Abstract
Nucleosome destabilization by histone variants and modifications has been implicated in the epigenetic regulation of gene expression, with the histone variant H2A.Z and acetylation of H3K56 (H3K56ac) being two examples. Here we find that deletion of SWR1, the major subunit of the SWR1 complex depositing H2A.Z into chromatin in exchange for H2A, promotes epigenetic white-opaque switching in Candida albicans. We demonstrate through nucleosome mapping that SWR1 is required for proper nucleosome positioning on the promoter of WOR1, the master regulator of switching, and that its effects differ in white and opaque cells. Furthermore, we find that H2A.Z is enriched adjacent to nucleosome-free regions at the WOR1 promoter in white cells, suggesting a role in the stabilization of a repressive chromatin state. Deletion of YNG2, a subunit of the NuA4 H4 histone acetyltransferase (HAT) that targets SWR1 activity through histone acetylation, produces a switching phenotype similar to that of swr1, and both may act downstream of the GlcNAc signaling pathway. We further uncovered a genetic interaction between swr1 and elevated H3K56ac with the discovery that the swr1 deletion mutant is highly sensitive to nicotinamide. Our results suggest that the interaction of H2A.Z and H3K56ac regulates epigenetic switching at the nucleosome level, as well as having global effects.
INTRODUCTION
Chromatin-level changes, such as histone modifications, variants, and nucleosome remodeling, have been implicated in the epigenetic regulation of gene expression in many systems. In recent years, focus has shifted toward the role of nucleosome dynamics in epigenetic regulation, in particular the importance of nucleosome destabilization in the establishment and inheritance of active chromatin states (1). Increasing evidence has shown that numerous histone modifications affect nucleosome stability and dynamics by altering the rate of nucleosome turnover and DNA access (2).
One such example of a histone modification altering nucleosome dynamics is the acetylation of H3 lysine 56. H3K56 acetylation (H3K56ac) occurs during S phase on newly synthesized histones and facilitates histone deposition onto replicating DNA as well as nucleosome assembly (3). Consequently, H3K56ac is found at high levels on newly synthesized chromatin and is involved in chromatin packing after DNA replication and repair in Saccharomyces cerevisiae (3–5). During transcriptional activation, H3K56ac promotes chromatin disassembly (6, 7). Replication-independent deposition of histones is also associated with H3K56ac (8). In addition, H3K56 is located near the DNA entry and exit points on the nucleosome core particle (9), further implicating its role in nucleosome dynamics. In yeast, the acetylation of H3K56 is performed by the histone acetyltransferase (HAT) Rtt109 (10, 11), and deacetylation occurs by the sirtuin class histone deacetylases (HDACs) Hst3 and Hst4 (12, 13).
H3K56 acetylation has been shown to regulate the epigenetic process of white-opaque switching in Candida albicans. A major fungal pathogen also found as a commensal with humans, C. albicans switches reversibly between two phenotypic states, white and opaque (14, 15). The two states differ in morphologies, transcriptional profiles, and responses to host defenses, and epigenetic switching may enhance the adaption of C. albicans to host environments (16–19). As in S. cerevisiae, H3K56 acetylation in C. albicans is regulated by Rtt109 and the sirtuin Hst3 (20, 21). Deletion of RTT109 in C. albicans, which prevents acetylation of H3K56, has been shown to impair white-opaque switching, while the HST3/hst3 mutant promotes switching. Treatment with nicotinamide (NAM), which can inhibit sirtuin activity noncompetitively (22, 23) and increases H3K56ac levels, also promotes switching to the opaque form, and this activity has also been shown to be dependent on RTT109 (24). Furthermore, hyperacetylation of H3K56 is toxic to C. albicans (21, 25).
In S. cerevisiae, high H3K56ac levels have been shown to reduce chromatin levels of the histone variant H2A.Z and increase the eviction of H2A.Z-containing nucleosomes (26). This is notable as H2A.Z itself has been implicated in altering nucleosome dynamics and stability. In addition, H3K56ac and H2A.Z have shared roles in genome stability through regulation of chromosome condensation (27, 28). H2A.Z is a highly conserved variant of H2A and is essential in a number of eukaryotic organisms but not in yeast (29–31). H2A.Z is enriched in euchromatin at the promoters of both actively expressed and repressed genes and prevents the spread of heterochromatin (32–34). Nucleosomes containing H2A.Z are enriched at the nucleosome-free regions (NFRs) at promoter regions, in particular those adjacent to the +1/−1 nucleosomes at the transcriptional start site of genes (32, 35). H2A.Z-containing nucleosomes have been noted to have altered stability; however, separate studies have shown H2A.Z nucleosomes to be either more or less stable than those with H2A (36, 37). Of particular note is the finding that nucleosomes containing both H3.3, the variant of H3 which is the canonical H3 in yeast, and H2A.Z are more labile (38). In multicellular organisms, H2A.Z has been associated with cellular differentiation and reprogramming (39). H2A.Z is deposited by the Swi/Snf family chromatin remodeling complex SWR1 through the ATP-dependent exchange of H2A.Z-H2B dimers with H2A-H2B (40, 41). SWR1 encodes the major subunit of the SWR1 complex. Neither the H2A.Z nor the SWR1 gene is essential in yeast; however, SWR1 activity causes genetic instability in H2A.Z-deleted cells (42). The acetylation of canonical histone H2 and H4 tails by the NuA4 HAT complex, in which Yng2 is a subunit of the HAT module, has been shown to recruit the SWR1 complex to deposit H2A.Z (43). In yeast, the NuA4 and SWR1 complexes share four subunits, whereas the two complexes function together as one in higher eukaryotes (44, 45). Recently, NFRs have been shown to be dominant over histone acetylation in targeting SWR1 activity (46). The localization and effect on nucleosomal dynamics of H2A.Z make it a promising subject for the study of epigenetic regulation.
In this study, we find that deposition of H2A.Z by the SWR1 complex is crucial in regulating the white-opaque transition in C. albicans. Abrogation of H2A.Z deposition through deletion of SWR1 results in a phenotype that favors switching to the opaque state and the stability of the opaque phase. We find that nucleosome dynamics on the promoter of WOR1, the master regulator of switching (47–49), differ between white and opaque phases and that deletion of SWR1 changes this dynamic. Furthermore, H2A.Z is present on the WOR1 promoter at higher levels in the white phase than in the opaque phase, suggesting that its absence destabilizes white cells. We find that deletion of YNG2 produces a white-opaque phenotype similar to that of the swr1 deletion mutant. Furthermore, we find that the swr1 deletion mutant displays a synthetic lethality with nicotinamide but no heightened sensitivity to DNA damage, suggesting a close functional relationship between H2A.Z and H3K56ac in C. albicans.
MATERIALS AND METHODS
Strains and culturing conditions.
Strains used in this study are listed in Table 1. Strains were maintained on YEP (1% yeast extract, 2% peptone) or SC (synthetic complete) medium with the appropriate carbon sources.
TABLE 1.
Strains used in this study
| Strain | Description | Genotype | Reference |
|---|---|---|---|
| JYC1 | WT | MTLa/a ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 68 |
| HLY4235 | SWR1/swr1 | MTLa/a ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG SWR1/swr1::HIS1 | This study |
| HLY4233 | swr1 | MTLa/a ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG swr1::HIS1/swr1::ARG4 | This study |
| HLY3555 | WT + pWOR1-GFP | MTLa/a ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG ADH1/adh1::pWOR1-GFP-URA3 | 47 |
| HLY4234 | swr1 + pWOR1-GFP | MTLa/a ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG swr1::HIS1/swr1::ARG4 ADH1/adh1::pWOR1-GFP-URA3 | This study |
| HLC54 | efg1 | MTLa/a ura3::1 imm434/ura3::1 imm434 efg1::hisG/efg1::hisG | 69 |
| HLY4238 | WT + HTA3-HA | MTLa/a ura3::imm434/ura3::imm434 HTA3/3×HA-HTA3-SAT1 ADH1/adh1::pMAL2-WOR1-URA3 | This study |
| HLY4236 | swr1 + HTA3-HA | MTLa/a ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG swr1::HIS1/swr1::ARG4 ADH1/adh1::pWOR1-GFP-URA3 HTA3/3′HA-HTA3-SAT1 ADH1/adh1::pMAL2-WOR1-URA3 | This study |
| HLY3993 | HST3/hst3 | MTLa/a ade2/ade2 ura3::ADE2/ura3::ADE2 HST3/hst3::FRT | 24 |
| HLY3997 | rtt109 | MTLa/a rtt109::FRT/rtt109::FRT | 24 |
| CLY4 | yng2 | ura3::1 imm434/ura3::1 imm434 yng2::hisG/yng2::hisG | 51 |
| HLY3883 | MTLa/a yng2 | MTLa/a ura3::1 imm434/ura3::1 imm434 yng2::hisG/yng2::hisG | This study |
| HLY4035 | yng2 + YNG2 | ura3::1 imm434/ura3::1 imm434 yng2::hisG/YNG2-13MYC-URA3 | 52 |
| HLY3886 | MTLa/a yng2 + YNG2 | MTLa/a ura3::1 imm434/ura3::1 imm434 yng2::hisG/YNG2-13MYC-URA3 | This study |
Strain construction.
SWR1 was deleted by using the deletion cassettes described previously by Noble and Johnson (50). PCR amplification of Candida dubliniensis HIS1 from plasmid pSN52 or of Candida dubliniensis ARG4 from plasmid pSN69 was performed by using long primers containing sequences flanking SWR1, such that the PCR product was flanked by 90-bp sequences homologous to flanking sequences of SWR1. The PCR product was then transformed into wild-type (WT) (JYC1) C. albicans to replace SWR1 by homologous recombination. Transformants were selected for survival on SC dextrose histidine or arginine dropout media, and deletion of SWR1 was verified by PCR.
H2A.Z (HTA3) was hemagglutinin (HA) tagged by PCR amplification of the HTA3 coding sequence with a 3×HA tag at the 5′ end of the sequence, with HindIII and PstI sites introduced at the 5′ and 3′ ends of the amplicon, respectively. The HTA3 promoter was amplified in a separate reaction, flanked by XhoI and HindIII sites. Subsequently, the HA-HTA3 and HTA3 promoter sequences were inserted into the pWOR1-GFP-SAT1 plasmid (24), replacing the GFP and WOR1 promoter sequences, respectively. The plasmid was digested with MscI prior to transformation into the WT or swr1 strain. All primers used for cloning are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer | Sequence | Description |
|---|---|---|
| SWR1D-F | CGATTTATTTAATTACAATTTCGGAAATTAAATCTCATCTTGCAGATCCAGCAAACACATCAAGCATATTCTCACAAGAAAACCAGAAATGCCAGAGAATGGAACCAGTGTGATGGATATCTGC | SWR1 deletion |
| SWR1D-R | AGACTTATCCTTGATGTCTGTACTAGTACCAACGATACGCAAGAGATTTTTATAAAAGAAAGGGCCATTTGCTTAATATGTAATACAAGGAGTGATACCATATAGCTCGGATCCACTAGTAACG | SWR1 deletion |
| SWR1outsideF | TGTTTACACTGTAATTTTCTGC | Check integration of deletion cassette at SWR1 locus |
| SWR1outsideR | ACTATACGATATGGTATCCGT | Check integration of deletion cassette at SWR1 locus |
| C.dubHIS-F | AACACAACTGCACAATCTGG | C. dubliniensis HIS1 check |
| C.dubHIS-R | ATTAGATACGTTGGTGGTTC | C. dubliniensis HIS1 check |
| C.dubARG-F | ACGGAGTACCACATACGATG | C. dubliniensis ARG4 check |
| C.dubARG-R | ACACAGAGATACCTTGTACT | C. dubliniensis ARG4 check |
| SWR1internalF | CTAATTCCAATATCCACCAAGC | SWR1 internal sequence check, 963 bp from 3′ end |
| SWR1internalR | GACCTTGTGATATAACTTTCG | SWR1 internal, 1,134 bp from 5′ end of coding sequence |
| HTA3_F3 | CCGCTCGAGTGATGTCAAAGTTCCTACAAAG | Amplification of HTA3 promoter |
| HTA3_R3 | CCCAAGCTTTGCATAGTCCGGGACGTCATAGGGATAGCCCGCATAGTCAGGAACATCGTATGGGTACATTGTAGTGTTTGTTGTGGTTTCG | Amplification of HTA3 promoter |
| HTA3_F4 | CCCAAGCTTGGATCCTATCCATATGACGTTCCAGATTACGCTGCTATGTCTGGGAAGGGAAAAGTGC | HA tagging of HTA3 |
| HTA3_R4 | GCAACTGCAGACCGCCACTAAATAGAGAGC | HA tagging of HTA3 |
| MTLa-F | TGAAGCGTGAGAGGCAGGAG | MTLa check |
| MTLa-R | AATTCCCTTTCTCTTCGATTAGG | MTLa check |
| MTLα-F | TTCGAGTACATTCTGGTCGCG | MTLα check |
| MTLα-R | TGTAAACATCCTCAATTGTACCCGA | MTLα check |
| WOR1-F | AAATAAACCATCGTCATCGGC | WOR1 qPCR |
| WOR1-R | AGGACCATTACCTAGACTCG | WOR1 qPCR |
| ACT1-F | TGGAAGCTGCTGGTATTGAC | ACT1 qPCR |
| ACT1-R | TTCAGCAATACCTGGGAACA | ACT1 qPCR |
Mating type conversion.
The MTLa/α yng2 deletion mutant (CLY4) (51) and the yng2+YNG2 strain (HLY4035) (52) were converted to MTLa/a by growth on solid SC medium with 2% sorbose at a density of 105 cells per plate for 6 days (53). The mating type locus was verified by PCR.
Switching assays.
Stable white or opaque cells from log-phase cultures were spread onto agarose plates of Lee's medium (modified from reference 54) supplemented with 25 μg/ml uridine and 50 μg/ml arginine, with 1.25% dextrose or N-acetyl-d-glucosamine (Spectrum Chemical) as the carbon source. For CO2 assays, plates were incubated in a Thermo Forma series II water-jacketed CO2 incubator at 25°C with 5% CO2. Colony morphology was scored after 7 days. Both whole-colony and sectored switching events were counted.
Reverse transcription-quantitative PCR (RT-qPCR).
RNA was isolated from log-phase cultures by using the Qiagen RNeasy kit. cDNA was synthesized from 2 μg total RNA by using the Bio-Rad iScript reverse transcription kit. Quantitative PCR (qPCR) was performed on the Bio-Rad iCycler instrument by using Bio-Rad SYBR green mix. Cycle parameters were 95°C for 1 min and then 39 cycles of 95°C for 10 s, 56°C for 45 s, and 68°C for 20 s. Oligonucleotides used for amplification are listed in Table 2.
Nucleosome mapping.
Micrococcal nuclease (MNase) digestion was performed with modifications of methods described previously (55, 56). Cells were grown to an optical density at 600 nm (OD600) of 2 and then cross-linked with 2% formaldehyde at a growth temperature for 30 min before treatment with zymolyase to spheroplast cells. Spheroplasts were then treated with a titration of 2 to 8 units of MNase, and digested samples were visualized by electrophoresis on a 2% agarose gel. Samples with distinct mononucleosome and dinucleosome band patterns were selected for further analysis. DNA from these samples was isolated by using phenol-chloroform extraction and used for qPCR for nucleosome mapping. Staphylococcus aureus MNase was acquired from Sigma-Aldrich. A full list of the primers used for mapping of the WOR1 promoter is available in Table S1 in the supplemental material. Chromatin immunoprecipitation was performed on mononucleosomal DNA as described previously (57). Anti-HA antibody (Abcam) was used for immunoprecipitation of HA-tagged H2A.Z. Nucleosomal values were normalized to values for the −1 nucleosome peak at the BUD2 promoter.
Spot assays.
C. albicans strains were grown in liquid yeast extract-peptone-dextrose (YPD) medium to log phase and diluted to an OD600 of 0.1, after which 5 additional 5-fold serial dilutions were made into double-distilled water (ddH2O) in a 96-well plate. An 8-channel multipipettor was used to transfer 2.5 μl of each dilution onto YPD agarose plates. For drug treatment plates, nicotinamide and methyl methanesulfonate (MMS) were obtained from Sigma-Aldrich. Plates were incubated for 3 days at 30°C, and colony growth was imaged with a Fujifilm LAS-4000 imager.
Fluorescence-activated cell sorting.
Fluorescence-activated cell sorting (FACS) was conducted by using the BD FACSCalibur system, using cells taken from actively growing liquid cultures. The FL1-H channel was used for green fluorescent protein (GFP) detection, and 10,000 cells in suspension were counted per sample. Results were analyzed with FlowJo cytometry analysis software.
RESULTS
The swr1 deletion mutant increases white-to-opaque switching frequency and stabilizes the opaque phase.
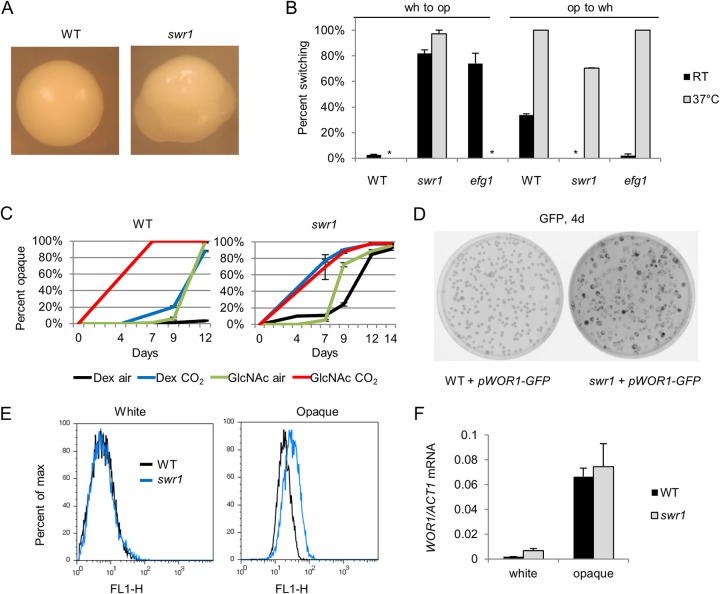
To investigate the role of chromatin remodeling by the SWR1 complex in white-opaque switching, we deleted the SWR1 gene in C. albicans using homologous recombination to replace it with auxotrophic markers as described previously by Noble and Johnson (50). In the resulting swr1 deletion mutant, chromatin immunoprecipitation confirmed that the deposition of H2A.Z into chromatin was abrogated (not shown). High rates of spontaneous switching to the opaque phase in white colonies of the swr1 mutant were observed at room temperature (RT), often occurring as multiple opaque sectors per white colony (Fig. 1A). The white-opaque switching percentage of the swr1 mutant at room temperature (25°C) was 82%, compared to 2 to 4% for the wild type. Opaque cells of the swr1 mutant are more stable than those of the wild type, with 0% switching to white after 7 days of growth at RT, compared to 34% for the WT (Fig. 1B). We next sought to determine the effect of temperature on the swr1 white-opaque phenotype. At 37°C, white-opaque switching of the swr1 mutant increased to 97%, while WT cells no longer switched. We also compared the phenotype of the swr1 mutant to that of the efg1 mutant, the deletion mutant of the transcriptional regulator EFG1 which is known to have high rates of white-opaque switching. Unlike the swr1 mutant, the efg1 mutant was not able to switch to opaque at high temperatures. At 37°C, opaque stability of the swr1 mutant was reduced but still greater than that of the WT, with 30% of opaque cells remaining opaque, while all WT opaque cells switched to white. On the other hand, efg1 opaque cells were less stable than swr1 cells and switched to white cells readily at 37°C (Fig. 1B). To test if SWR1 can restore function to the swr1 mutant, we assessed the white-opaque phenotype of the swr1/Tet-SWR1 mutant, a conditional mutant in which SWR1 expression is shut down in the presence of doxycycline (58). When grown on medium containing doxycycline, the strain displayed high levels of white-to-opaque switching and opaque stability, similar to the swr1 deletion mutant. In the absence of doxycycline, the strain did not readily switch to opaque and also had low opaque stability at 37°C (see Fig. S1 in the supplemental material). This demonstrates that the expression of SWR1 can restore the wild-type phenotype to an swr1 mutant. From these results, we find that white-opaque switching and opaque stability are enhanced in the swr1 mutant even at high temperature.
FIG 1.
Deletion of SWR1 leads to elevated white-opaque switching and opaque stability. (A) Single colonies of white WT (JYC1) and swr1 (HLY4233) cells after 7 days of growth at room temperature (25°C) on YPD. The swr1 colony displays multiple opaque sectors typical of the strain. (B) Spontaneous white-to-opaque and opaque-to-white switching rates of the WT, swr1, and efg1 (HLY3600) strains at RT (25°C) and at 37°C after 7 days of growth on SCD medium. The P value is <0.01 for swr1 opaque cells at 37°C compared to the wild type, and the P value is <0.005 for all others comparisons to the wild type, based on two-tailed Student's t test. * indicates that 0% switching occurred. (C) Percentage of spontaneous switching to opaque from white cells of the WT and swr1 strains after 7 days of growth at RT on Lee's medium with 1.25% dextrose (Dex) or 1.25% GlcNAc as the carbon source, in air or 5% CO2. (D) Fluorescence image of colonies of the WT carrying pWOR1-GFP (HLY3555) and the swr1 mutant carrying pWOR1-GFP (HLY4234) after 4 days of growth on SCD medium at RT. Opaque colonies and sectors, expressing more GFP, appear darker. (E) FACS profiles of GFP fluorescence in a population of 10,000 white or opaque cells of the WT carrying pWOR1-GFP (HLY3555) and the swr1 mutant carrying pWOR1-GFP (HLY4234). Cells were from a log-phase liquid culture at RT. (F) Levels of WOR1 mRNA transcript as assayed by RT-qPCR in white and opaque cells of the WT (JYC1) or the swr1 (HLY4233) deletion mutant. RNA was collected from log-phase cell cultures in YPD at room temperature. Transcript levels were normalized to ACT1 levels. All experiments were performed in triplicate.
We proceeded to investigate whether the deletion of SWR1 promotes switching downstream of known pathways. It has been documented that low concentrations of CO2 as well as utilization of N-acetylglucosamine (GlcNAc) as the main carbon source promote white-opaque switching, and these two pathways show a synergistic effect with each other (59, 60). To assay whether Swr1 acts downstream of either of these pathways, we cultured white cells of the WT and the swr1 mutant in air or 5% CO2 on Lee's medium with dextrose or GlcNAc as the carbon source and tracked switching to opaque over the course of 14 days. In the WT, as previously documented, both CO2 and GlcNAc promoted white-opaque switching and showed synergy with each other. In the swr1 mutant, we observed a higher level of spontaneous switching than in the WT, even under control conditions of growth in air on dextrose. Notably, switching of the swr1 mutant was elevated in the presence of CO2 regardless of the carbon source, showing a synergy similar to that of GlcNAc and CO2 (Fig. 1C). Unlike the swr1 mutant, the efg1 mutant readily switched to opaque without GlcNAc or CO2 with a timing similar to that of the wild type in the presence of both GlcNAc and CO2 (data not shown). Thus, SWR1 deletion appears to drive white-opaque switching in a pathway synergistic with the Flo8-mediated CO2-sensing pathway and may act downstream of the GlcNAc signaling pathway. Although wild-type opaque cells switch to white at 37°C under most conditions, it has been noted that GlcNAc as a carbon source increases switching in a manner synergistic with higher temperatures as well (60). This is consistent with the observation of white-to-opaque switching at high temperature in the swr1 mutant.
We then explored the mechanisms behind the white-opaque phenotype of the swr1 mutant through regulation of WOR1, the master regulator of switching. First, we investigated whether the swr1 mutant had a higher level of expression from the WOR1 promoter by using WT and swr1 strains transformed with the pWOR1-GFP plasmid, which expresses GFP from the native WOR1 promoter. After 4 days of growth on solid SCD medium, starting from white cells, the swr1 mutant showed numerous instances of darker colonies and sectors than those of the WT, indicative of the expression of pWOR1-GFP in cells that had become opaque (Fig. 1D). This further supports elevated white-to-opaque switching in swr1. To determine WOR1 expression levels in white and opaque cells, fluorescence-activated cell sorting (FACS) was performed on WT and swr1 cells carrying pWOR1-GFP. As seen in fluorescence histograms, transcription from the WOR1 promoter in white cells was not greater for the swr1 mutant than for the WT. In opaque cells, the level of GFP in the swr1 mutant was slightly elevated compared to that in the WT (Fig. 1E). Because the basal levels of WOR1 expression in white cells are extremely low, it could be difficult to detect a slight change in Wor1 levels by using FACS assays. Given the high white-to-opaque switching frequency in the swr1 mutant, it may at least be easier to reach the threshold to initiate the positive self-feedback loop of Wor1 in the mutant than in the WT. Indeed, RT-qPCR analysis of WOR1 mRNA levels in WT and swr1 white and opaque cells detected a slightly elevated WOR1 expression level in white swr1 cells compared to wild-type cells (Fig. 1F). This difference was not detected by FACS analysis. The WOR1 level in opaque swr1 cells was also slightly higher than that in opaque WT cells, consistent with the FACS data. While WOR1 expression was not greatly elevated in the swr1 mutant, a moderate increase may make it easier to reach the threshold to initiate the positive self-feedback loop of Wor1 and promote white-opaque switching or opaque stability through positive feedback.
Nucleosome positioning on the WOR1 promoter differs between white and opaque cells and is altered in the swr1 mutant.
We next sought to investigate whether deletion of SWR1 altered the white-opaque phenotype by changing the regulation of WOR1 through chromatin remodeling at the WOR1 promoter. As WOR1 has a long 8-kb promoter as well as a long 5′ untranslated region (UTR), nucleosomal dynamics upstream of WOR1 may be crucial to the regulation of its transcription. Since H2A.Z-containing nucleosomes have been shown to have altered stability and dynamics (36–38, 41), we proceeded with a nucleosome map of the WOR1 promoter in WT and swr1 cells, both of which carry HA-tagged HTA3 (H2A.Z), to facilitate further studies. To obtain nucleosomal DNA, Staphylococcus aureus micrococcal nuclease digestion was performed in order to cleave non-nucleosome-bound DNA while leaving nucleosome-bound DNA intact. qPCR was then performed on the nucleosomal DNA, amplifying staggered 100-bp regions along the WOR1 promoter and providing a map of nucleosomal enrichment in this region. We found that even in the WT, white and opaque cells showed a difference in nucleosome occupancy and positioning along the WOR1 promoter. While regular nucleosome peaks were observed at similar positions in both cell types, notably, nucleosome peaks were more defined in white cells. In particular, this was true of the +1/−1 nucleosomes adjacent to the transcriptional start site −1,997 bp upstream of the WOR1 ATG start codon. In white cells, there is a nucleosome-free region (NFR) between the +1 and −1 nucleosomes as well as one ∼2,550 to 2,350 bp upstream of the WOR1 ATG codon, absent in opaque cells. In the swr1 mutant, we saw a marked change in nucleosome positions on the WOR1 promoter compared to those in the WT. Nucleosome positioning was altered from that of the wild type, particularly in white cells, and the NFRs were abolished, with nucleosome occupancy being highly elevated in the region spanning bp −2550 to −2350 instead. The highly elevated levels of nucleosome occupancy at NFRs of the WOR1 promoter in white swr1 cells likely reflect slower nucleosome turnover, which is consistent with the known function of H2A.Z. In contrast, nucleosome occupancy at the NFRs of the WOR1 promoter is much lower in opaque swr1 cells than in wild-type white or opaque cells (Fig. 2). Overall, nucleosome mapping suggests that Swr1 is required for proper nucleosome positioning and stability at the WOR1 promoter and for the differential regulation of promoter nucleosomes in white and opaque states. Our data therefore support the importance of H2A.Z deposition in epigenetic regulation of transcription in cell fate determination.
FIG 2.
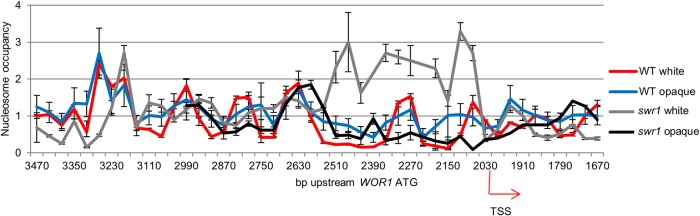
Nucleosome map of the WOR1 promoter shows white-opaque differences in the wild type and altered nucleosome positioning and occupancy in the swr1 mutant. Nucleosomal DNA was isolated from the WT strain carrying HTA3-HA (HLY4238) or the swr1 mutant carrying HTA3-HA (HLY4236) after removal of non-nucleosome-bound DNA with micrococcal nuclease digestion. Mapping was performed by qPCR using primers amplifying 100-bp regions along the WOR1 promoter. The error was calculated from the standard deviation of CT values from triplicate experiments. TSS, transcriptional start site.
H2A.Z is enriched at the WOR1 promoter in white cells.
H2A.Z is known to be enriched at developmentally regulated regions. To further investigate nucleosomal H2A.Z content, we mapped H2A.Z enrichment along the WOR1 promoter in white and opaque WT cells. In addition, swr1 cells were used as a control. H2A.Z was HA tagged in all strains used. H2A.Z enrichment was assayed by immunoprecipitation of H2A.Z-containing nucleosomes using anti-HA- on micrococcal nuclease-treated nucleosomes (Fig. 2), followed by qPCR using the same WOR1 primers as those used for nucleosome mapping to provide coverage along the WOR1 promoter. As expected, H2A.Z levels on promoter chromatin were very low in the swr1 mutant. In the wild type, notably, the H2A.Z level was higher in white cells than in opaque cells. Particularly of interest is that H2A.Z was enriched in the −2 and −1 nucleosomes at two NFRs on the WOR1 promoter (Fig. 3). The location of H2A.Z-enriched nucleosomes at NFRs is consistent with the role of H2A.Z in promoting nucleosome dynamics, as shown in yeast and high eukaryotes (61). The elevated H2A.Z deposition by the SWR1 complex at the WOR1 promoter in white cells indicates a role for H2A.Z in stabilizing the repressive chromatin state in white cells, potentially by maintaining NFRs and permitting the binding of repressive factors.
FIG 3.
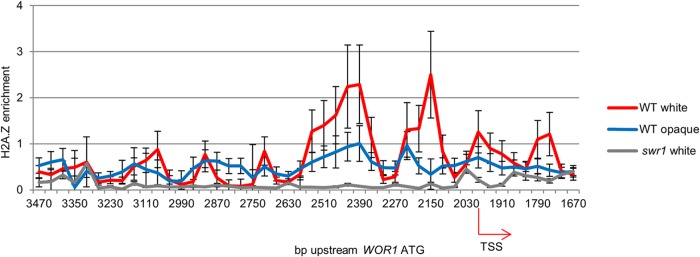
Differential enrichment of H2A.Z at the WOR1 promoter in white and opaque cells. Chromatin immunoprecipitation of HA-tagged H2A.Z was performed on nucleosomal DNA isolated from white and opaque cells of the WT strain carrying HTA3-HA and from white cells of the swr1 strain carrying HTA3-HA. qPCR was performed by using primers used to map the WOR1 promoter. Results were normalized to the total nucleosome level. TSS, transcriptional start site.
A deletion mutant of yng2, a subunit of the NuA4 complex, produces a white-opaque switching phenotype and synergy with CO2 similar to those of the swr1 mutant.
In exploring the regulation of H2A.Z deposition, we considered the recruitment of the SWR1 complex by NuA4 acetylation of H2 and H4 and the substantial overlap in subunits between the SWR1 and NuA4 complexes. Yng2 is a subunit of the NuA4 HAT module, the deletion of which, as we have shown, severely impairs H4 acetylation (51). Thus, we investigated whether deletion of YNG2 would produce a white-opaque phenotype similar to that of the swr1 mutant. We found that at room temperature, white cells of the yng2 mutant switched to opaque at a very high frequency compared to that of the wild type, while opaque cells of the yng2 mutant were highly stable. A complemented yng2+YNG2 strain showed a phenotype similar to that of the wild type (Fig. 4A). In addition, opaque yng2 cells were stable at 30°C but not at 37°C (not shown). Because of the relation between NuA4 acetylation of histones and SWR1 deposition of H2A.Z in the same nucleosome, we speculated that the yng2 and swr1 mutants produced their high-switching phenotype through the same pathways and would respond similarly to CO2 and GlcNAc. To test this, white cells of the yng2 mutant were grown at room temperature in air or 5% CO2, with dextrose or GlcNAc as a carbon source, and switching was assessed. We found that yng2 cells began switching to opaque sooner when incubated in CO2, preceding switching in air by several days. This behavior is similar to that of the swr1 mutant. Notably, the yng2 mutant switched more slowly when the carbon source was GlcNAc than when grown on dextrose, showing a more complete lack of synergy with GlcNAc than the swr1 mutant did (Fig. 4B). Therefore, like SWR1, Yng2/NuA4 also functions downstream of the GlcNAc signaling pathway, separate from the CO2-regulated pathway. This is consistent with the known function of NuA4 in histone acetylation, which in turn allows SWR1 recruitment and H2A.Z deposition.
FIG 4.
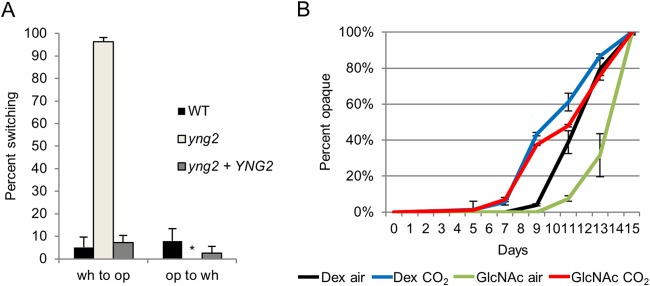
White-opaque switching of the yng2 mutant is elevated and shows synergy with CO2. (A) Spontaneous white-to-opaque and opaque-to-white switching rates of the yng2 strain (HLY3883) and the yng2+YNG2 complemented strain (HLY3886) compared to the WT after 14 days at room temperature on SCD medium. The P value is <0.005 for the yng2 and yng2+YNG2 strains compared to the WT. * indicates that 0% switching occurred. (B) Percentage of spontaneous switching of white yng2 cells to opaque after 7 days of growth on Lee's medium with 1.25% dextrose or 1.25% GlcNAc as the carbon source, in air or 5% CO2, at RT.
The swr1 mutant is highly sensitive to nicotinamide.
In addition to the epigenetic regulation of white-opaque switching, we also sought to investigate the effects of swr1 deletion on additional cellular functions. H2A.Z is enriched in nucleosomes surrounding transcriptional start sites and is involved in gene expression (62). Proper deposition of H2A.Z has been linked to numerous global effects, such as euchromatin-heterochromatin boundary establishment, centromeric functions, and DNA repair (33, 63–65). Recent work in S. cerevisiae has demonstrated that heightened acetylation of H3K56 is associated with reduced deposition and increased eviction of H2A.Z from chromatin by SWR1, with significant overlap in gene expression profiles between cells with constitutively acetylated H3K56 and htz1 (H2A.Z) deletion (26). It is known that nicotinamide (NAM) inhibits C. albicans growth by an elevation of H3K56ac levels through inhibition of the HDAC Hst3 (21, 24). To further explore the functional relationship between H3K56ac and H2A.Z deposition, we examined the sensitivities of the WT, SWR1/swr1, swr1, HST3/hst3, and rtt109 strains to NAM by a spot assay for growth on solid medium in the presence of nicotinamide. Cells were grown for 3 days at 30°C. The HST3/hst3 strain was highly sensitive to NAM and barely grew with 4 mM NAM, while the rtt109 strain was more resistant to NAM at 4 mM than the WT, consistent with previously reported results (24). Interestingly, the swr1 strain was even more sensitive to NAM than the HST3/hst3 strain, barely growing even on 2 mM NAM (Fig. 5). Unlike the HST3/hst3 strain, NAM sensitivity was not observed for the SWR1/swr1 strain. The lack of haploid insufficiency indicates that NAM probably does not directly act on Swr1. In contrast to NAM sensitivity, the swr1 and HST3/hst3 strains did not show any increased sensitivity to the DNA-damaging agent methyl methanesulfonate (MMS), while the rtt109 strain was sensitive (Fig. 5). In addition, the swr1 mutant showed no increased growth sensitivity to hydroxyurea or heat and osmotic stress (not shown). We have previously shown that the rtt109 mutant has an increased rate of generating aneuploidy (25). However, we did not observe any increase in aneuploidy formation in the swr1 mutant after growth on l-sorbose medium (not shown). Therefore, rtt109 (or blocking of H3K56 acetylation) is associated with aneuploidy and sensitivity to DNA damage, while H3K56ac and swr1 are associated with sensitivity to NAM. From our genetic studies, we suggest that the NAM sensitivity of swr1 and H3K56ac is associated with their roles in gene expression.
FIG 5.
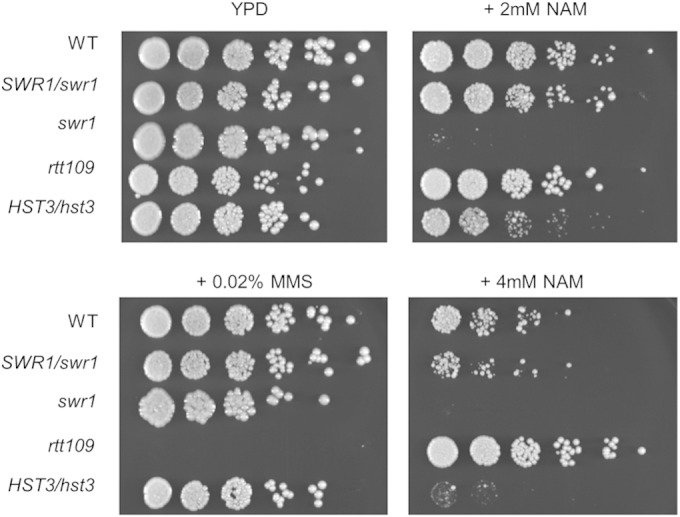
The swr1 mutant displays high sensitivity to nicotinamide but not to methyl methanesulfonate. Show are data from spot assays of the WT, SWR1/swr1 (HLY4235), swr1 (HLY4233), HST3/hst3 (HLY3993), and rtt109 (HLY3997) strains on YPD, YPD plus 2% MMS, or YPD plus 2 mM or 4 mM NAM. Fivefold serial dilutions from cultures with an OD600 of 0.1 were made on agar medium plates and incubated for 3 days at 30°C prior to imaging.
DISCUSSION
The role of histone variants and modifications on nucleosomal stability is a promising angle for the investigation of epigenetic regulation. In particular, the histone variant H2A.Z as well as the acetylation of H3K56 have been linked to changes in nucleosomal dynamics. Previous studies in our laboratory have demonstrated the effect of H3K56 acetylation on epigenetic regulation of white-opaque switching in C. albicans (24, 25). In this study, we show that epigenetic switching is also regulated by chromatin deposition of H2A.Z by SWR1. Both chromatin modifications affect the switching frequency and stability of cell fates but do not dramatically affect transcriptional levels of key cell fate regulators. A small increase in WOR1 expression was observed in the swr1 mutant, which may facilitate reaching the threshold for positive self-feedback regulation of Wor1. Additionally, we find synergy between the swr1 deletion and H3K56ac in C. albicans.
H2A.Z enrichment at promoter regions is often associated with both active and inactive transcription (32–34). Many studies have found H2A.Z nucleosomes enriched on the promoters of repressed genes, and H2A.Z deposition is important for proper induction of gene expression. Our study finds an example of these properties in demonstrating that H2A.Z is enriched at the WOR1 promoter in white cells and that white-to-opaque switching is enhanced in an swr1 mutant. Thus, H2A.Z is repressive to WOR1 expression or the opaque state. How does H2A.Z exert both positive and negative effects on transcription? One attractive model is that H2A.Z facilitates the binding of either activating or repressive complexes by keeping regions of promoters accessible (61). We find that nucleosome occupancy at the WOR1 promoter in white cells is elevated when SWR1 is deleted. Of particular note, a high H2A.Z level was found at nucleosome-free regions on the WOR1 promoter in WT cells, and in the swr1 mutant, these regions were no longer NFRs and instead had very high nucleosome occupancy. The existence of these NFRs in the white but not opaque state in the WT suggests a role for them in the repression of WOR1 transcription. Chromatin immunoprecipitation with microarray technology (ChIP-chip) studies of C. albicans have demonstrated that Efg1, a repressor of WOR1 and the opaque state, binds at the WOR1 promoter in white cells at only one major peak in the region between −3 kb and −2 kb, while it binds at multiple regions in opaque cells (66). This finding, in conjunction with our results, suggests that H2A.Z deposition in white cells maintains NFRs that facilitate the binding of repressive factors such as Efg1, favoring a repressive state of WOR1 transcription. Deletion of SWR1 may abolish these regions, preventing repressor binding and leading to a more permissive chromatin state for WOR1 transcription and an increased frequency of white-to-opaque switching. Future work is needed to examine this model of reduced accessibility of the WOR1 promoter to negative regulators of white-opaque switching in the swr1 mutant.
In exploring the functional relation of Swr1 with known white-opaque signaling pathways, we also find that the high-switching phenotype of the swr1 mutant showed synergy with CO2 in promoting white-opaque switching. This synergy with CO2 is similar to that of GlcNAc with CO2. Furthermore, the swr1 mutant shows no additional synergy with GlcNAc. Based on these data, we suggest that the SWR1 complex functions in the GlcNAc signaling pathway. A similar high-switching phenotype and synergy with CO2 are seen in the deletion mutant of yng2, a subunit of the NuA4 complex that shares subunits with the SWR1 complex and recruits it through histone H2 and H4 acetylation. Thus, it is likely that reduced histone acetylation in the yng2 mutant in turn reduces SWR1 deposition of H2A.Z, producing a phenotype similar to that of the swr1 mutant. Interestingly, swr1 opaque cells are more stable at high temperature than are those of the yng2 mutant, showing that there is some difference between the regulatory effects of the two. It has also been noted that the opaque-state-promoting effects of GlcNAc are synergistic with high temperature (56). The chromatin-level changes during temperature-induced opaque-to-white switching are a subject for further exploration. Our genetic data indicate that the GlcNAc signaling in white-opaque switching is likely to regulate WOR1 promoter chromatin in white cells by downregulating histone acetylation and H2A.Z deposition. Notably, the switching phenotype of the swr1 mutant differs from that of numerous other histone modifier mutants previously described to affect white-opaque switching. The swr1 mutant produces a higher switching frequency than many such mutants, such as those of the H3K4 methyltransferase set1; however, it does not produce complete switching as the efg1 set1 double mutant does (67). We have also noted that set1 shows no synergy with GlcNAc or CO2 (data not shown). Thus, the effects of histone modifications on white-opaque switching are manyfold, and their interactions with other pathways that regulate switching leave much to be explored.
A recent study has shown that a high level of H3K56ac reduces nucleosome incorporation of H2A.Z (26). Not only is H2A.Z deposition greatly reduced on H3K56ac-containing nucleosomes, the presence of nucleosomal H3K56ac also alters the activity of SWR1, increasing eviction of H2A.Z from nucleosomes. While nucleosomes containing both H3K56ac and H2A.Z are not less stable than those with H2A.Z alone, this increased exchange by H3K56ac can promote nucleosome turnover in H2A.Z-rich regions (26). Whether H2A.Z deposition can in turn promote the turnover of H3K56ac nucleosomes is an attractive prospect. We find in this study that the swr1 deletion mutant is highly sensitive to NAM. As hyperacetylation of H3K56 in chromatin is toxic to C. albicans, the high sensitivity of the swr1 mutant to NAM could be due to reduced nucleosome turnover and, consequently, the accumulation of H3K56ac on chromatin. In correlation, H2A.Z is enriched at the WOR1 promoter in white cells, while H3K56 acetylation is enriched in opaque cells (24). Future research is needed to examine if the reduced nucleosome occupancy at the WOR1 promoter in opaque swr1 cells is linked to the H3K56ac level. Genome-wide nucleosome mapping should determine whether NAM-treated swr1 cells show reduced nucleosome occupation at known H2A.Z-enriched regions. The interaction between H2A.Z deposition and H3K56ac may also exist in multicellular organisms, with implications for epigenetic regulation in developmental processes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Suzanne Noble, Terry Roemer, and Karl Kuchler for plasmids and C. albicans strains. We thank members of the Liu laboratory for helpful discussion, Yang Lu and John S. Stevenson for initial work with yng2, and Shelley Lane for constructing the HTA3-HA plasmid.
Z.G. was supported by a National Library of Medicine Biomedical Informatics training grant (5T15LM007443). This work was supported by National Institutes of Health grants R01GM/AI55155 and R01AI099190 to H.L.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00002-15.
REFERENCES
- 1.Henikoff S. 2008. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet 9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 2.Zentner GE, Henikoff S. 2013. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol 20:259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 3.Masumoto H, Hawke D, Kobayashi R, Verreault A. 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 4.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. 2008. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. 2008. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams SK, Truong D, Tyler JK. 2008. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci U S A 105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu F, Zhang K, Grunstein M. 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 8.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. 2007. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell 27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 10.Driscoll R, Hudson A, Jackson SP. 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 12.Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. 2006. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol 16:1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 13.Maas NL, Miller KM, DeFazio LG, Toczyski DP. 2006. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol Cell 23:109–119. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Rikkerink EH, Magee BB, Magee PT. 1988. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J Bacteriol 170:895–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol 169:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kvaal C, Lachke SA, Srikantha T, Daniels K, McCoy J, Soll DR. 1999. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect Immun 67:6652–6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lachke SA, Lockhart SR, Daniels KJ, Soll DR. 2003. Skin facilitates Candida albicans mating. Infect Immun 71:4970–4976. doi: 10.1128/IAI.71.9.4970-4976.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohse MB, Johnson AD. 2008. Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS One 3:e1473. doi: 10.1371/journal.pone.0001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuch BB, Mitrovich QM, Homann OR, Hernday AD, Monighetti CK, De La Vega FM, Johnson AD. 2010. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet 6:e1001070. doi: 10.1371/journal.pgen.1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopes da Rosa J, Boyartchuk VL, Zhu LJ, Kaufman PD. 2010. Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proc Natl Acad Sci U S A 107:1594–1599. doi: 10.1073/pnas.0912427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wurtele H, Tsao S, Lepine G, Mullick A, Tremblay J, Drogaris P, Lee EH, Thibault P, Verreault A, Raymond M. 2010. Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy. Nat Med 16:774–780. doi: 10.1038/nm.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem 277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 23.Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. 2001. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry 40:15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- 24.Stevenson JS, Liu H. 2011. Regulation of white and opaque cell-type formation in Candida albicans by Rtt109 and Hst3. Mol Microbiol 81:1078–1091. doi: 10.1111/j.1365-2958.2011.07754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevenson JS, Liu H. 2013. Nucleosome assembly factors CAF-1 and HIR modulate epigenetic switching frequencies in an H3K56 acetylation-associated manner in Candida albicans. Eukaryot Cell 12:591–603. doi: 10.1128/EC.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe S, Radman-Livaja M, Rando OJ, Peterson CL. 2013. A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science 340:195–199. doi: 10.1126/science.1229758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HS, Vanoosthuyse V, Fillingham J, Roguev A, Watt S, Kislinger T, Treyer A, Carpenter LR, Bennett CS, Emili A, Greenblatt JF, Hardwick KG, Krogan NJ, Bahler J, Keogh MC. 2009. An acetylated form of histone H2A.Z regulates chromosome architecture in Schizosaccharomyces pombe. Nat Struct Mol Biol 16:1286–1293. doi: 10.1038/nsmb.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka A, Tanizawa H, Sriswasdi S, Iwasaki O, Chatterjee AG, Speicher DW, Levin HL, Noguchi E, Noma K. 2012. Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation. Mol Cell 48:532–546. doi: 10.1016/j.molcel.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson JD, Gorovsky MA. 2000. Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucleic Acids Res 28:3811–3816. doi: 10.1093/nar/28.19.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Li B, Gorovsky MA. 1996. Essential and nonessential histone H2A variants in Tetrahymena thermophila. Mol Cell Biol 16:4305–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Daal A, Elgin SC. 1992. A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol Biol Cell 3:593–602. doi: 10.1091/mbc.3.6.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meneghini MD, Wu M, Madhani HD. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112:725–736. doi: 10.1016/S0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 34.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol 3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 36.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 37.Park YJ, Dyer PN, Tremethick DJ, Luger K. 2004. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J Biol Chem 279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 38.Henikoff S. 2009. Labile H3.3+H2A.Z nucleosomes mark ‘nucleosome-free regions’. Nat Genet 41:865–866. doi: 10.1038/ng0809-865. [DOI] [PubMed] [Google Scholar]
- 39.Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. 2008. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol 2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 42.Morillo-Huesca M, Clemente-Ruiz M, Andujar E, Prado F. 2010. The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A.Z. PLoS One 5:e12143. doi: 10.1371/journal.pone.0012143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altaf M, Auger A, Monnet-Saksouk J, Brodeur J, Piquet S, Cramet M, Bouchard N, Lacoste N, Utley RT, Gaudreau L, Cote J. 2010. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J Biol Chem 285:15966–15977. doi: 10.1074/jbc.M110.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Richardson DO, Roberts DN, Utley R, Erdjument-Bromage H, Tempst P, Cote J, Cairns BR. 2004. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol Cell Biol 24:9424–9436. doi: 10.1128/MCB.24.21.9424-9436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Auger A, Galarneau L, Altaf M, Nourani A, Doyon Y, Utley RT, Cronier D, Allard S, Cote J. 2008. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol Cell Biol 28:2257–2270. doi: 10.1128/MCB.01755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ranjan A, Mizuguchi G, FitzGerald PC, Wei D, Wang F, Huang Y, Luk E, Woodcock CL, Wu C. 2013. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell 154:1232–1245. doi: 10.1016/j.cell.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. 2006. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci U S A 103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, Gerstein M, Yi S, Snyder M, Soll DR. 2006. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell 5:1674–1687. doi: 10.1128/EC.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zordan RE, Galgoczy DJ, Johnson AD. 2006. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci U S A 103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu Y, Su C, Mao X, Raniga PP, Liu H, Chen J. 2008. Efg1-mediated recruitment of NuA4 to promoters is required for hypha-specific Swi/Snf binding and activation in Candida albicans. Mol Biol Cell 19:4260–4272. doi: 10.1091/mbc.E08-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Y, Su C, Solis NV, Filler SG, Liu H. 2013. Synergistic regulation of hyphal elongation by hypoxia, CO(2), and nutrient conditions controls the virulence of Candida albicans. Cell Host Microbe 14:499–509. doi: 10.1016/j.chom.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Janbon G, Sherman F, Rustchenko E. 1998. Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc Natl Acad Sci U S A 95:5150–5155. doi: 10.1073/pnas.95.9.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manning M, Mitchell TG. 1980. Strain variation and morphogenesis of yeast- and mycelial-phase Candida albicans in low-sulfate, synthetic medium. J Bacteriol 142:714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kent NA, Mellor J. 1995. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res 23:3786–3787. doi: 10.1093/nar/23.18.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bai L, Charvin G, Siggia ED, Cross FR. 2010. Nucleosome-depleted regions in cell-cycle-regulated promoters ensure reliable gene expression in every cell cycle. Dev Cell 18:544–555. doi: 10.1016/j.devcel.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu Y, Su C, Wang A, Liu H. 2011. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol 9:e1001105. doi: 10.1371/journal.pbio.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, Martel N, Veronneau S, Lemieux S, Kauffman S, Becker J, Storms R, Boone C, Bussey H. 2003. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol 50:167–181. doi: 10.1046/j.1365-2958.2003.03697.x. [DOI] [PubMed] [Google Scholar]
- 59.Huang G, Srikantha T, Sahni N, Yi S, Soll DR. 2009. CO(2) regulates white-to-opaque switching in Candida albicans. Curr Biol 19:330–334. doi: 10.1016/j.cub.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR. 2010. N-Acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog 6:e1000806. doi: 10.1371/journal.ppat.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teves SS, Weber CM, Henikoff S. 2014. Transcribing through the nucleosome. Trends Biochem Sci 39:577–586. doi: 10.1016/j.tibs.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 62.Weber CM, Henikoff S. 2014. Histone variants: dynamic punctuation in transcription. Genes Dev 28:672–682. doi: 10.1101/gad.238873.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Greaves IK, Rangasamy D, Ridgway P, Tremethick DJ. 2007. H2A.Z contributes to the unique 3D structure of the centromere. Proc Natl Acad Sci U S A 104:525–530. doi: 10.1073/pnas.0607870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. 2012. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell 48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kalocsay M, Hiller NJ, Jentsch S. 2009. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell 33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 66.Hernday AD, Lohse MB, Fordyce PM, Nobile CJ, DeRisi JL, Johnson AD. 2013. Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol Microbiol 90:22–35. doi: 10.1111/mmi.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hnisz D, Schwarzmuller T, Kuchler K. 2009. Transcriptional loops meet chromatin: a dual-layer network controls white-opaque switching in Candida albicans. Mol Microbiol 74:1–15. doi: 10.1111/j.1365-2958.2009.06772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen J, Lane S, Liu H. 2002. A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol Microbiol 46:1335–1344. doi: 10.1046/j.1365-2958.2002.03249.x. [DOI] [PubMed] [Google Scholar]
- 69.Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949. doi: 10.1016/S0092-8674(00)80358-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.