Abstract
Circadian pacemaker neurons in the Drosophila brain control daily rhythms in locomotor activity. These pacemaker neurons can be subdivided into early or late groups depending on whether rhythms in period (per) and timeless (tim) expression are initiated at the first instar (L1) larval stage or during metamorphosis, respectively. Because CLOCK-CYCLE (CLK-CYC) heterodimers initiate circadian oscillator function by activating per and tim transcription, a Clk-GFP transgene was used to mark when late pacemaker neurons begin to develop. We were surprised to see that CLK-GFP was already expressed in four of five clusters of late pacemaker neurons during the third instar (L3) larval stage. CLK-GFP is only detected in postmitotic neurons from L3 larvae, suggesting that these four late pacemaker neuron clusters are formed before the L3 larval stage. A GFP-cyc transgene was used to show that CYC, like CLK, is also expressed exclusively in pacemaker neurons from L3 larval brains, demonstrating that CLK-CYC is not sufficient to activate per and tim in late pacemaker neurons at the L3 larval stage. These results suggest that most late pacemaker neurons develop days before novel factors activate circadian oscillator function during metamorphosis.
Keywords: cell determination, circadian clock, clock proteins, gene expression, pacemaker neurons
Introduction
Circadian (∼24 h) clocks function to synchronize physiological, metabolic, and behavioral rhythms with daily environmental cycles. In animals, these clocks keep circadian time via transcriptional feedback loops, which use conserved transcriptional activators and repressors to drive rhythms in transcription. In Drosophila, CLOCK (CLK) and its heterodimeric partner CYCLE (CYC) initiate feedback loop function by activating expression of the repressors PERIOD (PER) and TIMELESS (TIM). PER-TIM complexes then inhibit CLK-CYC until they are degraded, thus enabling the next round of CLK-CYC transcription (Hardin, 2011).
In the brain, this feedback loop operates in ∼150 brain pacemaker neurons that function to drive locomotor activity rhythms in adults. Pacemaker neurons can be divided into multiple clusters based on their location, size, and neuropeptide expression, including eight pigment dispersing factor (PDF)-expressing small ventral lateral neurons (sLNvs), two PDF-negative sLNvs, eight large ventral lateral neurons (lLNvs), 12 dorsal lateral neurons (LNds), six lateral posterior neurons (LPNs), four anterior dorsal neuron 1 s (DN1as), ∼30 posterior dorsal neuron 1 s (DN1ps), four dorsal neuron 2 s (DN2s), and ∼80 dorsal neuron 3 s (DN3s; Helfrich-Förster, 2005; Nitabach and Taghert, 2008). These clusters of brain pacemaker neurons communicate via neuropeptides to form a network that determines the pattern of locomotor activity based on environmental inputs (Peschel and Helfrich-Förster, 2011; Taghert and Nitabach, 2012; Yoshii et al., 2012).
During development, CLK is detected in 8–10 sLNvs, a subset of four DN1as, and four DN2s several hours before PER is detected in late embryos and early first instar (L1) larvae (Helfrich-Förster et al., 2007; Houl et al., 2008). Rhythmic PER expression persists throughout larval and pupal development and in adults (Kaneko et al., 1997; Kaneko and Hall, 2000), demonstrating that circadian clocks operate continuously in “early” pacemaker neurons after embryogenesis. The remaining pacemaker neurons (e.g., LNds, DN1ps, LPNs, DN3s, lLNvs), which account for >85% of all brain pacemaker neurons, are not detected until 50–90% of pupal development, when they initiate rhythmic PER expression (Kaneko et al., 1997; Kaneko and Hall, 2000). This result suggests that “late” pacemaker neurons do not develop until the last half of metamorphosis.
To determine when late pacemaker neurons arise during metamorphosis, a Clk-GFP transgene was used because Clk is the first clock gene to be expressed and CLK-GFP can be detected with high sensitivity. Our initial experiments confirm CLK-GFP expression in ∼16 PER-positive early pacemaker neurons of L1 and L2 larvae. However, we were surprised to detect CLK-GFP in all groups of brain pacemaker neurons except the lLNvs in mid to late L3 larvae. These results demonstrate that late pacemaker neurons are present, but do not support circadian oscillator function until per expression is detected in late pupae several days later. This delay in per expression is not due to the lack of cyc expression as a GFP-cyc transgene is expressed exclusively in early and late pacemaker neurons. These results suggest that most late pacemaker neurons are present days before novel factors activate circadian oscillator function during metamorphosis.
Materials and Methods
Fly strains.
The following Drosophila strains were used in this study: w1118, w; Cyo/Sco; TM2/TM6B, PBac{yellow[+]-attP-9A}VK00018 (VK18) and y1 w67c23; P{CaryP}attP40 (attP40) (BestGene), cyc01 (Rutila et al., 1998), and Clkout (Mahesh et al., 2014).
Generating the Clk-GFP and GFP-CYC transgenes.
To detect CLK with high sensitivity, an attB-P[acman]-ClkV5-EGFP (Clk-GFP) transgene was generated that expresses a CLK-GFP fusion protein. To accomplish this, GFP was fused in frame to the C-terminal V5 epitope tag of CLK-V5 within the attB-P[acman]-ClkV5 plasmid (Mahesh et al., 2014). To introduce GFP, a 3′ genomic fragment of ClkV5 (from 351 bp upstream to 1580 bp downstream of the translation stop) was first cloned into the TA vector (Invitrogen) using SphI and NotI to form ClkV5-TA. An FseI site was introduced before the ClkV5 stop codon to form ClkV5Fse-TA using the QuikChange site directed mutagenesis kit (Stratagene). GFP coding sequence was amplified from PL-452-C-EGFP vector (Addgene) using primers containing FseI sites and inserted into the TA vector (Invitrogen). The GFP fragment was excised using FseI and inserted into ClkV5Fse-TA at the FseI site to form ClkV5GFP-TA. The 3′ Clk SphI-NotI genomic fragment in attB-P[acman]-Clk-V5 was swapped with the Clk SphI-NotI genomic fragment from ClkV5GFP-TA to form attB-P[acman]-ClkV5-GFP. This transgene, which we refer to as Clk-GFP, was inserted into the VK00018 attP site on chromosome 2 via PhiC31-mediated transgenesis (Groth et al., 2004; Venken et al., 2006). The Clk-GFP transgene was moved into Clkout for behavioral analysis and immunostaining.
An N-terminal eGFP tagged cycle (cyc) transgene (GFP-cyc) was constructed via recombineering (Venken et al., 2006). Phusion DNA polymerase (New England BioLabs) was used to amplify the eGFP-LoxP-kanamycin cassette from plasmid PL-452 N-eGFP (Addgene) using using primer cyc-L (5′-gtcggaaagggcttaatttttcataagcaaacgtcaccgattggcgATGGTGAGCAAGGGCGAGGAGCTG-3′), which contains 45 nt of cyc sequence upstream of the translation start (lowercase) and the first 24 nt of the GFP translated sequence (uppercase), and cyc-R (5′-gtaatttgcaatgcacttttccagtgaaactcaccagaactcctgaacttcACTAGTGGATCCCCTCGAGGGAC-3′), which contains 51 nt from cyc exon 1 (lowercase) and 23 nt from the 3′ end of the eGFP cassette (uppercase). This fragment was used to transform SW102 cells harboring the BAC clone C322–08N23 (BAC-PAC Resources Center), which contains the cyc genomic region, and recombinants containing the eGFP-LoxP-kanamycin cassette inserted into cyc were selected on plates containing kanamycin. The kanamycin gene was removed by inducing recombination at the LoxP sites (Venken et al., 2008, 2009), resulting in the chloramphenicol-resistant GFP-cyc p(ACMAN) clone. GFP-cyc was amplified in EPI 300 cells (Epicenter), and sequenced to confirm the N-terminal GFP-CYC fusion. The GFP-cyc transgene was inserted into attP40 on chromosome 2 via PhiC31-mediated transgenesis (Groth et al., 2004). The GFP-cyc transgene was moved into cyc01 for behavioral analysis and immunostaining.
Staging of larvae and pupae.
Clk-GFP; Clkout and GFP-cyc; cyc01 flies were maintained in a 12 h light/dark (LD) cycle for three cycles at 25°C. Larvae and pupae were staged as previously described (Helfrich-Förster, 1997; Kaneko et al., 1997). Larvae were collected at the following times: L1–L2, 24 h after hatching; L2, 36 h after hatching; L2–L3, 48 h after hatching; early L3, 60 h after hatching; late L3, 72 h after hatching. P20%, P40%, 50%, 60% of pupal (P) development: P20%, 24 h after pupa formation; P40%, 48 h after pupa formation; P50%, 60 h after pupa formation; P60%, 72 h after pupa formation.
Immunostaining larval CNSs and adult brains.
Antibody staining of adult brain and larval CNSs was performed as previously described (Houl et al., 2008). Larvae were staged and collected as above. Larval CNSs and adult brains were dissected and fixed with 3.7% formaldehyde. Samples were washed and incubated with primary and secondary antibodies in a solution containing 1× PBS, 5% BSA, 3% goat serum (Sigma-Aldrich) (3% donkey serum for primary antibodies raised in goat), 0.03% sodium deoxycholate, 0.03% Triton X-100. The following primary antibodies were used: guinea pig anti-CLK GP50 1:3000, goat anti-CLK dC-17 (Santa Cruz Biotechnology) 1:100, rabbit anti-GFP ab6556 (Abcam) 1:500, rabbit anti-GFP ab290 (Abcam) 1:2000, mouse anti-ELAV (DSHB) 1:100, preabsorbed rabbit anti-PER (gift from Michael Rosbash, Brandeis University, Waltham, MA) 1:15,000, rat anti-PER (gift from Orie Shafer, University of Michigan, Ann Arbor, MI) 1:500, and mouse anti-PDF (DSHB) 1:500. The following secondary antibodies were used at a 1:200 dilution: goat anti-rabbit AlexaFluor 647 (Invitrogen), donkey anti-rabbit Alexa 647 (Invitrogen), goat anti-guinea pig Cy-3 (Jackson ImmunoResearch Laboratories), goat anti-mouse Cy-3 (Jackson ImmunoResearch Laboratories), goat anti-rat Cy-5 (Jackson ImmunoResearch Laboratories), donkey anti-rat Cy-5 (Jackson ImmunoResearch Laboratories), goat anti-mouse AlexaFluor 488 (Invitrogen), and donkey anti-goat AlexaFluor 488 (Invitrogen).
Confocal microscopy.
Confocal stacks were imaged using an Olympus FV1000 confocal microscope equipped with 20 × /0.85 NA and 100 × 1.40 NA oil-immersion objectives. For double-labeling experiments, sequential scans of the argon ion 488 nm and HeNe (543 nm for Cy3, 633 nm for AlexaFluor 647 and Cy5) lasers were used to avoid bleed-through between channels. For imaging AlexaFluor 488 and Cy3, 488 and 543 nm lasers were used, with the 405/488/543 nm dichroic mirror for excitation. Fluorescence signals were separated by a beam splitter (560 nm long-pass) and recorded on spectral detectors set to 500–530 and 555–655 nm for AlexaFluor 488 and Cy3, respectively. For imaging AlexaFluor 488 and either AlexaFluor 647 or Cy5, Argon 488 and HeNe 633 nm lasers were used, with the 488/543/633 nm dichroic mirror for excitation. Fluorescence signals were separated by a dichroic beam splitter (560 nm long-pass). A spectral detector set to 500–555 nm was used for AlexaFluor 488 and a detector with 650 nm long-pass filter was used for AlexaFluor 647 or Cy5 signals. The Fluoview “Hi-Lo” look-up table was used to set the maximal signal below saturation and set the background to near zero using the high voltage and offset controls. Z-series were obtained at 2 μm step size, and Kalman-averaging was not used. Original Olympus images were saved as 12-bit oib format and processed using FV1000 confocal software to generate maximum intensity projections (Z-projections). Images were adjusted for brightness and contrast using Adobe Photoshop. For each genotype and developmental stage, brain images were acquired using the same settings (power, gain, offset) at the same time.
Assignment of pacemaker neuron subgroups.
Projected Z-series of CLK-GFP, CLK, PER, and GFP-CYC immunostaining were generated along the anterior–posterior (A–P) axis for dissected larval and adult brains. In larvae, DN3s and DN1 as could be readily distinguished from other pacemaker neurons by their locations along the dorsal–ventral (D–V) and lateral–contralateral (L–C) axis, whereas scans through the A–P axis were necessary to distinguish the more posterior DN2s from DN1ps, the more posterior LPNs from LNds, and the more posterior LNds from LNvs. Brains were only used for pacemaker neuron quantification if all subgroups could unambiguously be assigned. In adults, pacemaker neuron subgroups were assigned as described for larvae, except that there was no overlap between the more ventral sLNvs and LNds. The lLNvs often intermingled with LNds, but could typically be distinguished from LNds because they have larger cell bodies. When lLNvs and sLNvs were not marked with PDF, there was some ambiguity in discriminating between lLNvs and LNds.
Circadian locomotor activity monitoring.
Two- to 3-d-old male flies were entrained for 3 d in LD and transferred to constant darkness (DD) for 7 d at 25°C. Locomotor activity was monitored using the Drosophila Activity Monitor (DAM) system (Trikinetics). Locomotor activity during DD was analyzed using ClockLab (Actimetrics) software as described previously (Pfeiffenberger et al., 2010).
Results
Essentially all brain pacemaker neurons from adults are present in L3 larvae
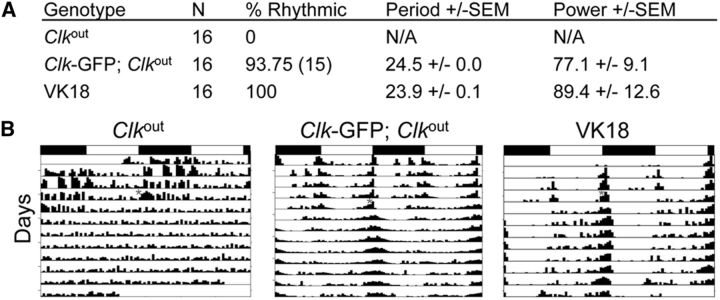
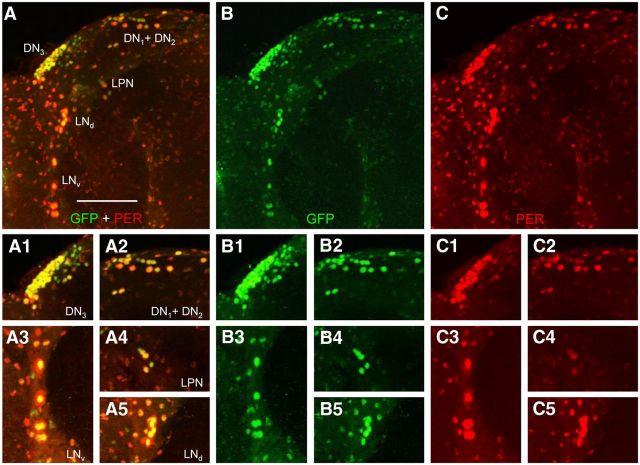
Given that Clk and cyc initiate circadian oscillator function (Hardin, 2011), we used CLK protein expression to mark developing brain pacemaker neurons. To detect CLK expression with high sensitivity, a ∼15 kb genomic DNA fragment was used to generate a transgene that expresses CLK fused to a C-terminal GFP tag (Clk-GFP; see Materials and Methods). The Clk-GFP transgene was integrated at the VK18 attP docking site by PhiC31 recombination (Groth et al., 2004; Venken et al., 2006), and used to generate Clk-GFP; Clkout flies. Behavioral analysis of Clk-GFP; Clkout flies shows that Clk-GFP effectively rescues locomotor activity rhythms of Clkout-null mutant flies with a period of ∼24 h in DD, which is similar to that of the wild-type control containing the VK18 attP docking site (Fig. 1). These results demonstrate that the CLK-GFP fusion protein supports circadian oscillator function. To determine whether CLK-GFP is expressed specifically in clock cells, brains from adults were dissected and immunostained with antibodies against GFP and the clock cell marker PER collected at Zeitgeber Time 22 (ZT22, where ZT0 is lights on and ZT12 is lights off in an LD cycle), when PER is localized to the nucleus (Hardin, 2011). Consistent with previous results, PER expression is detected in both brain pacemaker neurons and glial cells, with the highest intensity staining seen in pacemaker neurons (Shafer et al., 2006; Ng et al., 2011; Fig. 2C–C5). Strong CLK-GFP immunostaining is detected in all brain pacemaker neurons, with weaker immunostaining in most if not all PER-expressing glial cells (Fig. 2A–B5). These results demonstrate that CLK-GFP marks PER-expressing clock cells in adults, and thus accurately reflects CLK protein expression (Houl et al., 2008).
Figure 1.
The Clk-GFP transgene rescues activity rhythms and expression in brain pacemaker neurons. A, w1118; Clkout (Clkout), (w1118; Clk-GFP; Clkout), and PBac{yellow[+]-attP-9A}VK00018 (VK18, attP insertion site for Clk-GFP) flies were entrained in LD cycles for 3 d and transferred to constant darkness. Activity in constant darkness was analyzed as described in Materials and Methods. N, Number of animals tested; % Rhythmic, percentage of flies that showed significant rhythmicity; Period ± SEM, rhythm period ± SEM; Power ± SEM, rhythm power ± SEM. B, Representative actograms of flies tested in A. Flies of the indicated genotypes were entrained in LD cycles, then placed in DD for at least 7 d. The actograms show two LD cycles per line, where the second cycle on a line is the same as the first cycle on the following line. White boxes, Lights-on period; black boxes, lights-off period; gray asterisk, time when flies were placed in DD; vertical bars, fly activity. The height of vertical bars indicates relative level of activity.
Figure 2.
Spatial expression of CLK-GFP in brains from adults. Brains were dissected from Clk-GFP; Clkout adults collected at ZT22, immunostained with GFP and PER antisera, and imaged by confocal microscopy. A–C, A 102 μm projected Z-series image of the left brain hemisphere from an adult fly, where lateral is left and dorsal is top. GFP + PER (A), GFP (B), and PER (C) immunostaining is detected in DN1s, DN2s, DN3s, LNds, LPNs, and LNvs. Colocalization of GFP (green) and PER (red) is shown as yellow. A1–C5, Magnified view of pacemaker neuron subgroups from A (A1–A5), B (B1–B5), and C (C1–C5). A1, B1, C1, Images of a 46 μm projected Z-series of DN3s. A2, B2, C2, Images of a 38 μm projected Z-series of DN1s + DN2s. A3, B3, C3, Images of a 14 μm projected Z-series of LNvs. A4, B4, C4, Images of a 12 μm projected Z-series of LPNs. A5, B5, C5, Images of a 20 μm projected Z-series of LNds. Scale bar, 50 μm. All images are representative of 12 or more brain hemispheres.
Because Clk is required for per expression (Allada et al., 1998; Darlington et al., 1998), we immunostained brains from L3 larvae at ZT22 to confirm that CLK-GFP was expressed in approximately eight PER-positive pacemaker neurons per hemisphere. To our surprise, robust CLK-GFP signal was detected in ∼60 brain neurons per hemisphere, whereas robust PER signal was only detected in four to five sLNvs (Fig. 3A–C). Weaker PER immunostaining was detected in DN1as, and little or no PER was detected in DN2s because PER cycles with a peak at ZT12 in larval DN2s (Kaneko et al., 1997; Fig. 3A,C). Diffuse PER immunoreactivity is also detected in the vicinity of DN3s (Fig. 3A,C), but no PER immunostaining is detected in glial cells from L3 larval brains (Fig. 3A,C), consistent with previous results (Kaneko and Hall, 2000; Helfrich-Förster et al., 2007). The ∼60 CLK-GFP-positive brain pacemaker neurons encompassed almost all pacemaker neuron clusters that had previously been detected in late pupae and adults using PER as a marker (Kaneko et al., 1997; Kaneko and Hall, 2000; Shafer et al., 2006). In contrast, PER was expressed in a limited subset of CLK-GFP-expressing pacemaker neurons in L3 larvae, consistent with previous results (Kaneko et al., 1997; Kaneko and Hall, 2000).
Figure 3.
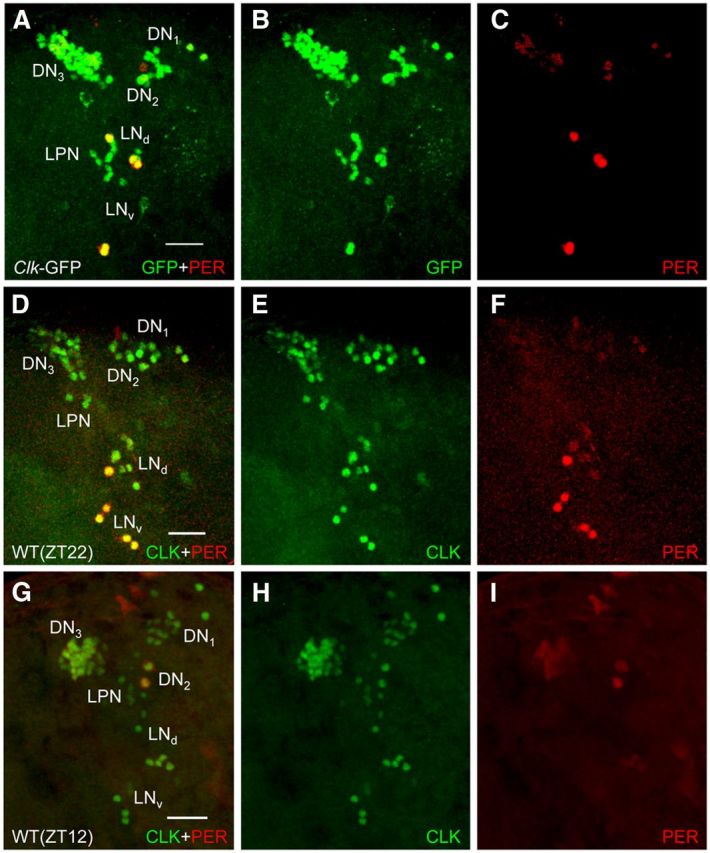
Spatial expression of CLK-GFP in brains from L3 larvae. CNSs were dissected from Clk-GFP; Clkout and WT L3 larvae collected at ZT22 (A–F) or WT L3 larvae collected at ZT12 (G–I), immunostained with GFP and PER antisera (for Clk-GFP; Clkout larvae) or CLK (dC-17) and PER antisera (for WT larvae), and imaged by confocal microscopy. A–C, A 52 μm projected Z-series image of the left hemisphere of a Clk-GFP late L3 larval brain, where lateral is left and dorsal is top. GFP + PER (A), GFP (B), and PER (C) immunostaining is shown for DN1s, DN2s, DN3s, LPNs, LNds, and LNvs. Colocalization of GFP (green) and PER (red) is shown as yellow. D–F, A 50 μm projected Z-series image of the left hemisphere of a WT late L3 larval brain, where lateral is left and dorsal is top. CLK + PER (D), CLK (E), and PER (F) immunostaining is shown for DN1s, DN2s, DN3s, LPNs, LNds, and LNvs. Colocalization of CLK (green) and PER (red) is shown as yellow. G–I, A 76 μm projected Z-series image of the left hemisphere of a WT late L3 larval brain, where lateral is left and dorsal is top. CLK + PER (G), CLK (H), and PER (I) immunostaining is shown for DN1s, DN2s, DN3s, LPNs, LNds, and LNvs. Scale bar, 20 μm. All images are representative of 12 or more brain hemispheres.
To confirm that the expanded pattern of CLK-GFP expression reflects that of endogenous CLK, wild-type L3 larvae were immunostained with CLK antiserum. Our previous analysis of CLK in L3 larvae reliably detected early, but not late, pacemaker neurons (Houl et al., 2008), thus a more sensitive CLK antiserum was used for these experiments (see Materials and Methods). CLK was consistently detected in ∼60 brain neurons per hemisphere that correspond spatially to the same clusters of pacemaker neurons seen with GFP-CLK (Fig. 3D,E). Likewise, CLK expression levels reflect those of CLK-GFP in that CLK immunostaining is strong in the different pacemaker neuron clusters. At ZT22 PER is present in only a small subset of CLK-expressing pacemaker neurons in each hemisphere corresponding to four to five LNvs that show strong PER immunostaining and 2 DN1as that show weaker PER immunostaining (Fig. 3F). Weak PER immunostaining is also detected in DN3s and LNds, which were previously observed to express low levels of TIM and tim-Gal4 driven GFP (Kaneko et al., 1997; Kaneko and Hall, 2000). To determine whether this weak PER staining was rhythmic, we collected wild-type L3 larvae at ZT12 and immunostained with CLK and PER. As expected, strong CLK expression was detected in all of the pacemaker neuron clusters observed at ZT22 (Fig. 3G,H), no PER expression was detected in LNvs and DN1as (Fig. 3G,I), and strong PER expression was detected in the DN2s (Fig. 3G,I). Weak PER expression was also detected in DN3s and LNds at ZT12 (Fig. 3G,I), suggesting that PER levels do not cycle in these two groups of pacemaker neurons. These results demonstrate that L3 larvae express CLK in nearly all brain pacemaker neurons seen in adults, but robust PER expression is limited to the approximately eight pacemaker neurons/hemisphere that support circadian oscillator function.
CLK-GFP expression in pacemaker neurons expands in L3 larvae
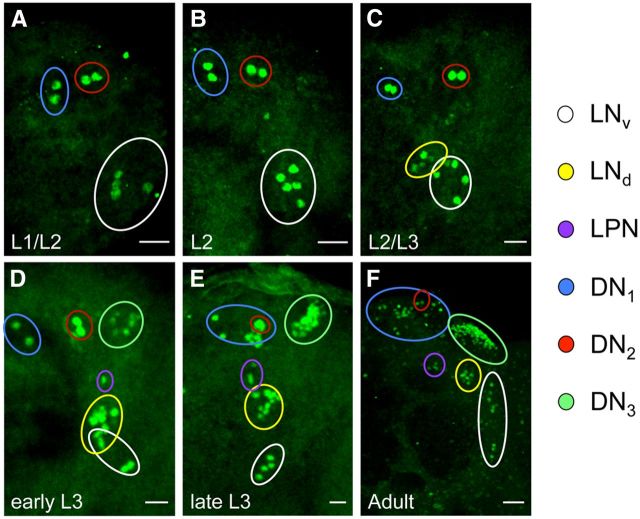
Because CLK is expressed in ∼60 neurons/hemisphere in L3 larvae, we wondered whether this expanded pattern of CLK expression was present at earlier larval stages. To test this possibility, dissected CNSs from L1 and L2 Clk-GFP larvae were immunostained with GFP antibody. In the CNS of both L1 and L2 larvae, CLK-GFP is expressed in approximately eight brain neurons per hemisphere. The location of these CLK-GFP-expressing neurons corresponds to sLNvs, DN1as, and DN2s (Fig. 4A,B; Table 1), consistent with previous reports of PER localization in L1 and L2 larvae (Kaneko et al., 1997; Kaneko and Hall, 2000).
Figure 4.
Expression of CLK-GFP in the brain during larval development. Clk-GFP; Clkout flies were grown to the indicated stage (see Materials and Methods) and collected at ZT22. CNSs from these larvae were dissected, immunostained with GFP antibody, and imaged by confocal microscopy. A–F, Images of 24 h L1–L2 (A), 36 h L2 (B), 48 h L2–L3 (C), 60 h early L3 (D), >72 h late L3 (E), and adult (F) brains. For these developmental stages, 42 μm (L1–L2), 46 μm (L2), 46 μm (L2–L3), 44 μm (early L3), 50 μm (late L3) or 78 μm (adult) projected Z-series images are shown. Scale bars: A–E, 20 μm; F, 10 μm. The colored circles denote pacemaker neuron subgroups according to the key on the right. The different groups of pacemaker neurons were distinguished as described in Materials and Methods. All images are representative of 12 or more brain hemispheres.
Table 1.
Quantifying CLK-GFP expression in brains during development
| Stage (h after hatching) | No. of stained cells |
||||||
|---|---|---|---|---|---|---|---|
| DN1a | DN1p | DN2 | DN3 | LNv | LNd | LPN | |
| L1–L2 (24) | 1.9 ± 0.3 | 0 | 2.0 ± 0.0 | 0 | 4.5 ± 0.5 | 0 | 0 |
| L2 (36) | 1.9 ± 0.3 | 0 | 1.9 ± 0.3 | 0 | 4.5 ± 0.5 | 0 | 0 |
| L2–L3 (48) | 2.0 ± 0.0 | 0 | 2.0 ± 0.0 | 0 | 4.5 ± 0.5 | 2.2 ± 1.1 | 0.2 ± 0.4 |
| L3 (60) | 2.0 ± 0.0 | 0 | 2.0 ± 0.0 | 6.2 ± 2.0 | 5.2 ± 0.8 | 5.7 ± 1.5 | 1.9 ± 0.6 |
| L3 (>72) | 1.9 ± 0.3 | 7.4 ± 1.5 | 1.9 ± 0.3 | 22.9 ± 3.9 | 4.9 ± 0.3 | 6.1 ± 1.0 | 3.1 ± 0.4 |
| Adult | 2.0 ± 0.0 | 15.4 ± 1.1 | 1.9 ± 0.4 | 32.4 ± 5.9 | 8.5 ± 1.9 | 5.0 ± 0.5 | 3.0 ± 1.0 |
Developing or adult flies carrying the Clk-GFP transgene were staged, processed, and imaged as described in Materials and Methods. Number of stained cells denotes the number of GFP-positive cells ± SEM for the different groups of pacemaker neurons. Different life cycle stages are indicated as: L1–L2 (24 h after hatching); mid–L2 (36 h after hatching); L2–L3 (48 h after hatching); mid–L3 (60 h after hatching); and late L3 (>72 h after hatching). PDF staining was used to identify LNvs in the adult brain. The different groups of pacemaker neurons were distinguished as described in Materials and Methods. Data are based on 12 or more brain hemispheres.
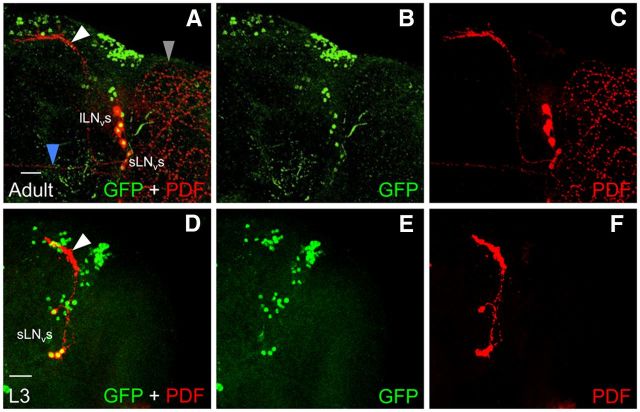
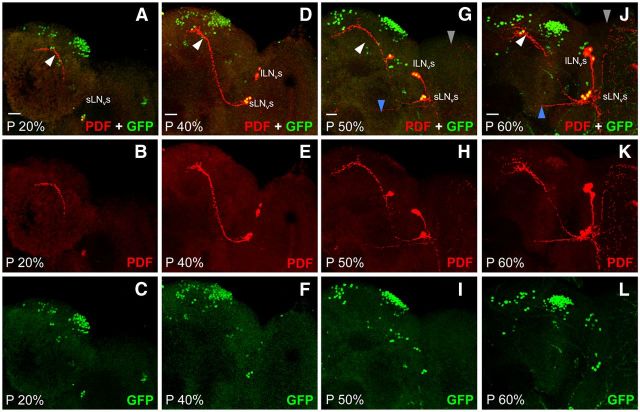
When Clk-GFP larvae are immunostained near the L2–L3 transition (48 h posthatching), additional CLK-GFP-positive neurons are detected in brain regions populated by LNds and LPNs (Fig. 4C; Table 1). Later on during the L3 stage (60 h after hatching), additional CLK-GFP neurons are detected in the LNd and DN3 brain regions (Fig. 4D; Table 1). By late L3 stage (72 h posthatching), CLK-GFP is detected in locations corresponding to all clusters of brain pacemaker neurons seen in adults except the lLNvs (Fig. 4E,F; Table 1). The absence of lLNvs was confirmed by immunostaining with an antibody to the neuropeptide PDF, which is expressed in dorsal projections from sLNvs and in posterior optic tract (POT) projections and medulla arborizations from lLNvs in adults (Fig. 5A–C; Helfrich-Förster, 2003). In late L3 larvae, PDF is only detected in dorsal projections (Fig. 5D–F), which argues that lLNvs are not yet present at this time during development. Indeed, lLNvs are not yet present at 20% pupal development (Fig. 6A–C), but by 40% pupal stage CLK-GFP is detected four additional PDF-positive neurons (Fig. 6D–F), which likely correspond to lLNvs even though they lack POT projections and medulla arborizations. By 50% pupal stage, these CLK-GFP and PDF-positive lLNvs show the characteristic POT projections and medulla arborizations (Fig. 6G–I), consistent with previous results (Helfrich-Förster, 1997; Kaneko et al., 1997), and the intensity of PDF immunostaining increases in POT projections and medulla arborizations by the 60% pupal stage (Fig. 6J–L). Although CLK-GFP is detected in all pacemaker neurons except lLNvs in late L3 larvae, there are fewer DN1ps and DN3s at this stage than in adults (Fig. 4E,F; Table 1), suggesting that all the neurons in these groups are either not yet present or that CLK is expressed in a subset of these neurons at this developmental stage. Analysis of CLK-GFP expression during larval development revealed that CLK expression expands beyond PER-expressing sLNv, DN1a, and DN2 neurons in L3 larvae to encompass all the clusters of brain pacemaker neurons except lLNvs.
Figure 5.
PDF expression in LNvs from L3 larvae and adults. Clk-GFP; Clkout L3 larvae and adults were collected at ZT2. CNSs from larvae and brains from adults were dissected, immunostained with GFP and PDF antibodies, and imaged by confocal microscopy. Colocalization of GFP (green) and PDF (red) is shown as yellow. A–C, an 88 μm projected Z-series image of the right hemisphere of a Clk-GFP adult fly brain, where lateral is right and dorsal is top. D–F, A 60 μm projected Z-series image of the right hemisphere of a Clk-GFP L3 larval fly brain, where lateral is right and dorsal is top. sLNv and lLNv denote the position of LNv cell bodies. White arrowhead, sLNv dorsal projection; blue arrowhead, lLNv posterior optic tract projection; gray arrowhead, lLNv medulla arborizations. Scale bar, 20 μm. All images are representative of 12 or more brain hemispheres.
Figure 6.
PDF expression in LNvs from pupae. Clk-GFP; Clkout pupae at the indicated developmental stages were collected at ZT2. Brains from pupae were dissected, immunostained with GFP and PDF antibodies, and imaged by confocal microscopy. A–C, A 66 μm projected Z-series image of GFP + PDF (A), PDF (B), and GFP (C) immunostaining. D–F, A 76 μm projected Z-series image of GFP + PDF (D), PDF (E), and GFP (F) immunostaining. G–I, A 56 μm projected Z-series image of GFP + PDF (G), PDF (H), and GFP (I) immunostaining. J–L, A 76 μm projected Z-series image of GFP + PDF (J), PDF (K), and GFP (L) immunostaining. A right hemisphere is shown in each image, where lateral is right and dorsal is top. sLNv and lLNv denote the position of LNv cell bodies. Colocalization of GFP (green) and PDF (red) is shown as yellow. White arrowhead, sLNv dorsal projection; blue arrowhead, lLNv posterior optic tract projection; gray arrowhead, lLNv medulla arborizations. Scale bar, 20 μm. All images are representative of 12 or more brain hemispheres.
CLK-GFP is expressed exclusively in postmitotic neurons of L3 larvae
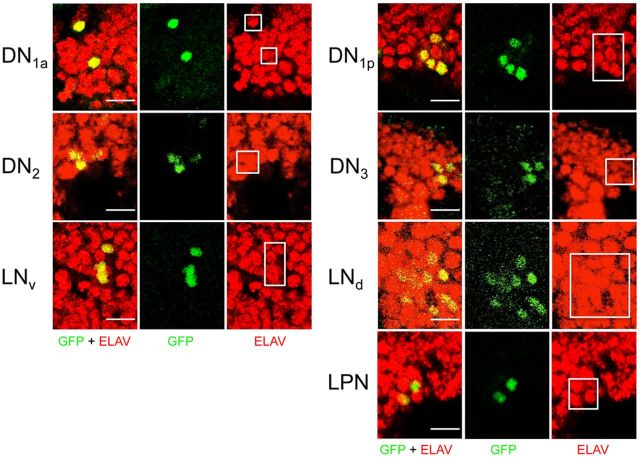
By the L3 larval stage early pacemaker neurons already extend projections and express the postmitotic neuronal marker embryonic lethal abnormal visual system (ELAV; Robinow and White, 1991; Kaneko et al., 1997; Kaneko and Hall, 2000), thus we reasoned that Clk-GFP is activated in preexisting groups of cells in L3 larvae that are fated to become late pacemaker neurons. To determine whether this is the case, early and late L3 larvae were immunostained with GFP and ELAV antibodies. In each case, we find that CLK-GFP is only detected in cells that express ELAV, including early brain pacemaker neurons (Fig. 7) and newly emerging DN1p, LNd, DN3, and LPN brain pacemaker neurons in L3 larval brains (Fig. 7). From these results, we conclude that CLK-GFP is expressed in postmitotic neurons, which suggests that DN1ps, LNds, DN3s, and LPNs derive from presumptive pacemaker neurons already present in L3 larvae.
Figure 7.
CLK-GFP is expressed in postmitotic neurons from L3 larvae. Clk-GFP; Clkout L3 larvae were collected at ZT22. CNSs from larvae were dissected, immunostained with GFP and ELAV antibodies, and imaged by confocal microscopy. GFP + ELAV, GFP, and ELAV immunostaining is shown for the indicated groups of pacemaker neurons. For each group of pacemaker neurons, a 2 μm (DN2, LNv, DN1p, DN3), 4 μm (DN1a), or 6 μm (LNd, LPN) projected Z-series image is shown. Colocalization of GFP (green) and ELAV (red) is shown as yellow. The white boxes indicate the region containing ELAV-positive cells that coimmunostain with GFP. Scale bar, 10 μm. All images are representative of 12 or more brain hemispheres.
GFP-CYC is expressed in all CLK-expressing pacemaker neurons in L3 larvae
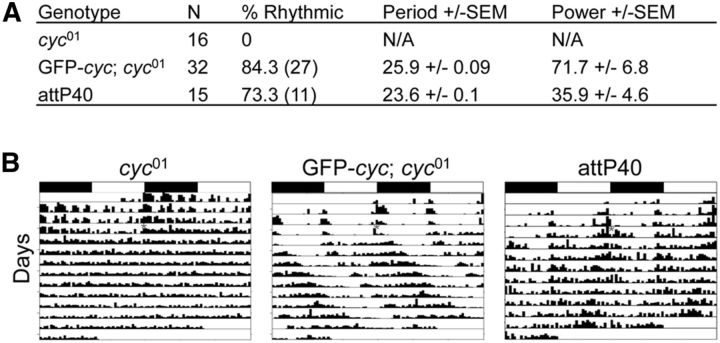
The lack of PER expression in late brain pacemaker neurons from L3 larvae was unexpected because CLK initiates circadian oscillator function by activating the feedback loop repressors per and tim even when expressed ectopically (Zhao et al., 2003; Kilman and Allada, 2009). Because CLK requires CYC to initiate oscillator function (Allada et al., 1998; Darlington et al., 1998; Rutila et al., 1998), the lack of PER expression in late brain pacemaker neurons from L3 larvae could be due to the absence of CYC expression. Unfortunately antibodies capable of detecting CYC in tissue are not available, so we constructed a transgene (GFP-cyc) that expresses CYC fused to a N-terminal GFP tag to determine CYC spatial expression (see Materials and Methods). The GFP-cyc transgene was integrated at the attP40 docking site by PhiC31 recombination (Groth et al., 2004; Venken et al., 2006), and used to generate GFP-cyc; cyc01 flies. Behavioral analysis of GFP-cyc; cyc01 flies shows that GFP-cyc rescues locomotor activity rhythms in cyc01-null mutants that are somewhat stronger and longer period (∼26 h) than the attP40 wild-type controls (Fig. 8). Because circadian period is also lengthened in flies containing only one copy of cyc (Rutila et al., 1998), the long period rhythms in GFP-cyc; cyc01 flies may result from reduced GFP-CYC expression or activity. Nevertheless, these results show that the GFP-CYC fusion protein is functional in the context of the circadian oscillator.
Figure 8.
The GFP-cyc transgene rescues activity rhythms and expression in brain pacemaker neurons. A, w1118; cyc01 (cyc01), w1118; GFP-cyc; cyc01 (GFP-cyc; cyc01), and y1 w67c23; P{CaryP}attP40 (attP40; attP insertion site for GFP-cyc) flies were entrained in LD cycles for 3 d and transferred to DD. Activity in DD was analyzed as described in Materials and Methods. N, Number of animals tested; % Rhythmic, percentage of flies that showed significant rhythmicity; Period ± SEM, rhythm period ± SEM; Power ± SEM, rhythm power ± SEM. B, Representative actograms of flies tested in A. Flies of the indicated genotypes were entrained in LD cycles, then placed in DD for at least 7 d. The actograms show two LD cycles per line, where the second cycle on a line is the same as the first cycle on the following line. White boxes, Lights-on period; black boxes, lights-off period; gray asterisk, time when flies were placed in DD; vertical bars, fly activity. The height of vertical bars indicates relative level of activity.
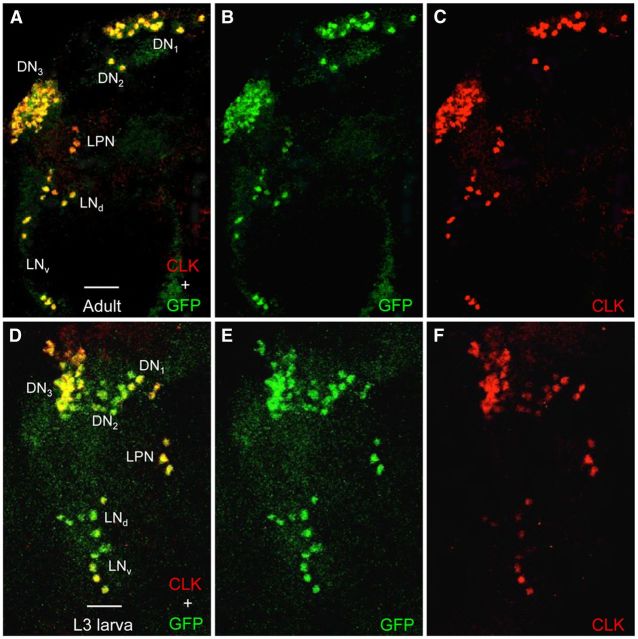
Because cyc is required for Clk to generate ectopic clocks in a wide array of brain neurons (Kilman and Allada, 2009), we expected GFP-CYC to be widely expressed in the adult brain. However, our results show that GFP-CYC is only expressed in brain pacemaker neurons that coexpress CLK (Figs. 9A–C). This result demonstrates that like the other core clock components (e.g., CLK, PER, and TIM), CYC is expressed exclusively in circadian oscillator cells, at least in brains from adults. In L3 larval brains, CYC is also expressed specifically in CLK-expressing neurons (Fig. 9D–F), indicating that the lack of oscillator function in all but the early pacemaker neuron subset of the ∼130 pacemaker neurons in L3 larval brains is not due to the absence of CYC expression. The lack of circadian oscillator function in late pacemaker neurons from L3 larvae may result from the inability of CLK-CYC to activate per and tim transcription. A previous study tested whether per is transcribed in L3 larvae using a per circadian regulatory sequence (CRS) enhancer driven lacZ reporter gene, which is activated by CLK-CYC and faithfully reports per spatial expression in adult brains (Darlington et al., 1998; Hao et al., 1999). This work found that β-galactosidase is only detected in early pacemaker neurons from L3 larvae (Hao et al., 1999), indicating that per is not transcribed in late pacemaker neurons at the L3 larval stage. The GFP-CYC and per CRS-lacZ reporter results indicate that CLK and CYC are not sufficient to initiate oscillator function in late pacemaker neurons, which implies that other factors are required to activate per and tim transcription, and thus oscillator function, in these neurons during metamorphosis.
Figure 9.
GFP-CYC expression is restricted to CLK-expressing neurons in adult and L3 larval brains. Brains were dissected from GFP-cyc; cyc01 adults and L3 larvae collected at ZT22, immunostained with GFP and CLK antisera, and imaged by confocal microscopy. Colocalization of GFP (green) and CLK (red) is shown as yellow. A–C, A 75 μm projected Z-series image of the left hemisphere of a GFP-cyc; cyc01 adult fly brain, where lateral is left and dorsal is top. GFP + CLK (A), GFP (B), and CLK (C) immunostaining is detected in the indicated groups of pacemaker neurons. D–F, A 60 μm projected Z-series image of the left hemisphere of a GFP-cyc; cyc01 L3 larval brain, where lateral is left and dorsal is top. GFP + CLK (D), GFP (E), and CLK (F) immunostaining is detected in the indicated groups of pacemaker neurons. Scale bar, 20 μm. All images are representative of 12 or more brain hemispheres.
Discussion
In this study, we show that the late developing DN1p, LNd, DN3, and LPN pacemaker neurons, which were previously detected during the second half of metamorphosis and in adults (Kaneko et al., 1997; Kaneko and Hall, 2000), are present >2 d earlier at the L3 larval stage. We initially detected late pacemaker neurons in L3 larvae using transgenic flies expressing CLK-GFP, then confirmed the presence of these neurons in wild-type L3 larvae using CLK antibody. One notable absence among the late pacemaker neurons in L3 larvae were the lLNvs, which do not appear until the last half of metamorphosis regardless of the clock neuron marker used. CLK-GFP expression extends beyond pacemaker neurons to include most, if not all, PER-expressing glial cells in adults (Fig. 2), but neither CLK-GFP nor PER are expressed in glial cells from L3 larvae (Fig. 3A–C). PER is robustly expressed with a peak at ZT22 (LNvs, DN1as) or ZT12 (DN2s) in early pacemaker neurons from L3 larvae (Kaneko et al., 1997; Fig. 3C,F,I), but PER is either absent (DN1ps, LPNs) or weakly expressed (DN3s, LNds) at both ZT22 and ZT12 in late pacemaker neurons from L3 larvae, indicating that the circadian oscillator in late pacemaker neurons is not functional. The lack of PER expression in late pacemaker neurons from L3 larvae is not due to the absence of CYC, which is expressed in all CLK-positive neurons in L3 larvae. These results suggest that CLK and CYC-positive late pacemaker neurons in L3 larvae remain in a “primed” state until the last half of metamorphosis, when other factors apparently activate oscillator function.
Our results show that all late pacemaker neurons except the lLNvs express CLK days before their circadian oscillators begin to function (Figs. 4–6; Table 1). Circadian oscillator function is initiated in these late pacemaker neurons during the second half of metamorphosis (Kaneko et al., 1997), which suggests that additional factors act during metamorphosis to promote robust CLK-CYC-dependent transcription of per, tim, and possibly other target genes. The ultimate triggers of transcriptional activity during metamorphosis are likely to be either the steroid hormone ecdysone, which binds to nuclear receptors to regulate target gene transcription, or the sesquiterpenoid Juvenile Hormone (JH), which binds to the bHLH-PAS transcription factors Methoprene-tolerant (Met) or germ-cell expressed (gce) to activate transcription of target genes (Di Cara and King-Jones, 2013). Ecdysone and JH titers rise and fall multiple times during larval and pupal development (Di Cara and King-Jones, 2013), thereby altering the transcriptional activity of their receptors. Because CLK-CYC is present well before metamorphosis in most late pacemaker neurons, the proximate triggering factor may activate CLK-CYC-dependent transcription via post-translational (i.e., adding or removing protein modifications) or transcriptional (i.e., chromatin remodeling or cooperative activator DNA binding) mechanisms. The only cluster of late pacemaker neurons that do not express CLK and CYC in L3 larvae are the lLNvs (Figs. 5, 6). Because these neurons begin to express CLK and PER around the same time PER starts to be expressed in other late pacemaker neurons (Kaneko et al., 1997; Fig. 6), the triggering mechanism may be the same. The significance of the difference in the timing of CLK expression in the lLNvs and other late brain pacemaker neurons is not clear.
We were surprised to see that CLK-CYC-expressing neurons lacked circadian oscillator function given that ectopic expression of CLK is capable of initiating oscillator function in many adult tissues (Zhao et al., 2003; Kilman and Allada, 2009). The ability of Clk to produce ectopic oscillators is dependent on cyc (Kilman and Allada, 2009), thus we predicted that cyc is broadly expressed. However, we found that GFP-CYC is highly expressed in CLK-positive brain neurons at the L3 larval and adult stages (Fig. 9), which suggests that CLK somehow promotes CYC expression. Given that CLK is a transcription activator, it is likely that CLK supports CYC expression by activating cyc transcription or by stabilizing CYC protein. If CLK activates cyc transcription, CLK could bind cyc regulatory sequences either as a homodimer or as a heterodimer with a more broadly expressed bHLH transcription activator. If CLK stabilizes CYC, then cyc transcription could be either broad or restricted to CLK-expressing cells. These mechanisms for CLK-dependent expression of CYC are not mutually exclusive; CLK could both activate cyc transcription and stabilize CYC. Other mechanisms for CLK regulation of CYC expression are possible, including translational control, RNA processing, and RNA stability. In any case, additional studies to assess where cyc is transcribed in vivo and to determine the stability of GFP-CYC will provide insight into how CYC expression is regulated.
Brain pacemaker neurons are first detected at three different stages of Drosophila development. The sLNv, DN1a, and DN2 early pacemaker neurons are detected in late embryos (Houl et al., 2008), and are present in all subsequent developmental stages and in adults (Helfrich-Förster, 1997; Kaneko et al., 1997). The sLNv cluster of pacemaker neurons extend projections and express PDF as early as the L1 larval stage (Helfrich-Förster, 1997; Kaneko et al., 1997; Kaneko and Hall, 2000), indicating that these cells are fully differentiated. The DN1as and DN2s extend projections by the L3 larval stage (Kaneko and Hall, 2000), but when these projections are first formed is not known. We show that CLK is coexpressed with the neuronal differentiation marker ELAV in early pacemaker neurons from L3 larvae (Fig. 7), strongly arguing that these pacemaker neurons do not give rise to late pacemaker neurons. CLK is also detected in ELAV-positive late pacemaker neurons from L3 larvae (Fig. 7), which shows that these neurons are already differentiated by the time CLK is expressed. This result implies that the circadian pacemaker neuron network is established well before CLK is expressed in late pacemaker neurons, which raises several important questions. When do late pacemaker neurons arise? What determines their fate? When and how are late pacemaker neurons integrated with early pacemaker neurons? The answers to these questions will provide insight into how this neural network is designed to ultimately control locomotor activity rhythms in adult flies.
Footnotes
This work was supported by National Institutes of Health Grant NS080638 to P.E.H. We thank Michael Rosbash for the cyc01 mutant and PER antibody raised in rabbit, Orie Shafer for PER antibody raised in rat, and Dr Stan Vitha from the Texas A&M Microscopy and Imaging Center for help with confocal imaging.
The authors declare no competing financial interests.
References
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/S0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Di Cara F, King-Jones K. How clocks and hormones act in concert to control the timing of insect development. Curr Top Dev Biol. 2013;105:1–36. doi: 10.1016/B978-0-12-396968-2.00001-4. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Glossop NR, Lyons L, Qiu J, Morrish B, Cheng Y, Helfrich-Förster C, Hardin P. The 69 bp circadian regulatory sequence (CRS) mediates per-like developmental, spatial, and circadian expression and behavioral rescue in Drosophila. J Neurosci. 1999;19:987–994. doi: 10.1523/JNEUROSCI.19-03-00987.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C. Development of pigment-dispersing hormone-immunoreactive neurons in the nervous system of Drosophila melanogaster. J Comp Neurol. 1997;380:335–354. doi: 10.1002/(SICI)1096-9861(19970414)380:3<335::AID-CNE4>3.3.CO%3B2-M. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc Res Tech. 2003;62:94–102. doi: 10.1002/jemt.10357. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. Neurobiology of the fruit fly's circadian clock. Genes Brain Behav. 2005;4:65–76. doi: 10.1111/j.1601-183X.2004.00092.x. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C, Shafer OT, Wülbeck C, Grieshaber E, Rieger D, Taghert P. Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. J Comp Neurol. 2007;500:47–70. doi: 10.1002/cne.21146. [DOI] [PubMed] [Google Scholar]
- Houl JH, Ng F, Taylor P, Hardin PE. CLOCK expression identifies developing circadian oscillator neurons in the brains of Drosophila embryos. BMC Neurosci. 2008;9:119. doi: 10.1186/1471-2202-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(SICI)1096-9861(20000619)422:1<66::AID-CNE5>3.0.CO%3B2-2. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Helfrich-Förster C, Hall JC. Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemaker candidates and novel features of clock gene product cycling. J Neurosci. 1997;17:6745–6760. doi: 10.1523/JNEUROSCI.17-17-06745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilman VL, Allada R. Genetic analysis of ectopic circadian clock induction in Drosophila. J Biol Rhythms. 2009;24:368–378. doi: 10.1177/0748730409343761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahesh G, Jeong E, Ng FS, Liu Y, Gunawardhana K, Houl JH, Yildirim E, Amunugama R, Jones R, Allen DL, Edery I, Kim EY, Hardin PE. Phosphorylation of the transcription activator CLOCK regulates progression through a approximately 24-h feedback loop to influence the circadian period in Drosophila. J Biol Chem. 2014;289:19681–19693. doi: 10.1074/jbc.M114.568493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng FS, Tangredi MM, Jackson FR. Glial cells physiologically modulate clock neurons and circadian behavior in a calcium-dependent manner. Curr Biol. 2011;21:625–634. doi: 10.1016/j.cub.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–R93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- Peschel N, Helfrich-Förster C. Setting the clock-by nature: circadian rhythm in the fruitfly Drosophila melanogaster. FEBS Lett. 2011;585:1435–1442. doi: 10.1016/j.febslet.2011.02.028. [DOI] [PubMed] [Google Scholar]
- Pfeiffenberger C, Lear BC, Keegan KP, Allada R. Sleep and circadian behavior monitoring in Drosophila. In: Zhang B, Freeman MR, Waddell S, editors. Drosophila neurobiology: a laboratory manual. New York: Cold Spring Harbor; 2010. pp. 483–504. [Google Scholar]
- Robinow S, White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/S0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Helfrich-Förster C, Renn SC, Taghert PH. Reevaluation of Drosophila melanogaster's neuronal circadian pacemakers reveals new neuronal classes. J Comp Neurol. 2006;498:180–193. doi: 10.1002/cne.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert PH, Nitabach MN. Peptide neuromodulation in invertebrate model systems. Neuron. 2012;76:82–97. doi: 10.1016/j.neuron.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Kasprowicz J, Kuenen S, Yan J, Hassan BA, Verstreken P. Recombineering-mediated tagging of Drosophila genomic constructs for in vivo localization and acute protein inactivation. Nucleic Acids Res. 2008;36:e114. doi: 10.1093/nar/gkn486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, Bellen HJ, Hoskins RA. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Rieger D, Helfrich-Förster C. Two clocks in the brain: an update of the morning and evening oscillator model in Drosophila. Prog Brain Res. 2012;199:59–82. doi: 10.1016/B978-0-444-59427-3.00027-7. [DOI] [PubMed] [Google Scholar]
- Zhao J, Kilman VL, Keegan KP, Peng Y, Emery P, Rosbash M, Allada R. Drosophila Clock can generate ectopic circadian clocks. Cell. 2003;113:755–766. doi: 10.1016/S0092-8674(03)00400-8. [DOI] [PubMed] [Google Scholar]