Abstract
The hemolytic uremic syndrome (HUS) whose main causative agent is enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a disease that mainly affects children under 5 years of age. Argentina is the country with the highest incidence of HUS in the world. Cattle are a major reservoir and source of infection with E. coli O157:H7. To date, the epidemiological factors that contribute to its prevalence are poorly understood. Single nucleotide polymorphism (SNP) typing has helped to define nine E. coli O157:H7 clades and the clade 8 strains were associated with most of the cases of severe disease. In this study, eight randomly selected isolates of EHEC O157:H7 from cattle in Argentina were studied as well as two human isolates. Four of them were classified as clade 8 through the screening for 23 SNPs; the two human isolates grouped in this clade as well, while two strains were closely related to strains representing clade 6. To assess the pathogenicity of these strains, we assayed correlates of virulence. Shiga toxin production was determined by an ELISA kit. Four strains were high producers and one of these strains that belonged to a novel genotype showed high verocytotoxic activity in cultured cells. Also, these clade 8 and 6 strains showed high RBC lysis and adherence to epithelial cells. One of the clade 6 strains showed stronger inhibition of normal water absorption than E. coli O157:H7 EDL933 in human colonic explants. In addition, two of the strains showing high levels of Stx2 production and RBC lysis activity were associated with lethality and uremia in a mouse model. Consequently, circulation of such strains in cattle may partially contribute to the high incidence of HUS in Argentina.
Introduction
Escherichia coli O157:H7 is a globally important zoonotic pathogen capable of causing hemorrhagic colitis and hemolytic uremic syndrome (HUS) in humans. HUS is widely distributed in the world and is described as an epidemy of low incidence rate in industrialized countries (1 to 3 cases per 100,000 children aged under 5 years) [1,2]. However, in Argentina, a country with the highest HUS incidence in the world [3,4], 12 to 14 cases were reported per 100,000 children aged under 5 years of age with about 400 new cases reported annually in the last decade. In Argentina, HUS is the leading cause of acute kidney failure in children and the second cause of chronic renal failure, and is also responsible for 20% of kidney transplants in children and adolescents [5].
A high percentage of domestic cattle in many countries are colonized by Shiga toxin-producing E. coli (STEC), which do not cause illness in cattle. Enterohemorrhagic E. coli (EHEC) [6,7,8,9] are STEC and can be differentiated by the presence of the 35.6 kb pathogenicity island called the locus of enterocyte effacement (LEE). This pathogenicity island encodes for a Type Three Secretion System (T3SS) [10,11] and gives strains the ability to produce a histopathological lesion in the intestinal epithelium known as an attaching and effacing (A/E) lesion. A/E lesions are characterized by the close adherence to enterocytes and depletion of intestinal microvilli.
The production of Shiga toxin (Stx), also known as verocytotoxin (VT), represents the most important virulence attribute in EHEC. After colonization, Stx is produced and released into the intestine. Most strains produce at least one Stx variant, which may be of type 1 (Stx1) and/or type 2 (Stx2) [12].
Isolates of EHEC O157:H7 have been shown to be genotypically diverse by different methods, including PFGE [13], octomer-based genome scanning [14], and multilocus variable number of tandem repeats analysis [13]. Variations in disease severity between outbreaks caused by E. coli O157:H7 are evident and depend on specific genotypes [15,16,17]. For example, the 1993 multistate outbreak in North America [18] and the 1996 outbreak in Sakai, Japan [19] had low rates of hospitalization and HUS [20]. In comparison, two outbreaks in the United States caused by contaminated lettuce and spinach had much higher frequencies of both hospitalization (mean, 63%) and HUS (mean, 13%) [21]. These outbreak strains along with more than 500 additional strains have been previously characterized by the analysis of 96 SNPs. By phylogenetic analyses, Manning et al [15] identified 39 SNP genotypes in this broad collection of E. coli O157 isolates, which differed at 20% of the SNP loci and were separated into nine different clades. The stx profiles varied among strains belonging to different clades and certain clades were associated with clinical symptoms. HUS patients, for example, were significantly more likely to be infected with strains of clade 8, which had increased in frequency over a 5-year period in Michigan [15]. Accordingly, it has been suggested that a subpopulation or more virulent lineage has emerged. These clade 8 strains have been identified in various clinical cases on multiple continents and countries, including Argentina, since 1984 [15]. To date, those factors that are important for enhancing the virulence of this subset of strains are not completely understood. The clade 8 strains have higher Stx2 expression than other clades [22] and have unique genetic features that may be important for the disease [23].
We and other groups have previously identified strains belonging to clade 8 in cattle from different provinces of Argentina [24,25]. For this reason, we sought to more closely examine the genetic relationships between strains by screening for previously described SNPs [15] and assessing their pathogenic potential following exposure to epithelial cells and infection in mice.
Materials and Methods
Bacterial isolates
Eight E. coli O157:H7 isolates recovered from cattle in the central humid Pampa region of Argentina from 2002 to 2011 (Fig 1 and Table 1) were examined along with 2 human local isolates from HUS cases. The cattle isolates were collected by rectal swab and obtained from private or INTA farms with permission from the farmer or manager. The standard E. coli O157:H7 EDL933 strain recovered from a patient in USA and the nonpathogenic E. coli DH5α were included in the study as positive and negative control, respectively, in virulence assays. Bacteria were grown aerobically on Luria-Bertani (LB, Difco Laboratories, USA) agar plates or in LB broth at 37°C. For functional studies, bacterial strains were grown in LB broth overnight at 150 rpm and then diluted 1/50 in Dulbecco’s modified Eagle’s medium/(DMEM)-F12 medium and grown to exponential phase (optical density (OD) at 600 nm of 0.3–0.4) at 37°C at 50 rpm.
Fig 1. Map showing the geographical location of cattle where Escherichia coli O157:H7 isolates were taken.
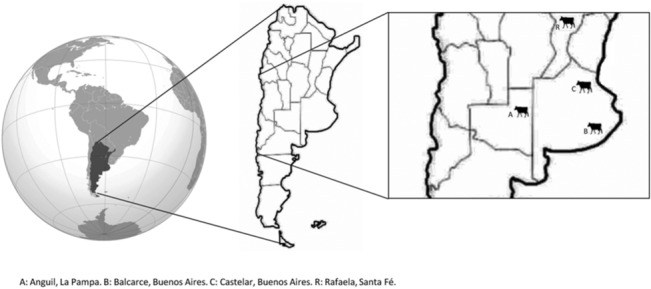
A: Anguil, La Pampa. B: Balcarce, Buenos Aires. C: Castelar, Buenos Aires. R: Rafaela, Santa Fe.
Table 1. Epidemiological, and genotypic characteristic of E. coli O157:H7 strains.
| Strain | Host | Location | Origin | Isolation year | Stx1 | Stx2 | Stx2 subtype | Clade as in [28] (nts SNP 539, 1060, 438, 630) 2 | Clade x 32 3 | Stx2 integration loci |
|---|---|---|---|---|---|---|---|---|---|---|
| EDL933 | Human, | patient | USA | + | + | stx2a | 3 (C-G-C-T) | clade 3 | wrbA | |
| 146N | Bovine | abattoir | ARG | + | + | stx2c | 3(C-G-C-T) | ND | yehV | |
| 438/99 | Bovine | abattoir | ARG | 1999 | - | + | stx2c | 3(C-G-C-T) | ND | ND* |
| 7.1 Anguil | Bovine | farm 1 | ARG | 2009 | - | + | stx2a-stx2c | none (C-T-C-T) | clade 6 | |
| 9.1 Anguil | Bovine | farm 1 | ARG | 2009 | - | + | stx2a-stx2c | none(A-T-C-T) | clade 8 | yehV |
| Vac 07–1 | Bovine | farm 1 | ARG | 2007 | - | + | stx2a-stx2c | none-(A-G-C-T) | clade 8 | |
| 125/99 | Human | patient | ARG | - | + | stx2a | none- (A-G-C-T) | clade 8 | yehV | |
| Rafaela II- 827 | Bovine | farm 1 | ARG | 2009 | - | + | stx2a-stx2c | none-(A-G-C-T) | clade 8 | ND* |
| Balcarce 14.2 | Bovine | farm 1 | ARG | 2009 | - | + | stx2a-stx2c | none- (A-G-C-T) | clade 6 | yehV |
| Balcarce 24.2 | Bovine | farm 1 | ARG | 2009 | - | + | stx2a-stx2c | none- (A-G-C-T) | clade 8 | |
| Neuquén 7562 | Human | patient | ARG | 2003 | - | + | stx2c | none-(A-G-C-T) | clade 8 | yehV |
Genotyping
Stx1 and Stx2 were detected using primers described by Olsvik and Strockbine [26] (Table 2). Subtyping to discriminate Stx2 variants was performed by PCR by using primers described by Scheutz et al [27] in Table 2.
Table 2. Primers used in this study.
| Primer | Sequence | Purpose | Tm (C°) | amplicon size (bp) | References |
|---|---|---|---|---|---|
| Vt1-a | CAGTTAATGTGGTGGCGAAG | Stx1 detection | 55 | 894 | [26] |
| Vt1-b | CTGCTAATAGTTCTGCGCATC | Stx1 detection | 55 | 894 | [26] |
| Vt2-a | CTTCGGTATCCTATTCCCGG | Stx2 detection | 45 | 478 | [26] |
| Vt2-b | GGATGCATCTCTGGTCATTG | Stx2 detection | 45 | 478 | [26] |
| stx2a-F2 | GCGATACTGRGBACTGTGGCC | Stx2 variant confirmedstx2a | 66 | 349 347 |
[27] |
| stx2a-R3 | CCGKCAACCTTCACTGTAAATGTG | Stx2 variant confirmedstx2a | 66 | 349 347 |
[27] |
| stx2a-R2 | GCCACCTTCACTGTGAATGTG | Stx2 variant confirmedstx2a | 66 | 349 347 |
[27] |
| stx2b-F1 | AAATATGAAGAAGATATTTGTAGCGGC | Stx2 variant confirmedstx2b | 65 | 251 | [27] |
| stx2b-R1 | CAGCAAATCCTGAACCTGACG | Stx2 variant confirmedstx2b | 65 | 251 | [27] |
| stx2c-F1 | GAAAGTCACAGTTTTTATATACAACGGGTA | Stx2 variant confirmed stx2c | 65 | 177 | [27] |
| stx2c-R2 | CCGGCCACYTTTACTGTGAATGTA | Stx2 variant confirmed stx2c | 65 | 177 | [27] |
| stx2d-F1 | AAARTCACAGTCTTTATATACAACGGGTG | Stx2 variant confirmedstx2d | 65 | 179 235 280 |
[27] |
| stx2d-R1 | TTYCCGGCCACTTTTACTGTG | Stx2 variant confirmedstx2d | 65 | 179 235 280 |
[27] |
| stx2d-O55-R | TCAACCGAGCACTTTGCAGTAG | Stx2 variant confirmedstx2d | 65 | 179 235 280 |
[27] |
| stx2d-R2 | GCCTGATGCACAGGTACTGGAC | Stx2 variant confirmedstx2d | 65 | 179 235 280 |
[27] |
| stx2e-F1 | CGGAGTATCGGGGAGAGGC | Stx2 variant confirmedstx2e | 66 | 411 | [27] |
| stx2e-R2 | CTTCCTGACACCTTCACAGTAAAGGT | Stx2 variant confirmedstx2e | 66 | 411 | [27] |
| stx2f-F1 | TGGGCGTCATTCACTGGTTG | Stx2 variant confirmedstx2e | 66 | 411 | [27] |
| stx2f-R1 | TAATGGCCGCCCTGTCTCC | Stx2 variant confirmedstx2e | 66 | 411 | [27] |
| stx2g-F1 | CACCGGGTAGTTATATTTCTGTGGATATC | Stx2 variant detectionstx2g | 62 | 573 | [27] |
| stx2g-R1 | GATGGCAATTCAGAATAACCGCT | Stx2 variant detectionstx2g | 62 | 573 | [27] |
| ECs2357f | TTAACCTTTTGTGGCGAACC | SNP ECs2357 detection | 58 | 253 | This study |
| ECs2357r | TACGGTTTGCCGCAGTTATT | SNP ECs2357 detection | 58 | 253 | This study |
| ECs2521f | CCGTAGCAGGTTCGGTAAAA | SNP ECs2521 detection | 60 | 200 | This study |
| ECs2521r | CGGTTCCAGTTCGTCGATAA | SNP ECs2521 detection | 60 | 200 | This study |
| ECs3881f | GAGAACGGCTACGCGTACAT | SNP ECs2521 detection | 60 | 200 | This study |
| ECs3881r | CGTTCCACACCTTTCTGGTT | SNP ECs2521 detection | 60 | 200 | This study |
| ECs4130f | GGGCTGCTGATTTTTGGTAT | SNP ECs4130 detection | 58 | 229 | This study |
| ECs4130r | CAGGCGACAGAATATCGTCA | SNP ECs4130 detection | 58 | 229 | This study |
| sbcB-F | GACAGCAGAAACAACGGATTTAAC | sbcB insertion site | 59 | 406 | [33] |
| sbcB-R2 | TCCAGGCGTAAGGATCGTAG | sbcB insertion site | 59 | 406 | [33] |
| A | AAGTGGCGTTGCTTTGTGAT | yehV insertion site | 60 | 340 | [55] |
| B | AACAGATGTGTGGTGAGTGTCTG | sbcB insertion site | 59 | 406 | [33] |
| C | AGGAAGGTACGCATTTGACC | wrbA insertion site | 60 | 314 | [33] |
| D | CGAATCGCTACGGAATAGAGA | wrbA insertion site | 60 | 314 | [33] |
| argW-A | CCGTAACGACATGAGCAACAAG | argW insertion site | 55 | 216 | [33] |
| argW-D | AATTAGCCCTTAGGAGGGGC | wrbA insertion site | 60 | 314 | [33] |
SNP typing for clade determination
The primers in Table 2 were initially used to generate amplicons to identify SNPs and classify strains into clades according to the algorithm developed by Riordan et al [28]. In addition, an expanded set of 23 of the 32 previously identified SNPs capable of differentiating the original SNP genotypes [15] were evaluated using the GoldenGate genotyping assay (Illumina; San Diego, CA) [29]. SNP loci evaluated in this assay are highlighted in SNP_info (S1 Table). Sequences specific for a set of 30 strains were included as controls to represent the original SNP genotypes identified by Manning et al. [15]; clades were defined using these previously defined control strains. All 23 SNPs were concatenated in MEGA6 [30] and the evolutionary history was inferred using the Neighbor-Joining method [31]. The bootstrap test was used with 1,000 replicates to identify clusters, or clades [32] and evolutionary distances were presented as the number of SNP differences per site. Sequences specific for a set of nine strains were included as controls to represent the original SNP genotypes identified previously [15]. The insertion sites of stx2 phages were determined according to Shringi et al [33] (Table 2).
Shiga-Toxin detection Kit
The expression of Stx was determined in filtered culture supernatants, with the addition or absence of mitomycin C, by using RIDASCREEN Kit Verotoxin enzyme immunoassay (R-Biopharm Latin America). Its expression was semi-quantified according to Beutin et. al [34].
Stx toxicity in Vero cells
Strains were incubated overnight at 37°C and 200 rpm in 5 ml of LB broth and subsequently the culture supernatant was obtained by centrifugation and filtration (0.22 μm filters). Filtered culture supernatants were assayed for cytotoxicity on Vero cells as previously described [35]. Briefly, Vero cell monolayers grown in 96-well plates were treated for 72 h under growth-arrested conditions (serum-free medium) with filtered culture supernatant from different strains. At the end of the incubation, plates were washed twice with PBS (145 mM NaCl, 10 mM NaH2PO4, pH 7.2) and incubated for 2 h with freshly diluted neutral red in PBS to a final concentration of 50 μg/ml. Cells were then washed with 1% CaCl2 and 4% formaldehyde twice and then were solubilized in 1% acetic acid and 50% ethanol. Absorbance at 546 nm was read in an automated plate spectrophotometer. Results were expressed as percent viability, with 100% represented by cells incubated under identical conditions but without treatment. The 50% cytotoxic dose (CD50) corresponded to the dilution required to kill 50% of Vero cells.
The results are the means of three experiments. The statistical difference was expressed as the P value determined by a two-way ANOVA and a Bonferroni post-test.
Adherence to Caco-2, HeLa and HEp-2 cells
The ability of E. coli strains to adhere to HeLa (ATCC-CCL2), Caco-2 (ATCC-HTB-37) and HEp-2 (ATCC-CCL-23) cell monolayers was assessed. Sterile glass coverslips (12 mm) were inoculated with 105 cells per well. The cells were grown to 90% confluence at 37°C in 5% CO2 in 24-well plates (Corning) in DMEM with 10% (vol/vol) heat-inactivated fetal bovine serum, 2 mM L-glutamine, penicillin (100,000 IU/liter), and streptomycin (100 mg/liter) [36]. Before use, the cells were washed with sterile phosphate-buffered saline (PBS; pH 7.4) and replenished with DMEM. The bacterial strains were grown in LB broth overnight at 37°C. For qualitative and quantitative assays, tissue culture cells were incubated with 107 bacteria per well (MOI 100) for 5 h at 37°C. The monolayers were washed, fixed, and stained with Giemsa solution for microscopic evaluation. For E. coli adherence quantification, the infected monolayers were washed three times with PBS and the adherent bacteria were recovered with 200 ul of 0.1% Triton X 100 in PBS buffer and plated on LB agar plates. Data are expressed as the CFU from adhered bacteria from triplicate wells and are the mean of at least three separate experiments. The statistical difference was expressed as the P value determined by a Tukey's Multiple Comparison Test analysis.
Red blood cell (RBC) lysis assay
The hemolytic activity exhibited by T3SS-encoding E. coli strains [37] was evaluated. Overnight grown cultures (1:100 dilution of EHEC O157:H7) were incubated with 5% suspension of red blood cells (RBC) in DMEM without phenol red for 4 h at 37°C under a 5% CO2 atmosphere as described by Larzábal, et al [38]. Briefly, the suspension was removed from the plates and centrifuged at 12,000g for 1 min. The supernatants were monitored for the presence of released hemoglobin by measuring OD543. The statistical difference was expressed as the P value determined by a Tukey's Multiple Comparison Test analysis. In order to eliminate any possible action of other virulence factors of E. coli strains on the RBCs lysis, such as secretory enterohemolysin (Ehly), or Shiga toxins (Stx), the bacteria were incubated in DMEM for 5 h to allow the secretion of these factors. The culture supernatant was filtered (0.22 micron filter) to have secretory virulence factors free of bacteria. The supernatant was incubated with a 5% suspension of RBCs in PBS at pH 7.4.
Functional test using human intestinal tissue
Colon fragments used in this study were obtained from surgeries performed on three adult cancer patients (informed consent was obtained) at the “Servicio de Cirugía Gastroenterológica, Hospital Churruca-Visca”, Buenos Aires, Argentina. The Ethics Committee of the Universidad de Buenos Aires approved the use of human tissues for research purposes. The colonic mucosa was obtained and mounted as a diaphragm on a modified Ussing chamber as previously described [39]. Transepithelial net water flux (Jw) and electrical measurements were used as functional test. Jw was recorded automatically across an Ussing chamber connected to a special electro-optical device [40]. The sensitivity of this instrument is approximately 50 nl. The spontaneous potential difference (PD) was simultaneously recorded in the other chamber across the micro-reference electrodes (Harvard Apparatus Inc, USA) placed adjacent to the epithelium under open-circuit conditions. The short circuit current (Isc) was measured with an automatic voltage clamp system that kept the PD at 0 mV. The transepithelial resistance (Rt) was calculated from the Isc and open-circuit PD values, according to Ohm’s law. The parameters were stabilized and then 200 μl (approximately 108 CFU) of bacterial culture of EDL933 or 7.1 Anguil were added to the mucosal side of each colon tissue (time 0). Then, both Jw and Isc were simultaneously recorded for 1 h. Because of tissue variability, data are analyzed as ∆Jw where ∆Jw = Jw (at a given time)—Jw (at time 0) and ∆Isc, where ∆Isc = Isc (at a given time)—Isc (at time 0). Each assay was carried out three times with colon fragments obtained from different patients. The results are reported as mean ± 1 standard error. Statistical significance between two mean values obtained for two experimental conditions were calculated using the "t" Student test. For statistical analysis of curves, ANOVA was used to evaluate changes in Jw of each of the strains according to the time.
Pathogenicity in murine model
BALB/c mice with normal intestinal microbiota of 21 days and an average weight of 10 g were orally inoculated with 100 μl PBS containing 109 bacteria [[41]. Five or ten mice were used per strain for this assay. The animals were housed in filtered air (HEPA filters) individual cages, with free access to food and sterile water. Survival and weight were recorded daily and morbidity was analyzed in duplicate by quantifying serum urea (g/l) on day three post-inoculation. Mice inoculated with PBS or E. coli DH5α were included as negative controls. Histological alterations in cecum and colon were analyzed by optical microscopy (magnification 400x). Statistical analyses were performed on uremia values by one-way ANOVA with the Tukey test for multiple comparisons. Lethality was analyzed using the Mantel-Cox test. The tests were conducted with the approval of Comite Institucional para el cuidado y uso de animales de experimentación (Institutional Committee for care and handling of experimental animals, CICUAE) of the Center for Veterinary and Agronomical Research (CICVyA) of Instituto Nacional de Tecnología Agropecuaria (INTA), Argentina (http://inta.gob.ar/documentos/cicuae-comite-institucional-para-el-cuidado-y-uso-de-animales-de-experimentacion). All animals were euthanized by CO2 inhalation before the experimental endpoint. We established the human endpoint when mice met one of the following signs: Loss of > 20% of the initial weight, lethargy, bristling coat, hemorrhagic diarrhea, and cry or howl when touched. Sixty-seven percent (n = 10) of animals were euthanized due to weight loss, 20% (n = 3) due to lethargy and 13% (n = 2) due to lethargy and weight loss
Results
SNP typing
SNP typing was used to genotype the strains with two methods. One proposed by Riordan et al [28] using 4 SNPs and another based on 23 SNPs that were shown to differentiate between the nine previously described clades [15]. With the method by Riordan et al. (28), six bovine isolates (7.1 Anguil, 9.1 Anguil, Vac 07–1, Rafaela II- 827, Balcarce 14.2, Balcarce 24.2) did not fit into any of the known clades, whereas two bovine strains (146N, 438/99) were classified as clade 3 (Table 1). The two local human strains were also unclassifiable using this method, which may be due to the low number of SNPs evaluated. In contrast to these results, with the method based on 23 SNPs, four of the six bovine isolates and both human isolates were classified as clade 8 in the Neighbor-Joining phylogeny (Fig 2). Specifically, one bovine and one human strain each clustered with control strains representing SNP genotypes (SG) 30 and 31 of clade 8. By contrast, the two bovine strains, 7.1 Anguil and Balcarce 14.2, had SNP profiles that clustered together with control strains belonging to clade 6 (Fig 2). In consequence, the 23 SNP based method allows a stronger assignment of clades as it uses more loci.
Fig 2. Neighbor-joining phylogeny of bovine-derived E. coli O157:H7 strains from Argentina and a subset of O157:H7 control strains representing single nucleotide polymorphism (SNP) genotypes (SGs) from eight of the nine previously defined clades [15].
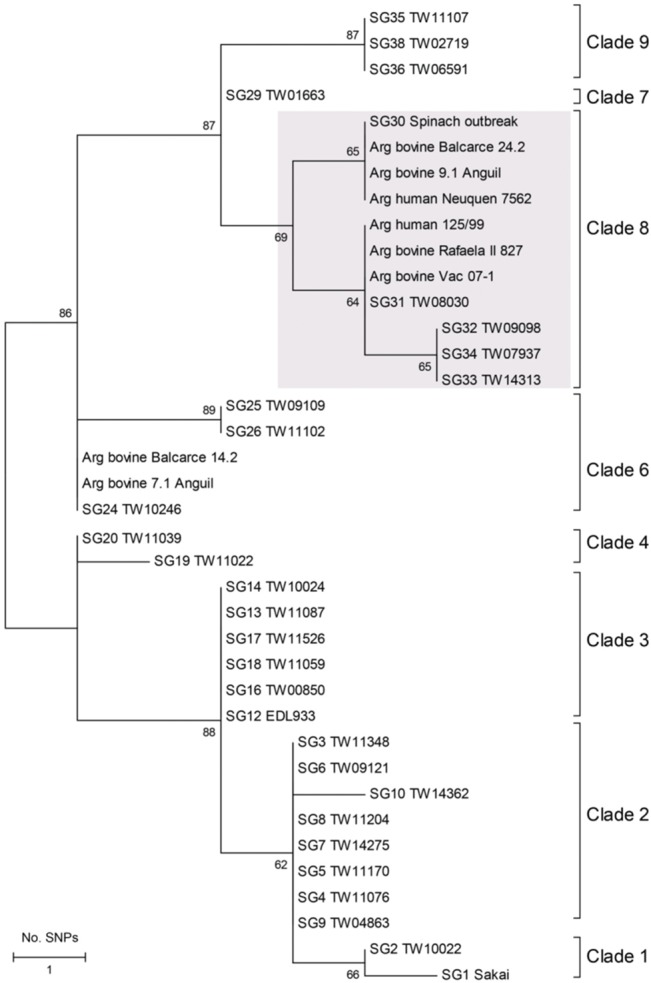
The phylogeny was constructed using 23 SNP loci. The numbers at the nodes represent the bootstrap support following 1,000 replications, while strains belonging to clade 8 are indicated by the gray box.
Stx typing
The strains were characterized by the stx type and variants of the stx2. All the cattle strains harbor stx2 but not stx1 and six out of eight strains had two copies of stx2 composed of the stx 2a and stx 2c variants.
Concerning the occupancy of classical insertion sites of Stx2 phages E. coli O157:H7 146N, 125/99, 9.1 Anguil, Balcarce 14.2 and Neuquén 7562 had these insertions in yehV. For the other strains, however, the insertion site was not identified, though wrbA, argW or sbcB remained intact (Table 1 ND*: not determined). Importantly, this method does not detect an insertion of the entire stx phage, but it detects any insertion with a phage that may or may not contain a stx gene.
Stx2 activity
As a correlate of virulence, we measured the Shiga toxin activity in bacterial culture supernatants in bovine-derived clade 8 isolates and in the two clade 6 strains (7.1 Anguil and Balcarce 14.2). Stx2 activity was measured in Vero cells (Fig 3). The CD50 was from 2.34 x105 (strain 7.1 Anguil, bovine, clade 6) to 1.33 x102 (strain 9.1 Anguil). The strain 7.1 Anguil presented a significantly higher level of activity (p<0.05) compared to all other strains. The standard EDL933 strain produced a CD50 of 6.46 x104.
Fig 3. Cytotoxic activity (dilutions 10−1 to 10−6) of culture supernatant isolates of E. coli O157: H7 in Vero cells (A) and CD50 table (B).
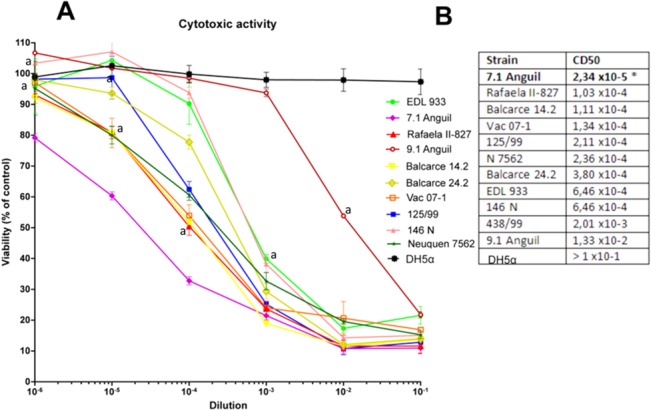
A:-induced cytotoxicity was observed with the analyzed supernatants at different dilutions. The results are expressed as percentage of detected neutral red (± 1 SEM); a 100% represents cells incubated without supernatant (control viability). DH5α strain represents a control devoided of Stx activity. Each assay was performed in triplicate, whereas each viability control was assessed by sextuplicate. a letters denoted statistically significant differences (p< 0.01) between the cytotoxic activity of E. coli O157:H7 7.1 Anguil and the rest of the strains in given dilution point. Only the closest point is marked as the other strains with lower activity has as expected also significantly lower activity B: CD50. The results are expressed as the required dose value of culture supernatant to present a Vero cell cytotoxicity of 50%. 100% represents cells incubated without supernatant (control viability). * = P <0.05.
We also evaluated, semiquantitatively, the toxin released in the culture medium by ELISA [34]. Strain Vac 07–1 (bovine, clade 8) produced around double the amount of Stx proteins compared to the other strains, When mitomycin C was added the induction effect was high for Rafaela II-827 Vac 07–1, 7.1 Anguil, Balcarce 24.2 (approximately ten-fold higher) and notably higher for Balcarce 14.2 (24 times). (S1 Fig). It is important to note the cytotoxic activity in Vero cells (Fig 3) was performed with bacteria not treated with mitomycin.
In general there was agreement between the levels of verocytotoxic activity and the Stx production. A clear relationship between stx2 copy number, and verocytotoxic activity in Vero cells or Stx production detected by ELISA could not be established. However. both assays showed that the top ranked isolates belongs to clade 8 or 6.
Adherence to epithelial cell lines
Adherence to intestinal epithelial cells is a crucial step in EHEC infection. Three epithelial cell lines were selected for the adherence studies: HeLa (human cervix adenocarcinoma), HEp-2 (human larynx carcinoma) and CaCo-2 (human colorectal adenocarcinoma). HeLa cells promoted the highest adherence, with Balcarce 24.2 (bovine, clade 8) and Balcarce 14.2 (bovine, clade 6) showing the highest scores (p<0.05) followed by EDL933 (Balcarce 24.2 vs EDL933, p < 0.05). The strain Balcarce 14.2 was the most adherent to CaCo-2 cells (p<0.05), followed by Balcarce 24.2.and EDL933. The strain 7.1 Anguil, 9.1 Anguil and Rafaela II-827 were the less adherent strains (in decreasing order) with CaCo2 cells. HEp-2 was the cell line with the lowest adherence to bacteria (Fig 4). A qualitative analysis based on micrographs showed (S2 Fig) that the adherence was localized (LA) or localized-like type (LAL) [42].
Fig 4. Adhesion of E. coli O157:H7 isolates to epithelial cells Caco-2, MOI:100; HeLa, MOI:100 and Hep-2 cells, MOI:200.
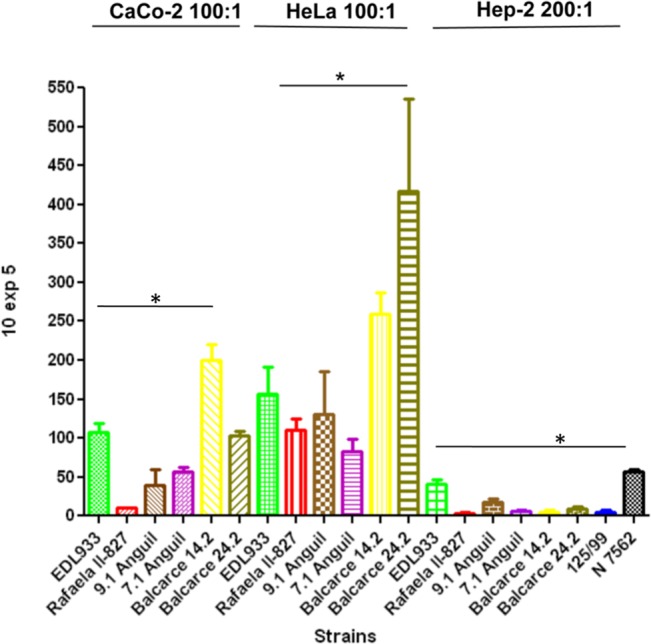
Each assay was performed in triplicate. * = p <0.05.
T3SS hemolytic activity
The RBC hemolysis caused by the T3SS is a correlate of T3SS physiological activity. For this reason, we performed hemolysis analyses. The cattle strain 7.1 Anguil was the most hemolytic and it was standardized to 100% hemolysis in ovine RBCs (Fig 5). Five of the isolates, 7.1 Anguil (bovine, clade 6), Rafaela II-827 (bovine, clade 8) and 146N, N7562 (human, clade 8) and 125/99 (human, clade 8), showed a high hemolytic activity (p <0.05) with values 80% higher than the other strains (Fig 5). EDL933 had a low activity, whereas the nonpathogenic DH5α strain that lacks a T3SS had no hemolytic activity. Supernatant of the bacterial isolates incubated in DMEM was then incubated with RBCs to verify if secreted hemolytic factors were present. Results were negative.
Fig 5. Hemolytic activity provoked by T3SS in of E. coli O157:H7 isolates from cattle from Argentina.
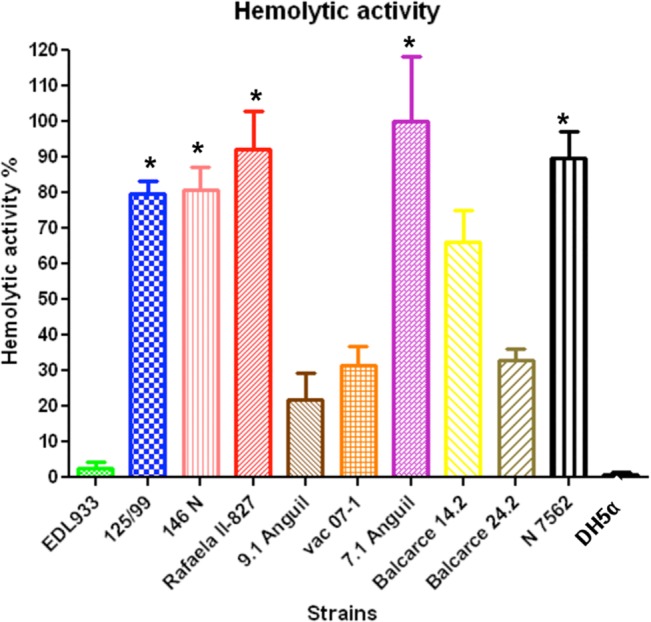
Sheep erythrocyte hemolysis was observed with a significant (* = P <0.05) lysis induced by strains 7.1 Anguil, Rafaela II-827, Neuquen 7562, 146N and 125/99 in relation to the other tested strains. The results are shown as the relative percentage of hemolysis (± 1 SEM) in relation to the strain with the highest values (7.1 Anguil). DH5α represents hemolysis caused by a non-pathogenic strain. Each assay was performed in triplicate.
Functionality evaluation of 7.1 Anguil strain in the human colon ex vivo
To observe the diarrheagenic potential of 7.1 Anguil (cattle, clade 6) with the highest Stx2 activity, we designed a functional test that allows the evaluation of 7.1 Anguil regarding to its capacity to modify fluid absorption across human colonic mucosa in vitro. This strain was compared to the standard strain EDL933. This experimental approach, previously employed to characterize the production of Stx2 in E. coli O157:H7 strain 125/99 [43] allows the simultaneous recording of the net water movement, electrical potential difference and short-circuit current across the intestinal barrier.
Absorptive Jw (0.24 ± 0.04 μl/min.cm2, n = 6) was observed in the human colonic mucosa placed between two identical Ringer solutions (2 ml each) in the Ussing chamber before the addition of bacteria. The electrical parameters tested simultaneously with water fluxes showed a PD of 1.6 ± 0.4 mV, Isc of 29.1 ± 4.2 μA/cm2 and Rt of 52.5 ± 11.9 Ω.cm2. Then, matched colonic mucosal obtained from the same patient were incubated with 200 μl (approximately 108 CFU) of bacterial culture of the 7.1 Anguil or EDL933 strains. A significant inhibition of Jw was observed with both strains, although the level of inhibition was higher with 7.1 Anguil (Fig 6, p < 0.05, n = 3). Isc simultaneously measured in colonic mucosa incubated with 7.1 Anguil or EDL933 remained unchanged for at least for 60 min (∆Isc (μAmp/cm2): 5.1 ± 8.0 vs 1.0 ± 3.0, respectively, NS, n = 3). This functional test indicates the high capacity of 7.1 Anguil to cause inhibition of the normal water absorption in human intestine ex vivo.
Fig 6. Effect of E. coli EDL933 and E. coli 7.1 Anguil strains on water absorption (Jw).
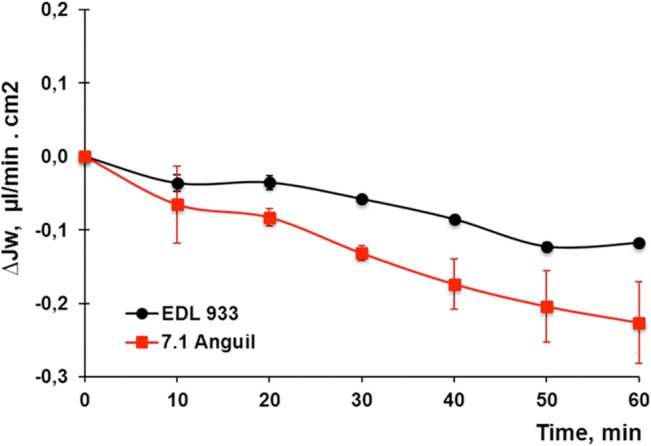
Fragments of human colon were incubated for 60 minutes. The results are expressed as mean ± SEM (n = 3 for each experimental situation).
Pathogenicity in a murine model
A mouse model of EHEC infection that reproduces some of the clinical observations of HUS (death, weight loss, and renal failure) was used [44] to evaluate pathogenicty of the EHEC strains. In the first test (five BALB/c mice per group), animals were inoculated with all strains studied here: bovine clade 8 isolates (Rafaela II- 827 and 9.1 Anguil), clade 6 bovine isolates (7.1 Anguil and Balcarce 14.2) and the standard EDL933 strain. As controls, one group was inoculated with E. coli DH5α and the other with PBS. Animals inoculated with 7.1 Anguil suffered an elevated uremia (1.1 g/l) compared to to EDL933 (0.7), DH5α (0.6 g/l), Rafaela II-827 (0.6 g/l), 9.1 Anguil (0.5 g/l), Balcarce 14.2 (0.5 g/l), and the negative control (PBS, 0.5 g/l) (p values <0.05). The strain 7.1 Anguil showed a lethality rate of 60%, whereas Rafaela II-827 showed a rate of 20% (S3 Fig). No lethality was observed with all of the other strains or with PBS. Animals inoculated with E. coli O157:H7 7.1 Anguil, Rafaela II-827 and 9.1 Anguil suffered weight loss in descendent order (S3 Fig).
In a second test, Rafaela II-827 (clade 8, bovine), 7.1 Anguil (clade 6, bovine) and EDL933 were compared using larger groups of mice (10). A lethality rate of 90% was observed in mice inoculated with Rafaela II-827 (p<0.01), whereas a 20% rate was observed for 7.1 Anguil (Fig 7). A significant weight loss (p<0.05) was also observed in mice inoculated with Rafaela II-827 on day 4 post inoculation (data not shown). Importantly, the histological examination of mice infected with both Rafaela II-827 and 7.1 Anguil strains demonstrated renal necrosis at both glomerular and tubular level and microhemorrhagic foci. In the cecum, a marked mucosal destruction with loss of surface epithelium was also observed (S4 Fig).
Fig 7. Mice infection with E. coli O157:H7 isolates.
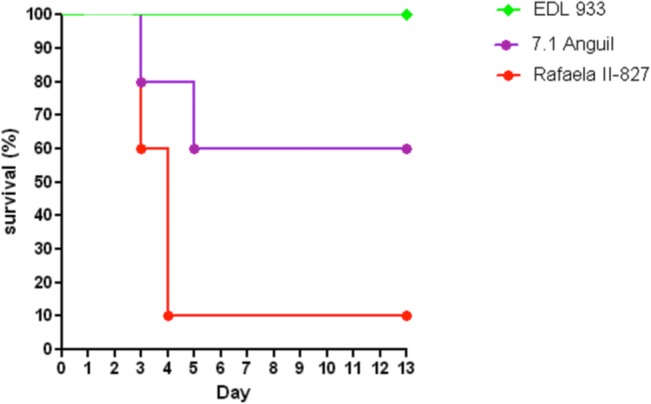
Animals were intragastrically infected with E. coli O157:H7 at inocula of 109 CFU in 200ul of PBS. Survival after intragastric inoculation of BALBc mice with EDL933, 7.1 Anguil, 9.1 Anguil, Rafaela II-827, Balcarce 14.2, Balcarce 24.2, DH5α or PBS.
Discussion
Argentina has the highest incidence of HUS in the world [3,4,45,46]; however, the factors that contribute to increased incidence in this region have not been clearly identified. It should be noted that cattle are the main reservoir of EHEC and that Argentina has more cattle than people (over 55 million cattle and 42 million inhabitants). In a prevalence study, Mellor et al [25] identified 72% of clade 8 E. coli O157:H7 isolates in Argentina and only 2% in Australia in a sample from 120 animals. Fifty percent of isolates from Argentinean cattle were clade 8. The isolates examined in the current study came from Pampa of Argentine, where the majority of cattle are concentrated [47], and originated from four different geographical locations.
Cattle does not suffer from the disease caused by E. coli O157:H7 and for this reason no clinical records are available to associate with different genotypes of EHEC. In addition, no bona-fide animal model is known to reproduce the infection pattern of EHEC and the clinical signs of HUS in humans, which makes the comparison difficult. We therefore sought to study the potential virulence of clade 8 isolates from Argentinean cattle through the use of biochemical in vitro correlates of virulence and a mouse infection model.
Although the relative virulence of the E. coli O157:H7 isolates, both from humans and cattle, has been previously analyzed, few studies have examined whether phylogenetic lineage, or clade assignment impacts virulence in vivo. Baker et al [48], for instance, compared the virulence of strains isolated from cattle and humans in gnotobiotic piglets and observed that those from humans were more virulent to mice. Eaton et al [49] studied the kidney pathogenesis of different human EHEC strains in mice. In turn Shringi et al [50] also compared the virulence of strains isolated from cattle and humans in a piglet and rabbit model and observed that the human strains were more virulent than the bovine strains. However these studies have not determined if lineage impacts E. coli O157:H7 virulence. Strains from clade 6and clade 8 tend to produce higher levels of Stx2 toxin. A high production of Stx2 by clade 8 strains has been previously reported [17] along with the ability of Stx2 to alter the normal water absorption across human colon in a dose-dependent manner without altering the electric parameters [51]. In consequence, the higher inhibition of water absorption caused by 7.1 Anguil compared to EDL933 may be attributed to a higher production of Stx2 by 7.1 Anguil. Although a reduced number of isolates were reported here, studies of more isolates from clade 8 and 6 are in progress to more strictly assess the hyprevirulence characteristics of these genotypes.
This study did not show a clear association between E. coli O157:H7 lineages and hemolysis. More isolates of different lineages should be studied to draw a definitive conclusion. The epithelial cell adherence assay was more conclusive in HeLa cells and a clade 8 isolate and a bovine isolate belonging to a novel lineage were the most adherent strains to this type of cell, although other clade 8 strains were less adherent than E. coli O157:H7 EDL933. Adherence of E. coli O157:H7 to epithelial and abiotic surfaces is a complex process. More than 10 tested or putative adhesins are described and those studied have been recently reviewed by McWilliams and Torres [52]. We have observed (unpublished observations) that there is no polymorphism in the structural genes of the adhesins described McWilliams and Torres [52] between E. coli TW14359 (a prototypic clade 8 strain) and EDL933 and Sakai strain. The differences in adherence may be attributed to differential expression patterns among the strains studied here or to other recently described putative adhesins [53] that may be involved in adherence to these cells. Other authors also have observed differences in adherence to different surfaces in spite of similar adhesin composition among E. coli O157:H7 strains.[54].
The behavior of Argentinean strains in this study were not uniform among strains belonging to clade 8. A similar finding was observed in previous studies [22]. Importantly, the clade 8 Rafaela II-827 strain and the 7.1 Anguil strain, which was most closely related to strains of clade 6, were the most lethal and pathogenic for mice. These data suggest that two strains recovered from Argentinean cattle with distinct genetic backgrounds may have an enhanced ability to cause disease in humans. Nonetheless, these strains were not evaluated by the complete set of 96 SNP loci [15] and not all of the original SNP genotypes were included in the current study. As a result, additional genomic characterization of each strain is necessary to better understand the evolutionary history and identify factors that are most important for virulence and disease severity.
Further investigations are in progress to evaluate the genetic basis of virulence that contributes to variation by clade as well as the assessment of the ecological differences that contribute to variation in transmission rates and linkage to food and waterborne disease.
Supporting Information
Supernatant was diluted 1:100 to have linear OD450 reads.
(TIF)
All observations were performed under a microscope. A: Adherence to Caco-2 cells 7.1 Anguil strain, MOI 200. B: Adhesion to Caco-2 cells of Balcarce 14.2 strain, MOI 200. C: Adherence to Caco-2 cells of strain EDL933 (positive control), MOI 200. D: Adherence to Caco-2 cells of DH5a strain (negative control), MOI 200. The monolayers were fixed and stained with 10% Giemsa stain.
(TIF)
Animals were intragastrically infected with E. coli O157:H7 at inocula of 109 CFU in 200ul of PBS. A. survival after intragastric inoculation of BALBc mice with EDL933, 7.1 Anguil, 9.1 Anguil, Rafaela II-827, Balcarce 14.2, Balcarce 24.2, DH5α or PBS. B. evolution of the weight of the mice before and after E. coli O157:H7 inoculation.
(TIF)
Kidney sections (A, C, E) and cecal mucosa sections (B, D, F) of BALB/c mouse infected with control strain DH5α (A, B), O157:H7 strain 7.1 Anguil (C, D) or with O157:H7 Rafaela II-827 (E, F). The mouse was necropsied on day 3 or 4 post-infection. Both strains, 7.1 Anguil and Rafaela II-827, caused glomerular and tubular renal necrosis level (arrows) and microhemorragc foci; in cecum mucosal destruction and necrosis was observed (triangles), with epithelial suface loss and inflammatory infiltration (asterisk). H&E stainning, 400x magnification.
(TIF)
(XLSX)
Acknowledgments
The present study was supported by PICT grant #0211 from FONCYT (Argentina). CI, ML and AC are CONICET fellows.
We thank Julia Sabio y García for her critical reading, Silvio Díaz for expert care and handling of animals, Alicia Araoz for histological procedures, and Valeria Rocha for her technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was partially funded by PICT grant #0211 from National Fund in Science and Technology (FONCYT), Argentina. Partially refers to the fact reagents from older finished grants were used. CI, ML and AC are CONICET fellows. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Noris M, Remuzzi G (2005) Hemolytic uremic syndrome. J Am Soc Nephrol 16: 1035–1050. [DOI] [PubMed] [Google Scholar]
- 2. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. (2011) Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17: 7–15. 10.3201/eid1701.091101p1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Palermo MS, Exeni RA, Fernandez GC (2009) Hemolytic uremic syndrome: pathogenesis and update of interventions. Expert Rev Anti Infect Ther 7: 697–707. 10.1586/eri.09.49 [DOI] [PubMed] [Google Scholar]
- 4. Rivas M, Padola NL, Lucchesi PMA, Massana M (2010) Diarrheagenic Escherichia coli in Argentina In: Torres AG, editor. Pathogenic Escherichia coli in Latin America. Oak Park (IL), Bentham Science Publishers Ltd. pp. 142–161. [Google Scholar]
- 5. Repetto HA (1997) Epidemic hemolytic-uremic syndrome in children. Kidney International 52: 1708–1719. [DOI] [PubMed] [Google Scholar]
- 6. Gioffre A, Meichtri L, Miliwebsky E, Baschkier A, Chillemi G, Romano MI, et al. (2002) Detection of Shiga toxin-producing Escherichia coli by PCR in cattle in Argentina. Evaluation of two procedures. Vet Microbiol 87: 301–313. [DOI] [PubMed] [Google Scholar]
- 7. Fernández D, Rodríguez E, Arroyo GH, Padola NL, Parma AE (2007) Excreción de Escherichia coli verocitotoxigénico (VTEC) en vacas de ordeño en diferentes estaciones del año. XI Congreso Argentino de Microbiología, Córdoba 10–12 Octubre Resumen en Revista Argentina de Microbiología 39: Supl. 1,178. [Google Scholar]
- 8. Meichtri L, Miliwebsky E, Gioffre A, Chinen I, Baschkier A, Chillemi G, et al. (2004) Shiga toxin-producing Escherichia coli in healthy young beef steers from Argentina: prevalence and virulence properties. Int J Food Microbiol 96: 189–198. [DOI] [PubMed] [Google Scholar]
- 9. Hussein HS, Bollinger LM (2005) Prevalence of Shiga toxin-producing Escherichia coli in beef cattle. J Food Prot 68: 2224–2241. [DOI] [PubMed] [Google Scholar]
- 10. McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB (1995) A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A 92: 1664–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McDaniel TK, Kaper JB (1997) A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol Microbiol 23: 399–407. [DOI] [PubMed] [Google Scholar]
- 12. O'Brien A, Newland J, Miller S, Holmes R, Smith H, Formal SB (1984) Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226: 694–696. [DOI] [PubMed] [Google Scholar]
- 13. Noller AC, McEllistrem MC, Stine OC, Morris JG Jr., Boxrud DJ, Dixon B, et al. (2003) Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J Clin Microbiol 41: 675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim J, Nietfeldt J, Benson AK (1999) Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc Natl Acad Sci U S A 96: 13288–13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manning SD, Motiwala AS, Springman AC, Qi W, Lacher DW, Ouellette LM, et al. (2008) Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc Natl Acad Sci U S A 105: 4868–4873. 10.1073/pnas.0710834105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laing CR, Buchanan C, Taboada EN, Zhang Y, Karmali MA, Thomas JE, et al. (2009) In silico genomic analyses reveal three distinct lineages of Escherichia coli O157:H7, one of which is associated with hyper-virulence. BMC Genomics 10: 287 10.1186/1471-2164-10-287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Neupane M A, Abu-Ali GS, Mitra A, Lacher DW, Manning SD, Riordan JT (2011) Shiga toxin 2 overexpression in Escherichia coli O157:H7 strains associated with severe human disease. Microb Pathog 51: 466–470. 10.1016/j.micpath.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. CDC (1993) Update: multistate outbreak of Escherichia coli O157:H7 infections from hamburgers—western United States, 1992–1993. MMWR Morb Mortal Wkly Rep 42: 258–263. [PubMed] [Google Scholar]
- 19. Michino H, Araki K, Minami S, Takaya S, Sakai N, Miyazaki M et al. (1999) Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am J Epidemiol 150: 787–796. [DOI] [PubMed] [Google Scholar]
- 20. Fukushima H, Hashizume T, Morita Y, Tanaka J, Azuma K, Mizumoto Y, et al. (1999) Clinical experiences in Sakai City Hospital during the massive outbreak of enterohemorrhagic Escherichia coli O157 infections in Sakai City, 1996. Pediatr Int 41:: 213–217. [DOI] [PubMed] [Google Scholar]
- 21.CDC (2006) Update on multi-state outbreak of Ecoli O157:H7 infections from fresh spinach, October 6,2006. MMWR Morbidity and Mortality Weekly Report 1045–1046.
- 22. Abu-Ali GS, Ouellette LM, Henderson ST, Lacher DW, Riordan JT, Whittam TS (2010) Increased adherence and expression of virulence genes in a lineage of Escherichia coli O157:H7 commonly associated with human infections. PLoS One 5: e10167 10.1371/journal.pone.0010167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kulasekara BR, Jacobs M, Zhou Y, Wu Z, Sims E, Saenphimmachak C, et al. (2009) Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect Immun 77: 3713–3721. 10.1128/IAI.00198-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larzábal M, Rabinovitz BC, Baldone V, Moreira AR, Elizondo AM, Vilte DA, et al Detección de Escherichia coli O157:H7 perteneciente al hipervirulento clado 8 en bovinos de Argentina. In: Microbiología AAM, editor; 2010; Buenos Aires.
- 25. Mellor GE, Sim EM, Barlow RS, D'Astek BA, Galli L, Chinen I, et al. (2012) Phylogenetically related Argentinean and Australian Escherichia coli O157 isolates are distinguished by virulence clades and alternative Shiga toxin 1 and 2 prophages. Appl Environ Microbiol 78: 4724–4731. 10.1128/AEM.00365-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Olsvik O, Rimstad E, Hornes E, Strockbine N, Wasteson Y, Lund A, et al. (1991) A nested PCR followed by magnetic separation of amplified fragments for detection of Escherichia coli Shiga-like toxin genes. Mol Cell Probes 5: 429–435. [DOI] [PubMed] [Google Scholar]
- 27. Scheutz F, Teel L, Beutin L, Piérard D, Buvens G, Karch H, et al. (2012) Multicenter evaluation of a sequence-based protocol for subtyping shiga toxins and standardizing stx nomenclature. J Clin Microbiol 50: 2951–2963. 10.1128/JCM.00860-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Riordan JT, Viswanath SB, Manning SD, Whittam TS (2008) Genetic differentiation of Escherichia coli O157:H7 clades associated with human disease by real-time PCR. J Clin Microbiol 46: 2070–2073. 10.1128/JCM.00203-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fan JB, Gunderson KL, Bibikova M, Yeakley JM, Chen J, Wickham Garcia E, et al. (2006) Illumina universal bead arrays. Methods Enzymol 410: 57–73. [DOI] [PubMed] [Google Scholar]
- 30. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 32. Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 33. Shringi S, Schmidt C, Katherine K, Brayton KA, Hancock DD, Besser TE, et al. (2012) Carriage of stx2a differentiates clinical and bovine-biased strains of Escherichia coli O157. PLoS One 7: e51572 10.1371/journal.pone.0051572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Beutin L, Steinruck H, Krause G, Steege K, Haby S, Hultsch G, et al. (2007) Comparative evaluation of the Ridascreen Verotoxin enzyme immunoassay for detection of Shiga-toxin producing strains of Escherichia coli (STEC) from food and other sources. J Appl Microbiol 102: 630–639. [DOI] [PubMed] [Google Scholar]
- 35. Karmali MA, Petric M, Lim C, Fleming PC, Arbus GS, Lior H (1985) The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli . J Infect Dis 151: 775–782. [DOI] [PubMed] [Google Scholar]
- 36. Torres A, Giron J, Perna N, Burland V, Blattner F, Avelino-Flores F, et al. (2002) Identification and characterization of lpfABCC´DE, a fimbrial opreron of enterohemorrhagic Escherichia coli O157:H7. Infect Immun 70: 5416–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Warawa J, Finlay BB, Kenny B (1999) Type III secretion-dependent hemolytic activity of enteropathogenic Escherichia coli . Infect Immun 67: 5538–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Larzabal M, Mercado EC, Vilte DA, Salazar-Gonzalez H, Cataldi A, Navarro-Garcia F (2010) Designed coiled-coil peptides inhibit the type three secretion system of enteropathogenic Escherichia coli . PLoS One 5: e9046 10.1371/journal.pone.0009046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gerhardt E, Masso M, Paton AW, Paton JC, Zotta E, Ibarra C. (2013) Inhibition of water absorption and selective damage to human colonic mucosa are induced by subtilase cytotoxin produced by Escherichia coli O113:H21. Infect Immun 81: 2931–2937. 10.1128/IAI.00287-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dorr RA, Kierbel A, Vera J, Parisi M (1997) A new data-acquisition system for the measurement of the net water flux across epithelia. Comput Methods Programs Biomed 53: 9–14. [DOI] [PubMed] [Google Scholar]
- 41. Wadolkowski EA, Burris JA, O'Brien AD (1990) Mouse model for colonization and disease caused by enterohemorrhagic Escherichia coli O157:H7. Infect Immun 58: 2438–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mora A, Blanco M, Yamamoto D, Dahbi G, Blanco JE, López C, et al. (2009) HeLa-cell adherence patterns and actin aggregation of enteropathogenic Escherichia coli (EPEC) and Shiga-toxin-producing E. coli (STEC) strains carrying different eae and tir alleles. Int Microbiol 12: 243–251. [PubMed] [Google Scholar]
- 43.Albanese A, Gerhardt E, Garcia H, Amigo N, Cataldi A, Zotta E, et al. (2015) Inhibition of water absorption and selective damage to human colonic mucosa induced by Shiga toxin-2 are enhanced by Escherichia coli O157:H7 infection. Int J Med Microbiol. [DOI] [PubMed]
- 44. Brando RJ, Miliwebsky E, Bentancor L, Deza N, Baschkier A, Ramos MV, et al. (2008) Renal damage and death in weaned mice after oral infection with Shiga toxin 2-producing Escherichia coli strains. Clin Exp Immunol 153: 297–306. 10.1111/j.1365-2249.2008.03698.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rivero MA, Padola NL, Etcheverria AI, Parma AE (2004) [Enterohemorrhagic Escherichia coli and hemolytic-uremic syndrome in Argentina]. Medicina (B Aires) 64: 352–356. [PubMed] [Google Scholar]
- 46. Rivas M, Caletti MG, Chinen I, Refi SM, Roldan CD, Chillemi G, et al. (2003) Home-prepared hamburger and sporadic hemolytic uremic syndrome, Argentina. Emerg Infect Dis 9: 1184–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shimizu E, Macías A, Paolicchi F, Magnano G, Zapata L, Fernández A, et al. (2014) Genotyping Mycobacterium bovis from cattle in the Central Pampas of Argentina: temporal and regional trends. Mem Inst Oswaldo Cruz 109: 236–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baker DR, Moxley RA, Steele MB, Lejeune JT, Christopher-Hennings J, Chen DG, et al. (2007) Differences in virulence among Escherichia coli O157:H7 strains isolated from humans during disease outbreaks and from healthy cattle. Appl Environ Microbiol 73: 7338–7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eaton KA, Friedman DI, Francis GJ, Tyler JS, Young VB, Haeger J, et al. (2008) Pathogenesis of renal disease due to enterohemorrhagic Escherichia coli in germ-free mice. Infect Immun 76: 3054–3063. 10.1128/IAI.01626-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shringi S, Garcia A, Lahmers KK, Potter KA, Muthupalani S, Swennes AG, et al. (2012) Differential virulence of clinical and bovine-biased enterohemorrhagic Escherichia coli O157:H7 genotypes in piglet and Dutch belted rabbit models. Infect Immun 80: 369–380. 10.1128/IAI.05470-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fiorito P, Burgos JM, Miyakawa MF, Rivas M, Chillemi G, Berkowski D, et al. (2000) Effect of Shiga toxin 2 on water and ion transport in human colon in vitro. Dig Dis Sci 45: 480–486. [DOI] [PubMed] [Google Scholar]
- 52. McWilliams BD, Torres AG (2014) EHEC Adhesins. Microbiol Spectr 2: EHEC00032013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kudva IT, Griffin RW, Krastins B, Sarracino DA, Calderwood SB, John M (2012) Proteins other than the locus of enterocyte effacement-encoded proteins contribute to Escherichia coli O157:H7 adherence to bovine rectoanal junction stratified squamous epithelial cells. BMC Microbiol 12: 103 10.1186/1471-2180-12-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Matheus-Guimaraes C, Goncalves EM, Cabilio Guth BE (2014) Interactions of O157 and non-O157 Shiga toxin-producing Escherichia coli (STEC) recovered from bovine hide and carcass with human cells and abiotic surfaces. Foodborne Pathog Dis 11: 248–255. 10.1089/fpd.2013.1653 [DOI] [PubMed] [Google Scholar]
- 55. Besser TE, Shaikh N, Holt NJ, Tarr PI, Konkel ME, Malik-Kale P, et al. (2007) Greater diversity of Shiga toxin-encoding bacteriophage insertion sites among Escherichia coli O157:H7 isolates from cattle than in those from humans. Appl Environ Microbiol 73: 671–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supernatant was diluted 1:100 to have linear OD450 reads.
(TIF)
All observations were performed under a microscope. A: Adherence to Caco-2 cells 7.1 Anguil strain, MOI 200. B: Adhesion to Caco-2 cells of Balcarce 14.2 strain, MOI 200. C: Adherence to Caco-2 cells of strain EDL933 (positive control), MOI 200. D: Adherence to Caco-2 cells of DH5a strain (negative control), MOI 200. The monolayers were fixed and stained with 10% Giemsa stain.
(TIF)
Animals were intragastrically infected with E. coli O157:H7 at inocula of 109 CFU in 200ul of PBS. A. survival after intragastric inoculation of BALBc mice with EDL933, 7.1 Anguil, 9.1 Anguil, Rafaela II-827, Balcarce 14.2, Balcarce 24.2, DH5α or PBS. B. evolution of the weight of the mice before and after E. coli O157:H7 inoculation.
(TIF)
Kidney sections (A, C, E) and cecal mucosa sections (B, D, F) of BALB/c mouse infected with control strain DH5α (A, B), O157:H7 strain 7.1 Anguil (C, D) or with O157:H7 Rafaela II-827 (E, F). The mouse was necropsied on day 3 or 4 post-infection. Both strains, 7.1 Anguil and Rafaela II-827, caused glomerular and tubular renal necrosis level (arrows) and microhemorragc foci; in cecum mucosal destruction and necrosis was observed (triangles), with epithelial suface loss and inflammatory infiltration (asterisk). H&E stainning, 400x magnification.
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


