into differences in breast tumor biology between American women of African and European ancestry. Oncotarget. 2014;5:237–48. doi: 10.18632/oncotarget.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman B, Moorman PG, Millikan R, Qaqish BF, Geradts J, Aldrich TE, et al. The Carolina Breast Cancer Study: integrating population-based epidemiology and molecular biology. Breast Cancer Res Treat. 1995;35:51–60. doi: 10.1007/BF00694745. [DOI] [PubMed] [Google Scholar]
- 17.Barnholtz-Sloan JS, Shetty PB, Guan X, Nyante SJ, Luo J, Brennan DJ, et al. FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African-American and younger women. Carcinogenesis. 2010;31:1417–23. doi: 10.1093/carcin/bgq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conway K, Edmiston SN, May R, Kuan PF, Chu H, Bryant C, et al. DNA methylation profiling in the Carolina Breast Cancer Study defines cancer subclasses differing in clinicopathologic characteristics and survival. Breast Cancer Research. 2014;16:450. doi: 10.1186/s13058-014-0450-6. DOI: 10.1186/s13058-014-0450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, et al. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet. 2009;18:4808–17. doi: 10.1093/hmg/ddp445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wedderburn, Robert WM. Quasi-likelihood functions, generalized linear models, and the Gauss-Newton method. Biometrika. 1974;61:439–47. [Google Scholar]
- 21.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bardowell SA, Parker J, Fan C, Crandell J, Perou CM, Swift-Scanlan T. Differential methylation relative to breast cancer subtype and matched normal tissue reveals distinct patterns. Breast Cancer Res Treat. 2013;142:365–80. doi: 10.1007/s10549-013-2738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heyn H, Moran S, Hernando-Herraez I, Sayols S, Gomez A, Sandoval J, et al. DNA methylation contributes to natural human variation. Genome Res. 2013;23:1363–72. doi: 10.1101/gr.154187.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas NE, Slater NA, Edmiston SN, Zhou X, Kuan PF, Groben PA, et al. DNA methylation profiles in primary cutaneous melanomas are associated with clinically significant pathologic features. Pigment Cell & Melanoma Research. 2014;27:1097–1105. doi: 10.1111/pcmr.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu S, Wang Y, Roe B, Pearson WR. Characterization of the human class Mu glutathione S-transferase gene cluster and the GSTM1 deletion. J Biol Chem. 1998;273:3517–27. doi: 10.1074/jbc.273.6.3517. [DOI] [PubMed] [Google Scholar]
- 26.Rønneberg JA, Fleischer T, Solvang HK, Nordgard SH, Edvardsen H, Potapenko I, et al. Methylation profiling with a panel of cancer related genes: association with estrogen receptor, TP53 mutation status and expression subtypes in sporadic breast cancer. Mol Oncol. 2011;5:61–76. doi: 10.1016/j.molonc.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fackler MJ, Umbricht CB, Williams D, Argani P, Cruz LA, Merino VF, et al. Genome-wide methylation analysis identifies genes specific to breast cancer hormone receptor status and risk of recurrence. Cancer Res. 2011;71:6195–207. doi: 10.1158/0008-5472.CAN-11-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koestler DC, Christensen B, Karagas MR, Marsit CJ, Langevin SM, Kelsey KT, et al. Blood-based profiles of DNA methylation predict the underlying distribution of cell types: a validation analysis. Epigenetics. 2013;8:816–26. doi: 10.4161/epi.25430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Moen EL, Liu C, Mu W, Gamazon ER, Delaney SM, et al. 2014 doi: 10.1093/hmg/ddu313. pii: ddu313. PMID: 24943591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20:883–9. doi: 10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moen EL, Zhang X, Mu W, Delaney SM, Wing C, McQuade J, et al. Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics. 2013;194:987–96. doi: 10.1534/genetics.113.151381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terry MB, Ferris JS, Pilsner R, Flom JD, Tehranifar P, Santella RM, et al. Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol. Biomarkers Prev. 2008;17:2306–2310. doi: 10.1158/1055-9965.EPI-08-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adkins RM, Krushkal J, Tylavsky FA, Thomas F. Racial differences in gene-specific DNA methylation levels are present at birth. Birth Defects Research Part A – Clinical and Molecular Teratology. 2011;91:728–736. doi: 10.1002/bdra.20770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang WK, Gu W, Li EM, Wu ZY, Shen ZY, Shen JH, et al. Reduced membranous and ectopic cytoplasmic expression of DSC2 in esophageal squamous cell carcinoma: an independent prognostic factor. Hum Pathol. 2010;41:1456–65. doi: 10.1016/j.humpath.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Hamidov Z, Altendorf-Hofmann A, Chen Y, Settmacher U, Petersen I, Knösel T. Reduced expression of desmocollin 2 is an independent prognostic biomarker for shorter patients survival in pancreatic ductal adenocarcinoma. J Clin Pathol. 2011;64:990–4. doi: 10.1136/jclinpath-2011-200099. [DOI] [PubMed] [Google Scholar]
- 37.Culhane AC, Quackenbush J. Confounding effects in "A six-gene signature predicting breast cancer lung metastasis". Cancer Res. 2009;69:7480–5. doi: 10.1158/0008-5472.CAN-08-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell DA, Taylor JA, Paulson DF, Robertson CN, Mohler JL, Lucier GW. Genetic risk and carcinogen exposure: a common inherited defect of the carcinogen-metabolism gene glutathione S-transferase M1 (GSTM1) that increases susceptibility to bladder cancer. J Natl Cancer Inst. 1993;85:1159–64. doi: 10.1093/jnci/85.14.1159. [DOI] [PubMed] [Google Scholar]
- 39.Tobias ES, Hurlstone AF, MacKenzie E, McFarlane R, Black DM. The TES gene at 7q31.1 is methylated in tumours and encodes a novel growth-suppressing LIM domain protein. Oncogene. 2001;20:2844–53. doi: 10.1038/sj.onc.1204433. [DOI] [PubMed] [Google Scholar]
- 40.Zhu J, Li X, Kong X, Moran MS, Su P, Haffty BG, et al. Testin is a tumor suppressor and prognostic marker in breast cancer. Cancer Sci. 2012;103:2092–101. doi: 10.1111/cas.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pils D, Horak P, Vanhara P, Anees M, Petz M, Alfanz A, et al. Methylation status of TUSC3 is a prognostic factor in ovarian cancer. Cancer. 2013;119:946–54. doi: 10.1002/cncr.27850. [DOI] [PubMed] [Google Scholar]
- 42.Choufani S, Shapiro JS, Susiarjo M, Butcher DT, Grafodatskaya D, Lou Y, et al. A novel approach identifies new differentially methylated regions (DMRs) associated with imprinted genes. Genome Res. 2011;21:465–76. doi: 10.1101/gr.111922.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breton CV, Salam MT, Gilliland FD. Heritability and role for the environment in DNA methylation in AXL receptor tyrosine kinase. Epigenetics. 2011;6:895–8. doi: 10.4161/epi.6.7.15768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gjerdrum C, Tiron C, Høiby T, Stefansson I, Haugen H, Sandal T, et al. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc Natl Acad Sci USA. 2010;107:1124–9. doi: 10.1073/pnas.0909333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vajkoczy P, Knyazev P, Kunkel A, Capelle HH, Behrndt S, von Tengg-Kobligk H, et al. Dominant-negative inhibition of the Axl receptor tyrosine kinase suppresses brain tumor cell growth and invasion and prolongs survival. Proc Natl Acad Sci USA. 2006;103:5799–804. doi: 10.1073/pnas.0510923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mudduluru G, Allgayer H. The human receptor tyrosine kinase Axl gene—promoter characterization and regulation of constitutive expression by Sp1, Sp3 and CpG methylation. Biosci Rep. 2008;28:161–76. doi: 10.1042/BSR20080046. [DOI] [PubMed] [Google Scholar]
- 47.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–7. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.