Abstract
Anophthalmia/microphthalmia (A/M) is a genetically heterogeneous birth defect for which the etiology is unknown in more than 50% of patients. We used exome sequencing with the ACE Exome™ (Personalis, Inc; 18 cases) and UCSF Genomics Core (21 cases) to sequence 28 patients with A/M and four patients with varied developmental eye defects. In the 28 patients with A/M, we identified de novo mutations in three patients (OTX2, p.(Gln91His), RARB, p.Arg387Cys and GDF6, p.Ala249Glu) and inherited mutations in STRA6 in two patients. In patients with developmental eye defects, a female with cataracts and cardiomyopathy had a de novo COL4A1 mutation, p.(Gly773Arg), expanding the phenotype associated with COL4A1 to include cardiomyopathy. A male with a chorioretinal defect, microcephaly, seizures and sensorineural deafness had two PNPT1 mutations, p.(Ala507Ser) and c.401-1G>A, and we describe eye defects associated with this gene for the first time. Exome sequencing was efficient for identifying mutations in pathogenic genes for which there is no clinical testing available and for identifying cases that expand phenotypic spectra, such as the PNPT1 and COL4A1-associated disorders described here.
Keywords: Anophthalmia/microphthalmia, exome sequencing, PNPT1, COL4A1, FBLN1
Introduction
Anophthalmia, microphthalmia and coloboma are important structural birth defects because of the medical significance of severely reduced vision.1,2 A minority of individuals with A/M receive an identified genetic cause for their birth defect. Array comparative genomic hybridization (array CGH) found pathogenic chromosome aberrations in 3–13% of A/M patients in two small studies of 32 and 37 individuals.3,4 Loss of function mutations in SOX2, the most commonly mutated gene, have been identified in 10–20% of A/M patients and ALDH1A3 mutations have been estimated to occur in up to 10% of patients with A/M.5,6 Screening of 150 A/M patients for mutations in SOX2, OTX2, RAX, FOXE3, VSX2, GDF6 and PAX6 by direct sequencing and semi-quantitative multiplex PCR detected mutations in 21% patients.7 One study used exome sequencing to examine 11 unrelated patients with developmental eye disorders, including cataracts/microcornea, Peter’s anomaly and microphthalmia, and found causative mutations in GJA8, CRYGC, PAX6 and CYP1B1 in four individuals.8 In this study, we provide the results of exome sequencing in 25 trios (affected patient with unaffected biological parents), two duos with affected parent and child, and five single probands with A/M or other developmental eye defects.
Materials and Methods
Patient MCA399 (COL4A1)
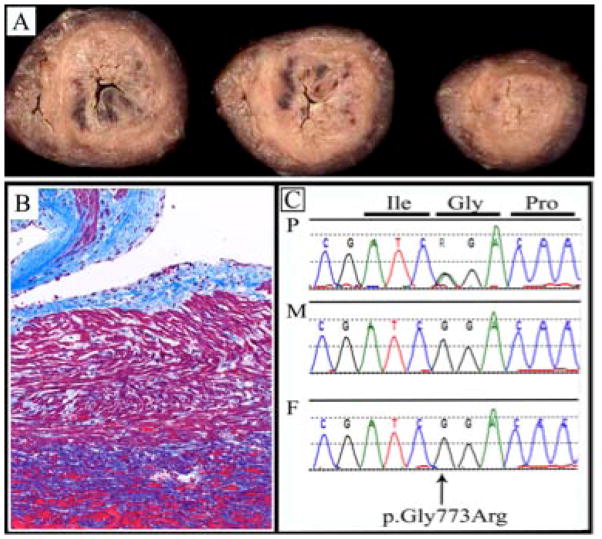
The pregnancy was complicated by mild pre-eclampsia. Growth retardation was noted from 28 weeks of gestation. A female baby was delivered by a C-section for premature labor at 34 weeks of gestation and weighed 1,300 g (<10th percentile). She had respiratory distress, requiring continuous positive airway pressure. She was diagnosed with congenital cataracts and microcornea at five weeks of age. She was scheduled for elective cataract extraction at ten weeks of life, but in surgery, she suffered ventricular fibrillation and asystole. After resuscitation, she was placed on extracorporeal membrane oxygenation. An echocardiogram on the day following her arrest showed poor left and right ventricular function. Cranial ultrasound showed patchy periventricular white matter echogenicity consistent with hemorrhage or ischemic injury. She developed status epilepticus and multiple organ dysfunction and her parents opted for comfort care. TORCH screening was negative. Array CGH revealed a small, maternally-inherited deletion of unknown significance [arr 7q21.1(87,530,062–87,804,618)x1]. At autopsy, congenital cataracts were confirmed and the brain showed multifocal, intraparenchymal hemorrhages up to two cm in size that were interpreted as diffuse hypoxic ischemic encephalopathy. Examination of the heart revealed a hypertrophic cardiomyopathy involving both ventricles (Figs. 1A and 1B) and microscopy showed abnormal mitochondria with collapsed cristae. The appearance of the skeletal muscle was consistent with a chronic metabolic myopathy, with endomysial fibrosis, inflammation and abnormal mitochondria.
Fig. 1. A female with congenital cataracts and cardiomyopathy with p.(Gln773Arg) in COL4A1.
(a) Photograph of gross pathology of heart muscle from a female with p.(Gln773Arg) in COL4A1, showing hypertrophy of the left and right ventricles consistent with cardiomyopathy. (b) Photograph of histology of heart muscle stained with trichrome to show fibrosis (indicated by blue staining), indicating long-standing cardiomyopathy. (c) Chromatogram showing c.2317G>A, predicting p.(Gly773Arg) in COL4A1. The mutation is present in the patient and not inherited from either parent.
Patient MCA350 (PNPT1)
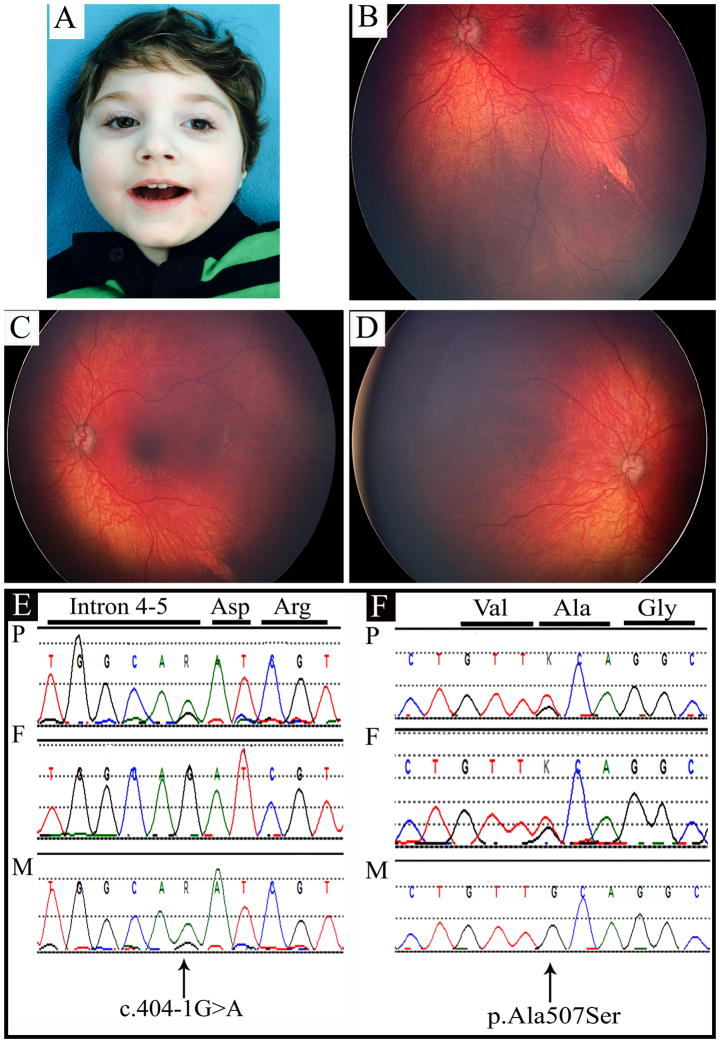
Intrauterine growth retardation was noted at 35 weeks of gestation and a male infant weighing 1,900 g (3rd-10th percentile), was delivered by C-section. His early medical problems included hypotonia and developmental delays. He had a left-sided peripheral chorioretinal defect (Figs. 2B and 2C), optic atrophy and an electroretinogram showed a mild reduction in outer retinal function affecting rods. He was diagnosed with cortical visual impairment. He manifested infantile spasms and his electroencephalogram revealed hypsarrhythmia. He developed myoclonic epilepsy with daily seizures that were uncontrolled despite medications and a ketogenic diet. He had bilateral severe to profound sensorineural hearing loss. At at 13 months of age (Fig. 2A), length was 69.5 cm (just below 3rd percentile), weight was 7.8 kg (3rd percentile) and occipitofrontal circumference was 39.5 cm (<3rd centile). He had a sloping forehead with a ridged metopic suture, reduced central tone and he was unable to visually fix, follow or track. His investigations included normal metabolic testing and array CGH revealed a normal male karyotype. A magnetic resonance imaging (MRI) scan of the brain at six months of age showed diminished white matter, simplified gyri and thinning of the corpus callosum and optic tracts.
Fig. 2. A Male with Developmental Delays, Seizures, Sensorineural Hearing Loss and a Unilateral Chorioretinal Defect with Two Inherited PNPT1 mutations.
Fig. 2A. Frontal photograph of the patient at twenty two months of age.
(b,c) Photographs of left retina, showing chorioretinal defect.
Fig. 2D. Photograph of right retina showing normal morphology.
Fig. 2E. Chromatogram showing splice mutation c.401-1G>A in PNPT1. The mutation is inherited from the patient’s unaffected mother.d
Fig. 2F. Chromatogram showing c.1519G>T, predicting p.(Ala507Ser) in PNPT1. The mutation was inherited from the patient’s unaffected father.
We used exome sequencing with the ACE Exome™ (Personalis, Inc; 18 cases with 12 trios, one autosomal dominant pedigree and 5 single probands) and UCSF Genomics Core (21 cases with 20 trios and one autosomal dominant pedigree) to sequence 28 patients with A/M and four patients with developmental eye defects. For the exomes processed by the UCSF Core, sequencing and analysis were performed as described.8
Additional analyses were undertaken to search for de novo mutations. Raw sequence data was processed using the FastX (http://hannonlab.cshl/edu/fastx_toolkit) suite of utilities, BWA_MEM algorithm and SAMtools mpileup utility.10 The DeNovoGear program 11 was used to calculate the likelihood of de novo mutation at each site in the proband’s exome. Sites that had a likelihood of being de novo >0.75 and had a mapping quality score >30 were selected for further analysis. Variants were then assessed for their potential to be deleterious and mutations that had a SIFT score <0.05 or a PolyPhen-2 score >0.909 were retained. Variants were excluded from consideration if they were not in exons, were present in dbSNP135, or had a minor allele frequency of greater than 0.01 in the 1000 Genomes database.
For the exomes sequenced by Personalis, coverage across all medically relevant genes was improved through high depth augmented exome sequencing (Personalis, Inc., Menlo Park, CA, USA) using their ACE (Accuracy and Content Enhanced) whole exome sequencing methods. Sequencing was performed to high depth, attaining a mean sequence read depth of ~200X. ACE Exome data was aligned and variants were called and annotated using the Personalis Pipeline and the trio data were analyzed through the Personalis Disease Variant Discovery Service. For both methods of exome sequencing, mean coverage for SOX2 was greater than 30X for all samples with bilateral anophthalmia.
Results
We identified mutations that we classified as pathogenic in eight individuals (Table 1). A female with bilateral anophthalmia, growth delays, intellectual disability and autism had a de novo, novel mutation in OTX2, c.273G>C, predicting p.(Gln91His) (MIM 600037; NM_001270525; Fig. S1A). A Hispanic female with bilateral severe microphthalmia and unilateral coloboma, left diaphragmatic hernia, cleft palate and an Arnold Chiari I malformation had a published, de novo mutation in RARB, c.1159C>T, predicting p.Arg387Cys (MIM 180220; NM_000965; Fig. S1B). 12 A Caucasian male with bilateral anophthalmia had a de novo mutation in GDF6, c.746C>A, predicting p.Ala249Glu (MIM 613094; NM_001001557.2; Fig. S1C) that was described in three probands with coloboma, microphthalmia, post-axial polydactyly and Klippel-Feil syndrome. 13
Table 1.
Clinical Details and Sequencing Results in 12 Patients with Eye Defects and Pathogenic Mutations
| Number | Sex; Ethnicity | Right eye | Left eye | Other Clinical Findings | Prior Testing | Gene | Mutation/Zygosity/Inheritance |
|---|---|---|---|---|---|---|---|
| 1/11-022 | F; C | AN | AN | ID, autism, growth delay | Array CGH | OTX2 | p.(Gln91His)/Het./de novo |
| 2/12-026 | F; H | MICRO, COL | Severe AN/MICRO | L diaphragmatic hernia, cleft palate, Chiari I malformation | Array CGH | RARB | p.Arg387Cys/Het./de novo |
| 3/09-125 | M; C | AN | AN | - | - | GDF6 | p.Ala249Glu/Het/de novo |
| 4/13-007 | M; C | MICRO | MICRO | - | Array CGH | STRA6 | p.Tyr18Thrfs*54/Het./Mat c.1684+1G>A/Het./Pat. |
| 5/11-035 | F; A/C | MICRO, COL, retinal detachment | MICRO, COL | Vascular ring, R aortic arch, ID | SOX2 | STRA6 | p.(Arg408Gln )/Het./Mat. |
| 6/MCA363 | F; H | CAT | CAT | R microtia | Array CGH | GCNT2 | p.Tyr347Cys/HZ/ Mat. and Pat. |
| 7/MCA399 | F; C | CAT | CAT | Cardiomyopathy | Array CGH | COL4A1 | p.Gln773Arg/Het./de novo |
| 8/MCA350 | M; C | - | Chorioretinal defect | Microcephaly, seizures, sensorineural deafness | Array CGH | PNPT1 | p.(Ala507Ser)/Het./Pat. And c.401-1G>A/Het./Mat. |
| 9/11-033 | M; C | MICRO, CAT, PHPV | - | ID | - | ABCB6 | p.(Ala43Asn)/Het./Mat. |
| 10/MCA355 | F; C | ASD | Pulmonic stenosis | - | VSX2 | p.(Arg274Cys)/Het./Pat. | |
| 11/09-152 | M; U | COL | COL | Enlarged lateral/3rd ventricles, brachydactyly | - | SIX6 | p.(Glu129Lys)/Het./Pat. |
| 12/MCA319 | M; H | AN | AN | Facial cleft, syndactyly | Array CGH | FBLN1 | p.(Thr566Ile)/HZ/Mat. and Pat. |
F = female; M = male; C = Caucasian; H = Hispanic; A = Asian; U = unknown; AN = Anophthalmia; MICRO = Microphthalmia; COL = Coloboma; CAT = Cataracts; PHPV = Persistent Hyperplasia of the Primary Vitreous; ID = Intellectual Disability; L = left; R = right; Array CGH = array comparative genomic hybridization; Het = heterozygous; HZ = homozygous; Mat. = maternally inherited; Pat. = paternally inherited.
A male with bilateral microphthalmia was a compound heterozygote for two STRA6 (MIM610745) mutations – a maternally inherited, frameshift mutation, c.52delT, predicting p.(Tyr18Thrfs*54), and a paternally inherited, splice-site mutation, c.1684+1G>A (NM_022369.3; data not shown). In a patient with bilateral microphthalmia and coloboma, unilateral retinal detachment, right-sided aortic arch, vascular ring and intellectual disability, a single STRA6 mutation was identified, c.1223G>A, predicting p.(Arg408Gln) (data not shown). This mutation was inherited from the patient’s mother, who also had bilateral microphthalmia. A second mutation was not found.
In patients without A/M, two patients with cataracts had previously reported mutations in GCNT2 (MIM110800) and COL4A1 (MIM120130), whereas one patient with a unilateral chorioretinal defect was compound heterozygous for two novel mutations in PNPT1 (MIM 176885). In a Hispanic female with congenital cataracts and unilateral microtia (Fig. 3A), we detected homozygosity for c.1040A>C, predicting p.(Tyr347Cys) in GCNT2 (NM_001491.2; Fig. 3B). 14 A female infant with congenital cataracts and cardiomyopathy had a de novo mutation, c.2317G>A, predicting p.(Gly773Arg) (NM_001845.4; Fig. 1C) in COL4A1 that has also previously been described. 15–17 In a male with severe delays, seizures, microcephaly, sensorineural deafness and a chorioretinal defect, we found compound heterozygosity for two PNPT1 sequence variants – a mutation affecting the acceptor splice-site for exon 5, c.401-1G>A, that was maternally inherited (NM_033109.4; Fig. 2E) and c.1519G>T, predicting p.(Ala507Ser) (Fig. 2F) that was paternally inherited. Although eye manifestations have not been described in patients with PNPT1 gene mutations, the remaining phenotypic features were consistent with the clinical presentations seen in other patients 18,19 and as both sequence variants were predicted to be deleterious, we consider them to be disease-causing.
We found four variants in known A/M genes that were of uncertain significance because of non-penetrance or inheritance in a state not known to be associated with disease (Table 1). In an Hispanic male with bilateral anophthalmia, a lateral facial cleft and syndactyly born to consanguineous parents, we found homozygosity for a Fibulin-1 (FBLN1 MIM135820) mutation, c.1607C>T, predicting p.(Thr566Ile) (NM_006486.2; Fig. S2D). This mutation has been detected in the NHLBI exome variant server cohort, but the frequency of the mutant allele is low at 0.002 and the allele was predicted to be possibly damaging by PolyPhen-2 (probability of 0.507). Haploinsufficiency for FBLN1 was described in a pedigree with complex synpolydactyly 20, but the significance for eye development is unknown.
Discussion
We report eight pathogenic mutations and four sequence variants of unknown significance in 32 patients with developmental eye defects, demonstrating the utility of exome sequencing to efficiently identify mutations in pathogenic genes, several of which currently have no clinical genetic testing available.
PNPT1 mutations have previously been described. Two siblings with severe sensorineural hearing loss without any other associated features were homozygous for p.(Glu475Gly) in the PNPase residue 18 and two siblings with severe encephalopathy, choreoathetotic movements and dystonia with a presentation similar to a respiratory-chain deficiency were homozygous for p.(Gly387Arg).19 PNPT1 is a polynucleotide phosphorylase (PNPase) that is located in the mitochondrial intermembranous space and facilitates the importation of mRNAs, such as 5SRNA, MRP RNA, RNAse P and RNA, into cells. No patient has previously been reported with eye findings.
In a female with congenital cataracts, hypertrophic cardiomyopathy, skeletal myopathy and evidence of abnormal mitochondria, we found a reported de novo, COL4A1 mutation, c.2317G>A, predicting p.(Gly773Arg) as found in three case reports.15–17 COL4A1 mutations have primarily been associated with cerebral malformations. The phenotype can be broad and supraventricular arrhythmias, structural heart defects and skeletal muscle myopathy have been recorded, although we found no prior mention of cardiomyopathy as found in this child.21 The eye phenotype in patients with COL4A1 mutations includes retinal arteriolar tortuosity, congenital or juvenile cataracts, Axenfeld Rieger syndrome, glaucoma, microcornea and microphthalmia.21
We became interested in FBLN1 because haploinsufficiency for this gene resulting from an autosomal chromosome translocation was described in a family with synpolydactyly, a limb malformation related to the syndactyly seen in our patient.20 FBLN1 was expressed in sclerae obtained from human adult (35–68 years) donor eyes and is also found at the sites of epithelial-mesenchymal interaction and in mesenchymal tissue in the central nervous system, the endocardium of the heart, perichondrium and perineurium of peripheral nerves. FBLN1 expression in cultured human scleral fibroblasts is regulated by retinoic acid, a molecule that is involved in the regulation of eye growth.22 However, the relationship with anophthalmia in our patient is unknown. In conclusion, exome sequencing efficiently identified the genetic etiology of eye defects in patients for whom it would have otherwise been difficult to arrive at a diagnosis, due to mutations in genes that are not included in clinical panels or available as single gene tests. The genetic etiologies of the eye defects identified through exome sequencing in this study expands the phenotypic spectra of PNPT1, COL4A1 and FBNL1-associated disorders. This study highlights the genetic heterogeneity and broad phenotypes underlying developmental eye defects and the challenges associated with genetic testing for these conditions.
Supplementary Material
Fig. S1. Chromatograms Showing Mutations in Pathogenic Genes in Developmental Eye Disorders.
Fig. S1A. Chromatogram showing c.273G>C, predicting p.Gln91His in OTX2 in a patient with bilateral anophthalmia. The mutation is not present in the mother or father and thus is de novo. Fig. S1B. Chromatogram showing c.1159C>T, predicting p.Arg387Cys in RARB in a patient with severe microphthalmia and coloboma. The mutation is not present in the mother or father and thus is de novo.
Fig. S1C. Chromatogram showing c.746C>A, predicting p.Ala249Glu in GDF6 in a patient with bilateral anophthalmia. The mutation is not present in the mother or father and thus is de novo.
Fig. S1D. Chromatogram showing c.128C>A, predicting p.Ala43Asn in ABCB6 in a patient with unilateral microphthalmia, cataract and persistent hyperplasia of the primary vitreous. The mutation was inherited from the unaffected mother.
Fig. S2. Chromatograms Showing Mutations and Sequence Variants of Unknown Significance in Developmental Eye Disorders.
Fig. S2A. Chromatogram showing c.128C>A, predicting p.Ala43Asn in ABCB6 in a patient with unilateral microphthalmia, cataract and persistent hyperplasia of the primary vitreous. The mutation was inherited from the unaffected mother. Fig. S2B. Chromatogram showing c.820C>T, predicting p.(Arg274Cys) in VSX2 in a patient with unilateral anterior segment dysgenesis. The mutation was inherited from the unaffected father. Fig. S2C. Chromatogram showing c.385G>A, predicting p.(Glu129Lys) in SIX6 in a patient with bilateral coloboma. The mutation was inherited from the unaffected father. Fig. S2D. Chromatogram showing c.1607C>T, predicting p.(Thr566Ile) in FBLN1 in a patient with bilateral anophthalmia. The mutation was homozygous and inherited from both parents.
Acknowledgments
We thank Jessie Cameron (Hospital for Sick Children, Toronto) for research sequencing of the AGK gene. This work was supported by grant R21EY022779-01 to Anne Slavotinek from the National Eye Institute, National Institutes of Health and by a Research Allocation Program grant, Clinical Application of Exome Sequencing to Developmental Eye Disorders, from the Academic Senate, University of California, San Francisco, to Anne Slavotinek. Ehsan Ullah was financially supported by the Higher Education Commission of Pakistan under the IRSIP Fellowship program. We are very grateful to all of the patients and their families.
Footnotes
Conflict of Interest
Some authors, as indicated, are employed by and receive a salary and/or are shareholders in Personalis, Inc. Personalis’ commercial services include an augmented clinical exome sequencing test: The ACE Clinical Exome™ Test.
References
- 1.Bardakjian T, Weiss A, Schneider AS. Anophthalmia/Microphthalmia Overview. In: Pagon RA, Bird TC, Dolan CR, Stephens K, editors. GeneReviews [Internet] Seattle (WA): University of Washington, Seattle; 1993–2004. [Google Scholar]
- 2.Slavotinek AM. Eye development genes and known syndromes. Mol Genet Metab. 2011;104:448–56. doi: 10.1016/j.ymgme.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balikova I, de Ravel T, Ayuso C, et al. High frequency of submicroscopic chromosomal deletions in patients with idiopathic congenital eye malformations. Am J Ophthalmol. 2011;151:1087–94. doi: 10.1016/j.ajo.2010.11.025. [DOI] [PubMed] [Google Scholar]
- 4.Raca G, Jackson CA, Kucinskas L, et al. Array comparative genomic hybridization analysis in patients with anophthalmia, microphthalmia, and coloboma. Genet Med. 2011;13:437–42. doi: 10.1097/GIM.0b013e318204cfd2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider A, Bardakjian T, Reis LM, Tyler RC, Semina EV. Novel SOX2 mutations and genotype phenotype correlation in anophthalmia and microphthalmia. Am J Med Genet. 2009;149A:2706–15. doi: 10.1002/ajmg.a.33098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fares-Taie L, Gerber S, Chassaing N, et al. ALDH1A3 mutations cause recessive anophthalmia and microphthalmia. Am J Hum Genet. 2013;92:265–70. doi: 10.1016/j.ajhg.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chassaing N, Causse A, Vigouroux A, et al. Molecular findings and clinical data in a cohort of 150 patients with anophthalmia/microphthalmia. Clin Genet. 2014;86:326–34. doi: 10.1111/cge.12275. [DOI] [PubMed] [Google Scholar]
- 8.Prokudin I, Simons C, Grigg JR, et al. Exome sequencing in developmental eye disease leads to identification of causal variants in GJA8, CRYGC, PAX6 and CYP1B1. Eur J Hum Genet. 2014;22:907–15. doi: 10.1038/ejhg.2013.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yahyavi M, Abouzeid H, Gawdat G, et al. ALDH1A3 loss of function causes bilateral anophthalmia/microphthalmia and hypoplasia of the optic nerve and optic chiasm. Hum Mol Genet. 2013;22:3250–8. doi: 10.1093/hmg/ddt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramu A, Noordam MJ, Schwartz RS, et al. DeNovoGear: de novo indel and point mutation discovery and phasing. Nat Methods. 2013;10:985–7. doi: 10.1038/nmeth.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srour M, Chitayat D, Caron V, et al. Recessive and dominant mutations in retinoic acid receptor beta in cases with microphthalmia and diaphragmatic hernia. Am J Hum Genet. 2013;93:765–72. doi: 10.1016/j.ajhg.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asai-Coakwell M, French CR, Ye M, et al. Incomplete penetrance and phenotypic variability characterize Gdf6-attributable oculo-skeletal phenotypes. Hum Mol Genet. 2009;18:1110–21. doi: 10.1093/hmg/ddp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aldahmesh MA, Khan AO, Mohamed JY, et al. Genomic analysis of pediatric cataract in Saudi Arabia reveals novel candidate disease genes. Genet Med. 2012;14:955–62. doi: 10.1038/gim.2012.86. [DOI] [PubMed] [Google Scholar]
- 15.Shah S, Ellard S, Kneen R, et al. Childhood presentation of COL4A1 mutations. Dev Med Child Neurol. 2012;54:569–74. doi: 10.1111/j.1469-8749.2011.04198.x. [DOI] [PubMed] [Google Scholar]
- 16.Colin E, Sentilhes L, Sarfati A, et al. Fetal intracerebral hemorrhage and cataract: think COL4A1. J Perinatol. 2014;34:75–7. doi: 10.1038/jp.2013.135. [DOI] [PubMed] [Google Scholar]
- 17.Deml B, Reis LM, Maheshwari M, et al. Whole exome analysis identifies dominant COL4A1 mutations in patients with complex ocular phenotypes involving microphthalmia. Clin Genet. 2014;86:475–81. doi: 10.1111/cge.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Ameln S, Wang G, Boulouiz R, et al. A mutation in PNPT1, encoding mitochondrial-RNA-import protein PNPase, causes hereditary hearing loss. Am J Hum Genet. 2012;91:919–27. doi: 10.1016/j.ajhg.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vedrenne V, Gowher A, De Lonlay P, et al. Mutation in PNPT1, which encodes a polyribonucleotide nucleotidyltransferase, impairs RNA import into mitochondria and causes respiratory-chain deficiency. Am J Hum Genet. 2012;91:912–8. doi: 10.1016/j.ajhg.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Debeer P, Schoenmakers EF, Twal WO, et al. The fibulin-1 gene (FBLN1) is disrupted in a t(12;22) associated with a complex type of synpolydactyly. J Med Genet. 2002;39:98–104. doi: 10.1136/jmg.39.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo DS, Labelle-Dumais C, Gould DB. COL4A1 and COL4A2 mutations and disease: insights into pathogenic mechanisms and potential therapeutic targets. Hum Mol Genet. 2012;21(R1):R97–110. doi: 10.1093/hmg/dds346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young TL, Guo XD, King RA, et al. Identification of genes expressed in a human scleral cDNA library. Mol Vis. 2003;9:508–14. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Chromatograms Showing Mutations in Pathogenic Genes in Developmental Eye Disorders.
Fig. S1A. Chromatogram showing c.273G>C, predicting p.Gln91His in OTX2 in a patient with bilateral anophthalmia. The mutation is not present in the mother or father and thus is de novo. Fig. S1B. Chromatogram showing c.1159C>T, predicting p.Arg387Cys in RARB in a patient with severe microphthalmia and coloboma. The mutation is not present in the mother or father and thus is de novo.
Fig. S1C. Chromatogram showing c.746C>A, predicting p.Ala249Glu in GDF6 in a patient with bilateral anophthalmia. The mutation is not present in the mother or father and thus is de novo.
Fig. S1D. Chromatogram showing c.128C>A, predicting p.Ala43Asn in ABCB6 in a patient with unilateral microphthalmia, cataract and persistent hyperplasia of the primary vitreous. The mutation was inherited from the unaffected mother.
Fig. S2. Chromatograms Showing Mutations and Sequence Variants of Unknown Significance in Developmental Eye Disorders.
Fig. S2A. Chromatogram showing c.128C>A, predicting p.Ala43Asn in ABCB6 in a patient with unilateral microphthalmia, cataract and persistent hyperplasia of the primary vitreous. The mutation was inherited from the unaffected mother. Fig. S2B. Chromatogram showing c.820C>T, predicting p.(Arg274Cys) in VSX2 in a patient with unilateral anterior segment dysgenesis. The mutation was inherited from the unaffected father. Fig. S2C. Chromatogram showing c.385G>A, predicting p.(Glu129Lys) in SIX6 in a patient with bilateral coloboma. The mutation was inherited from the unaffected father. Fig. S2D. Chromatogram showing c.1607C>T, predicting p.(Thr566Ile) in FBLN1 in a patient with bilateral anophthalmia. The mutation was homozygous and inherited from both parents.