Abstract
Aspergillus nidulans (Pal) ambient pH signaling takes place in cortical structures containing components of the ESCRT pathway, which are hijacked by the alkaline pH-activated, ubiquitin-modified version of the arrestin-like protein PalF and taken to the plasma membrane. There, ESCRTs scaffold the assembly of dedicated Pal proteins acting downstream. The molecular details of this pathway, which results in the two-step proteolytic processing of the transcription factor PacC, have received considerable attention due to the key role that it plays in fungal pathogenicity. While current evidence strongly indicates that the pH signaling role of ESCRT complexes is limited to plasma membrane-associated structures where PacC proteolysis would take place, the localization of the PalB protease, which almost certainly catalyzes the first and only pH-regulated proteolytic step, had not been investigated. In view of ESCRT participation, this formally leaves open the possibility that PalB activation requires endocytic internalization. As endocytosis is essential for hyphal growth, nonlethal endocytic mutations are predicted to cause an incomplete block. We used a SynA internalization assay to measure the extent to which any given mutation prevents endocytosis. We show that none of the tested mutations impairing endocytosis to different degrees, including slaB1, conditionally causing a complete block, have any effect on the activation of the pathway. We further show that PalB, like PalA and PalC, localizes to cortical structures in an alkaline pH-dependent manner. Therefore, signaling through the Pal pathway does not involve endocytosis.
INTRODUCTION
In Aspergillus nidulans, the alkaline ambient pH-sensing plasma membrane module involves three proteins, the 7-transmembrane domain (TMD) receptor PalH, the arrestin-like PalF, and the 3-TMD helper PalI (1–5). This sensing module transduces a signal generated by alkaline ambient pH to a downstream module containing the Snf7 interactors PalA and PalC and the signaling protease PalB (3, 6–11). The downstream module also involves components of endosomal sorting complexes required for transport I (ESCRT-I), ESCRT-II, and Snf7 and Vps20 of ESCRT-III (10, 12). PalB, the final Pal participant in the pathway, almost certainly catalyzes the C-terminal proteolysis of the 72-kDa form of the transcription factor PacC (PacC72) to yield PacC53 and, following a second proteasome-mediated limited proteolysis, PacC27 (4, 13–15).
The pH signaling involvement of ESCRT complexes, which have a key role in multivesicular body (MVB) sorting in endosomes, led to a model in which the pH-sensing module and the ESCRT-associated proteins would be spatially separated, with the former located at the plasma membrane and the latter located on the membrane of endosomes (16). The two complexes would be connected by endocytosis, such that the activated plasma membrane sensor would be sorted in endocytic vesicles that would traffic to endosomes to engage downstream components and ultimately activate PalB-mediated PacC72 proteolysis in this locale. This model was challenged when localization experiments demonstrated that rather than being recruited to endosomes, PalA and PalC were recruited to cortical plasma membrane-associated foci in an alkaline ambient pH- and ESCRT-dependent manner (8, 10). This and the observation that the key ESCRT-I component Vps23 was recruited to similar membrane-associated foci (10) strongly indicated that the PalH/PalF sensor module hijacks ESCRTs to the plasma membrane to scaffold pH signaling complexes. These data also were combined with evidence in both A. nidulans and Saccharomyces cerevisiae showing that the toggle that triggers the signaling pathway is the PalH-dependent ubiquitination of PalF/Rim8 (2, 4, 17). Thus, in the revised and most recent model (5), PalH sensing of alkaline pH leads to ubiquitination of its PalF arrestin partner (PalF-Ub). PalF-Ub next recruits Vps23 to cortical spots, and this results in the recruitment of ESCRT-II and the subsequent polymerization of ESCRT-III at cortical foci (10). This model also assumes that PalB, which is an ESCRT-III interactor (9), is recruited to these foci, implying that PacC72 cleavage takes place in them, which has not been demonstrated previously.
While it seems unquestionable that pH signaling complexes assemble on these foci, it cannot be ruled out that proteolysis requires endocytosis of the signaling complexes, a possibility circumstantially suggested by the case of mammalian arrestins, whose ubiquitination promotes the endocytic internalization of their cognate receptors (18). Ambient pH-driven PalH internalization cannot be tested directly, because PalH detection by green fluorescent protein (GFP) tagging requires overexpression, which artificially results in predominance of PalH in the endovacuolar system (3), rather than PalH localization in the plasma membrane.
Cooverexpression of PalI or PalF with overexpressed PalH-GFP prevents its localization to endovacuoles and results in plasma membrane predominance, strongly indicating that the sensing module must be assembled for its correct localization (3, 4). Thus, imaging whether PalH/PalF does or does not remain in the plasma membrane after exposure to alkaline pH would require overexpression of PalH, PalF, and PalI, which may in turn make any downstream component limiting, casting doubts on any conclusion obtained with this experimental design.
A direct test of endocytic involvement in pH signaling would be to determine whether PacC72 processing proceeds in the absence of endocytosis. However, endocytosis is essential for A. nidulans, because it plays a key role in hyphal growth (19–22); thus, the degree to which mutations preventing endocytosis permit hyphal growth is expected to correlate inversely with the extent to which they impair endocytosis, with a complete endocytic block being lethal (21–23). Here, we demonstrate that PalB also is recruited to cortical structures. In addition, we make use of available mutations impairing endocytosis to show that none results in any detectable impairment in PacC72 proteolytic processing. Together, these data strongly suggest that, in A. nidulans, endocytosis is not required for pH signaling.
MATERIALS AND METHODS
Strains.
Unless otherwise indicated, Aspergillus complete (MCA) and synthetic complete medium (SC) (24), containing 1% glucose and, unless otherwise indicated, 5 mM ammonium tartrate (i.e., 10 mM NH4+) as carbon and nitrogen sources, respectively, were used for growth tests and strain maintenance, except for the slaB1 mutant, which usually was cultured on SC or MCA containing 10 mM nitrate. Strains, which carried markers in standard use, are listed in Table 1.
TABLE 1.
Strains used in this work
| Strain code | Genotype |
|---|---|
| MAD991 | wA3; pyroA4; myoAS371E |
| MAD1269 | yA2; inoB2; argB2::[argB*-alcAp::palB-gfp]; palB38 |
| MAD1329 | yA2; inoB2; argB2; palB38 |
| MAD1350 | pacC900; palH72 |
| MAD1362 | inoB2; palB38; pacC900 |
| MAD1739 | pyrG89; pyroA4; nkuAΔ::bar |
| MAD2335 | pyrG89; pyroA4; riboB2; nkuAΔ::argB; arfB::Tn::pyr-4 |
| MAD2384 | pyroA4; slaB1; pyrG89; nkuAΔ::bar |
| MAD2467 | argB2::[argB*-alcAp::gfp-synA]; nkuAΔ::bar; pyroA4 |
| MAD2556 | yA2; pabaA1; pyroA4; gfp-synA::pyrGfum; slaB1; pyrG89?; nkuAΔ::bar? |
| MAD2560 | wA2; pabaA1; pyroA4::[pyroA*-gpdAmini::palH-gfp] |
| MAD2673 | gfp-synA::pyrGfum; pyrG89?; nkuAΔ::bar? |
| MAD2725 | slaB1; pyroA4::[gpdAmini-palH::gfp-pyroA*]; nkuAΔ::bar?; pyrG89? |
| MAD3492 | yA2; pabaA1; argB2::[argB*-alcAp::gfp-synAen−] |
| MAD3802 | pyroA4; palB-gfp::pyrGfum; pyrG89; nkuAΔ::bar |
| MaD3803 | pyroA4; palB-gfp::pyrGfum; pyrG89; nkuAΔ::bar |
| MAD3877 | pyroA4; pacC900 |
| MAD3964 | slaB1; pyrG89; pyroA4; pacC900 |
| MAD3968 | nkuAΔ::argB; argB2; pyrG89; pyroA4; riboB2; sagAΔ::riboBfum |
| MAD4209 | fimA::Tn431::pyr-4; pyrG89; pyroA4; nkuAΔ::argB; riboB2; veA1 |
| MAD4251 | pyroA4; fimA::Tn431::pyr-4; pyrG89?; nkuAΔ::argB?; gfp-synA::pyrGfum |
| MAD4269 | fimA::Tn431::pyr-4; pacC900; pyroA4; inoB2; pyrG89?; nkuAΔ::argB? |
| MAD4270 | fimA::Tn431::pyr-4; pacC900; pyroA4; pyrG89?; nkuAΔ::argB? |
| MAD4298 | wA2; fimA::Tn431::pyr-4; pyrG89?; nkuAΔ::argB?; pyroA4::[pyroA*-gpdAmini::palH-gfp] |
| MAD4384 | wA4; pyrG89?; inoB2; nkuAΔ::argB?; arfB::Tn::pyr-4; pacC900 |
| MAD4386 | pyrG89?; nkuAΔ::argB?; arfB::Tn::pyr-4; pyroA4::[pyroA*-gpdAmini::palH-gfp] |
| MAD4387 | pyrG89?; pyroA4; nkuAΔ::argB?; arfB::Tn::pyr-4; gfp-synA::pyrGfum |
| MAD4546 | nkuAΔ::argB?; pyrG89?; pyroA4::[pyroA*-gpdAmini::palH-gfp]; riboB2?; sagAΔ::riboBfum |
| MAD4549 | wA4; nkuAΔ::argB; argB2?; pyrG89?; pyroA4; riboB2?; sagAΔ::riboBfum; gfp-synA::pyrGfum |
| MAD4616 | wA4; myoAS371E; gfp-synA::pyrGfum |
| MAD4618 | wA4; myoAS371E; pyroA4::[pyroA*-gpdAmini::palH-gfp] |
| MAD4627 | wA4; myoAS371E; pacC900 |
Microscopy.
With the exception of PalB-GFP images, all imaging was made using a Leica DMI6000 inverted microscope driven by Metamorph essentially as described previously (25, 26). Alkaline pH-mediated recruitment of PalB-GFP to cortical puncta was made as described for PalC-GFP (8), using a Nikon Eclipse E90 microscope, a Hamamatsu ORCA ER camera, a 1.40-numeric-aperture 100× objective, and a Semrock Brightline-GFP filter combination. The setup was driven by Metamorph, which was used to manipulate all images. To determine the recruitment of PalB-GFP to cortical structures, cells were precultured for 20 h at 25°C in 25 mM NaH2PO4 with watch minimal medium, pH 5.2 (27), containing 1% (vol/vol) ethanol and 5 mM ammonium tartrate as carbon and nitrogen sources, respectively. Germlings were shifted to acidic (pH 5.2) or alkaline (pH 8.2, with 25 mM Na2HPO4) medium and incubated for a further 30 min before being photographed. Cortical structures were counted in at least 32 germlings for each pH condition, and the counts were normalized per micron of hyphal length. All images were converted to 8 bit and annotated with Corel Draw.
Constructs and genetic engineering.
GFP-SynA transgenes have been described (25, 28, 29). For moderately high expression of PalH-GFP, we used the constitutive gpdAmini promoter, contained within a pgpd003 construct that targets integration to the pyroA locus (30). Briefly, a chimera consisting of the PalH coding region with GFP attached in frame to its C terminus was obtained from pALCargB::PalH-GFP (p1475 in our collection) (12) and cloned as a HindIII-XmaI fragment in pgpd003, which was used for transformation into A. nidulans using pyridoxine prototrophy for selection. Single-copy transformants were identified by Southern blotting and used for all subsequent crosses. MAD1269, used to photograph PalB-GFP recruitment to cortical puncta, carried a single-copy integration (determined by Southern blotting) of p1398 into the argB locus. This plasmid consisted of a genomic copy of PalB with a C-terminal S65T GFP (sGFP) tag, expressed under the control of the alcAp promoter, and contains a frameshifted copy of argB targeting integration to the mutant argB2 allele of the recipient strain. This strain carried the null palB38 allele such that palB-GFP was the only source of PalB. Endogenously tagged palB-GFP was constructed by gene replacement using a palB-(GA)5-GFP::pyrGAf::palB (3′-untranslated region [UTR]) cassette assembled by PCR.
Cultures for pH shift experiments.
A. nidulans strains were cultured in 200 ml minimal medium (MM; inoculum, 3 × 107 conidiospores) containing 1% glucose, 5 mM ammonium tartrate, and appropriate supplements, adjusted to pH 4.3 with 50 mM sodium citrate. slaB1 was cultured in MM containing 50 mM sodium nitrate or 25 mM ammonium sulfate (permissive and restrictive conditions, respectively). After 15 h of incubation at 30°C under these acidic conditions, mycelia were collected by filtration using Miracloth (Calbiochem) and transferred to fresh medium, buffered to pH 8 with HEPES-NaOH. Samples were taken before and, at regular times, after the alkaline pH shift. Mycelia were pressed dry, quick-frozen in dry ice, and lyophilized to obtain 200 to 300 mg of dry weight per sample in the case of strains showing relatively normal growth. A variation of this procedure was used for arfBΔ, fimAΔ, and slaB1 (ammonium conditions) strains owing to the low biomass yield. In these cases, mycelia were collected by filtration using a 0.45-μm-pore-size filter. The yield of lyophilized mycelia was 20 to 30 mg.
Protein extraction and Western blots.
The protein extraction procedure was a modification of a reported protocol (22). Lyophilized mycelia were homogenized using an FP120 Fast Prep cell disruptor and a 0.5-mm ceramic bead with a 10-s pulse at a setting of 4.0. Aliquots (6 mg) of powdered biomass were transferred to a 2-ml Eppendorf tube. Proteins were solubilized after the addition of 1 ml of lysis solution (0.2 M NaOH and 0.2% [vol/vol] β-mercaptoethanol) per tube and vigorous vortexing. Proteins were recovered after precipitation with 7.5% (vol/vol) trichloroacetic acid (TCA) followed by centrifugation at 14,000 × g for 5 min at 4°C. The resulting pellets were resuspended with 1 M Tris base (0.1 ml), mixed with 2 ml of Laemmli buffer, and incubated at 100°C for 2 min. Proteins (5 to 10 μl of each sample) were resolved in 8% SDS-polyacrylamide gels and electrotransferred to nitrocellulose membranes. These were reacted either with mouse monoclonal anti-c-myc (1/5,000) (clone 9E10; Sigma-Aldrich), rat monoclonal anti-hemagglutinin (HA; 1/1,000) (3F10; Roche), or, for a loading control, mouse anti-actin monoclonal antibody (1/20,000) (clone 4; MP Biomedicals, LLC) or rabbit anti-hexokinase polyclonal antibody (1/20,000) (AB1807; Chemicon International). Peroxidase-conjugated goat anti-rat IgM+G (3010-05; Southern Biotech) at 1/4,000, goat anti-mouse IgG immunoglobulin (Jackson) at 1/5,000 (anti-Myc) or 1/8,000 (anti-actin), and donkey anti-rabbit IgG (NA934; GE Healthcare) at 1/10,000 were used as appropriate. Peroxidase activity was detected with Amersham Biosciences ECL.
RESULTS
SynA as a reporter of endocytosis.
To assess the extent to which any given mutation impairs endocytosis, we used a well-characterized cargo, the secretory v-SNARE SynA. SynA is an integral membrane protein of secretory carriers that are targeted to the apex by MyoE (31). Once SynA reaches the apical plasma membrane, it undergoes limited basipetal diffusion because it is very efficiently taken up by endocytosis mediated by a highly active ring of actin patches localized subapically. The combined action of targeted exocytosis and efficient endocytosis results in the typical steady-state localization of SynA to an apical crescent (21, 28) (Fig. 1A).
FIG 1.
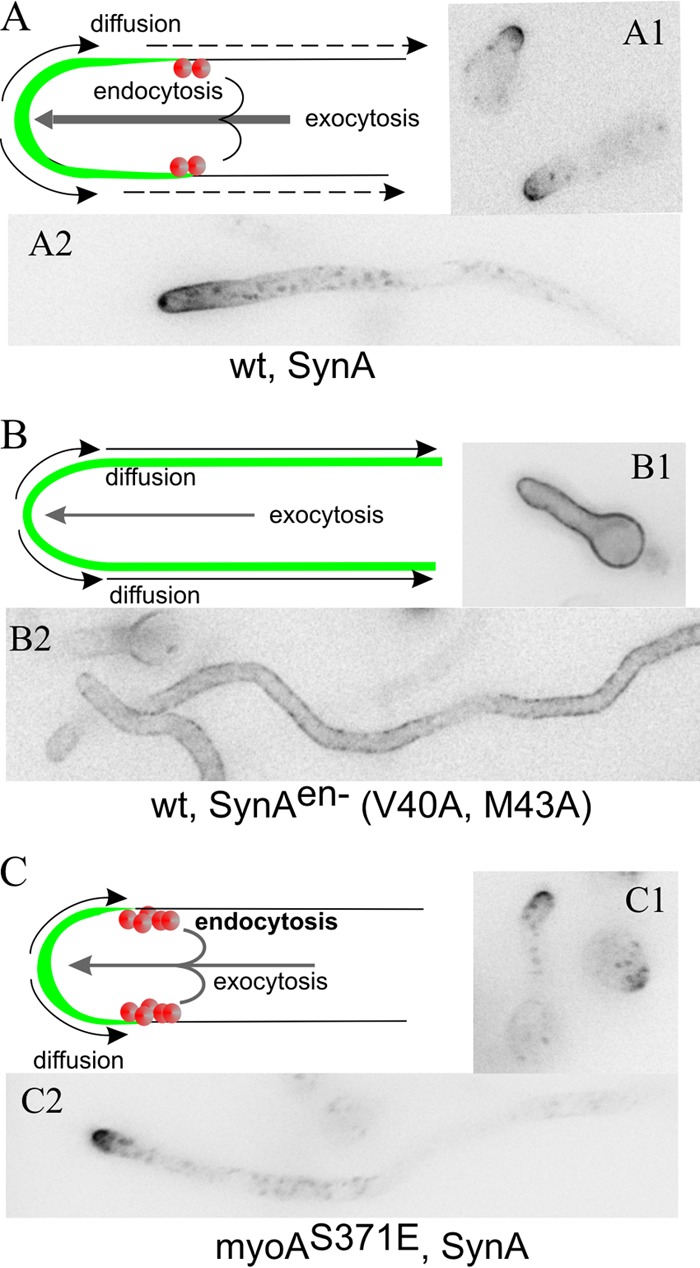
SynA as a reporter of endocytosis. (A) SynA is a single-pass v-SNARE delivered to the apex by exocytic carriers. Upon reaching the plasma membrane, it undergoes basipetal diffusion until captured by the subapical ring of endocytic patches (red circles), which acts as a diffusion barrier. (B) SynAen− is not taken up by endocytosis; therefore, it labels the plasma membrane uniformly (25). (C) In myoAS371E, endocytosis is accelerated (34).
Consistent with the fact that endocytosis prevents SynA diffusion across the plasma membrane (32), a double Val40Ala Met43Ala amino acid substitution in SynA, preventing its endocytic sorting but having no effect on diffusion, results in uniform SynA distribution along the plasma membrane (25, 33) (Fig. 1B). Therefore, this uniform SynA distribution may be used to diagnose impairment of endocytosis. To validate further the diagnostic value of SynA localization, we photographed SynA in a strain carrying the myoAS371E mutation resulting in accelerated endocytosis (34). SynA was strongly polarized, more so than in the wild type, consistent with it being taken up more rapidly after reaching the plasma membrane (Fig. 1C). Thus, we conclude that SynA is a faithful and sensitive reporter of endocytosis.
Classification of A. nidulans endocytic mutations according to effectiveness.
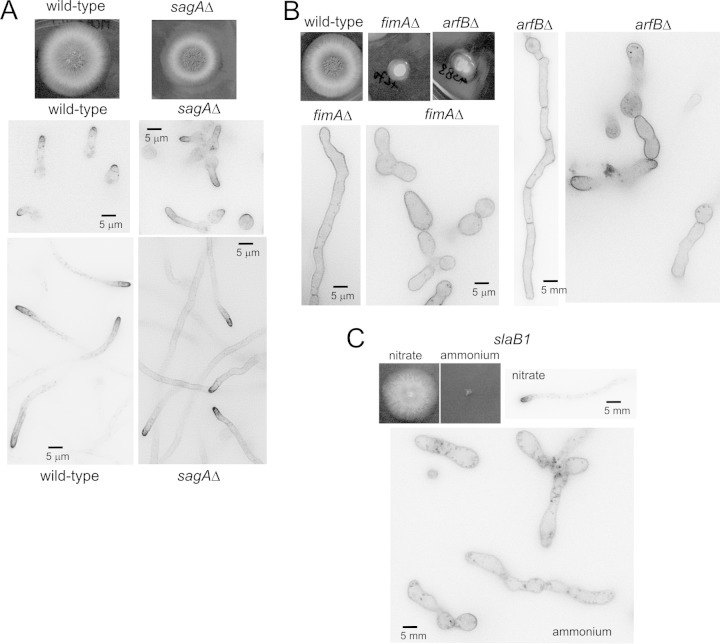
As endocytosis is essential, the extent to which a mutation impairs endocytosis was expected to correlate inversely with colony growth. The A. nidulans sagAend3 gene encodes the S. cerevisiae End3 orthologue (35). A sagAend3 null mutant displays a minor colony growth defect (Fig. 2A), indicating that sagAend3 is not crucial for endocytosis. Indeed, SynA still is polarized in this mutant background. However, both in germlings and in long sagAΔ hyphae, SynA clearly localizes to the plasma membrane of tip-distal regions more so than in the wild type, demonstrating that the mutation leads to an endocytic deficit, albeit minor (Fig. 2A). ArfB and FimA are the A. nidulans orthologues of yeast Arf3 (36) and Sac6 (37), respectively. As judged by the severe defects in morphogenesis (19, 20) and colony growth (Fig. 2B) displayed by the corresponding null mutants, these proteins were expected to be crucial for endocytosis. By microscopy, both arfBΔ and fimAΔ spores gave rise to heterogeneous cell populations (19, 20), with some germlings showing yeast-like morphology and some cells progressing to hyphae, in many cases morphologically aberrant. In both cases SynA was depolarized, indicating a major endocytic deficit (Fig. 2B).
FIG 2.
Growth and SynA localization phenotypes of endocytosis mutants. (A) Growth tests (at 37°C) and GFP-SynA localization in germlings and hyphae in the sagAΔ mutant and in the wild type. (B) Growth phenotypes and SynA localization in fimAΔ and arfBΔ strains. (C) Growth and SynA localization phenotypes of slaB1 cells (22) cultured on ammonium (20 mM ammonium sulfate) or nitrate (10 mM sodium nitrate), which are strongly repressing and inducing conditions, respectively, for the niiAp-driven expression of SlaB. All microcopy images are displayed at the same magnification to facilitate interpanel comparisons.
slaB is the A. nidulans orthologue of S. cerevisiae SLA2, a key regulator of endocytic patches (38, 39). We have previously demonstrated, using heterokaryon rescue, that slaB deletion is lethal (21), a conclusion that was further buttressed using slaB1, a conditional expression allele having the niiA (encoding nitrite reductase) promoter, which is repressible by ammonium and inducible by nitrate (22) (Fig. 2C). On ammonium, slaB1 spores germinate but do not maintain polarity beyond yeast-like cells and do not internalize SynA, forming characteristic pits in the plasma membrane (Fig. 2C) (22). Contrary to the heterogeneous populations observed with arfBΔ and fimAΔ strains, slaB1 yeast-like cells represented the sole cellular type found under ammonium conditions. Altogether, these data indicate that the sagAΔ strain results in weak impairment of endocytosis, arfBΔ and fimAΔ strains result in stronger impairment, and the slaB1 strain (under ammonium conditions) essentially abolishes endocytosis.
PalH has a tendency to be endocytosed if not stabilized in the plasma membrane.
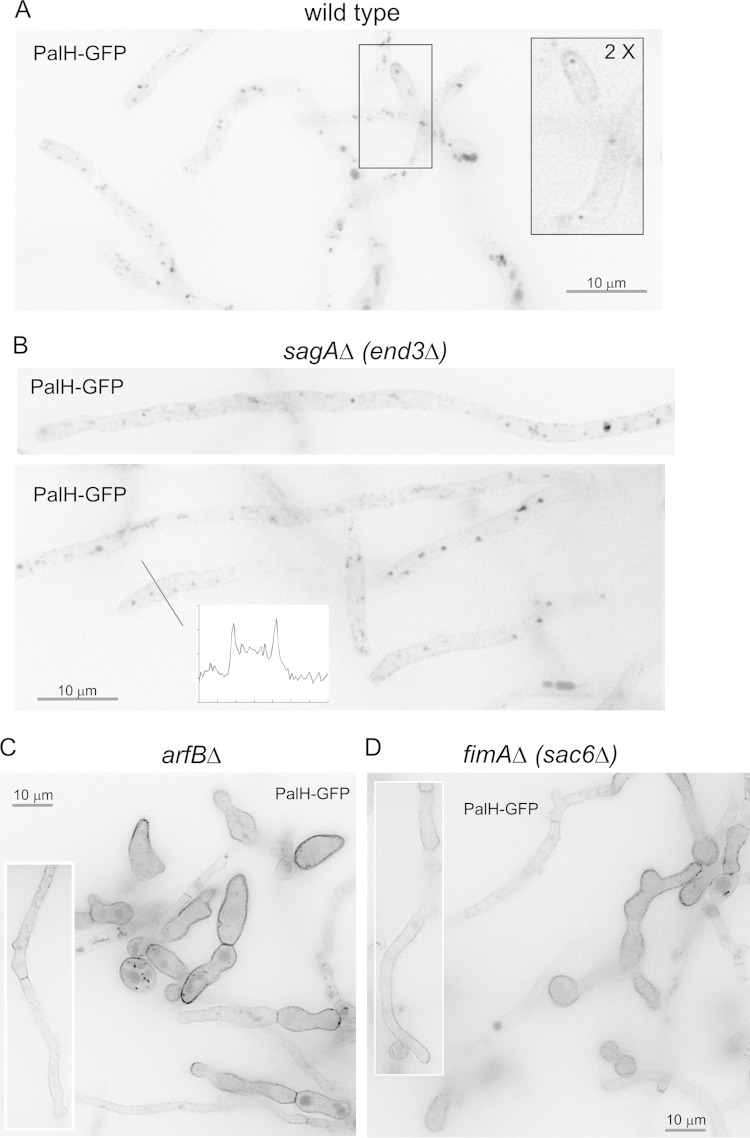
To confirm that mutations preventing SynA endocytosis have similar effects on the ambient pH receptor PalH, we crossed transgenes encoding PalH-GFP into the mutant backgrounds. Figure 3A shows that, when overexpressed alone, PalH-GFP predominates in internal structures corresponding to endosomes and vacuoles, staining the plasma membrane only very faintly. Endocytosis is crucially dependent on F-actin polymerization (40). The subcellular distribution of PalH-GFP was markedly shifted toward the plasma membrane after treating cells with a sublethal concentration of the anti-F-actin drug latrunculin B to impair endocytosis (data not shown). Therefore, when overexpressed alone, PalH-GFP reaches the plasma membrane but has a strong tendency to be endocytosed.
FIG 3.
Effect of endocytosis-impairing mutations on the localization of PalH-GFP. (A) Wild-type strain carrying a single-copy integration of a transgene driving the expression of PalH-GFP under the control of the moderately strong gpdAmini promoter (30). The boxed inset is shown at double magnification to better illustrate the faint GFP staining of the plasma membrane. Internal structures are endosomes and vacuoles. (B) sagAΔ strain expressing PalH-GFP; note the faint yet consistently greater staining of the plasma membrane compared to that of the wild-type control. (C) arfBΔ strain expressing PalH-GFP. Note the heterogeneous population consisting of cells with strong and moderate labeling of the plasma membrane, in both cases uniformly. The boxed hypha shows a cell of the moderate labeling and “more hyphal” class. (D) fimAΔ strain expressing PalH-GFP. Note the heterogeneous population, as in the case of the arfBΔ mutant. Fields in panels C and D are shown at half the magnification of those in panels A and B to illustrate population heterogeneity. As the gpdAmini::palH-gfp transgene was, in all cases, a single-copy integration of the construct targeted to the pyroA locus, levels of PalH-GFP expression are equivalent in all cases.
We next determined the effects of the different endocytosis mutations on PalH-GFP. These strictly correlated with those observed with SynA. The sagAΔ mutant weakly increased the localization of PalH to the plasma membrane (Fig. 3B). fimAΔ and arfBΔ strains showed a similar, markedly stronger effect. In fimAΔ and arfBΔ strains, in both hyphae and abnormal giant (yeast-like) cells, PalH largely predominated and exclusively localized, respectively, to the plasma membrane (Fig. 3C and D), which incidentally suggests that hyphae progress beyond the stage of giant cells because, although strongly impaired, they are incompletely blocked in endocytosis.
Endocytosis mutations do not prevent pH signaling.
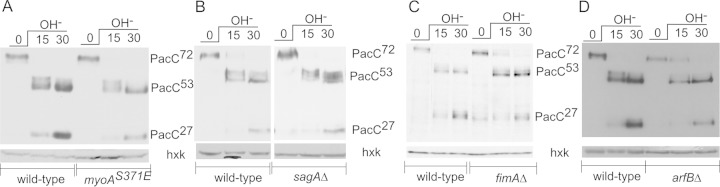
Having determined the degree to which the above-described mutations prevent PalH endocytosis, we examined, by Western blotting, the proteolytic processing of PacC in the different mutant backgrounds following a shift from acidic to alkaline conditions. We focused our analysis on the initial 30-min period after the pH shift, when PacC72 is converted to PacC53 by pH-dependent proteolysis, which is the readout of the activation of the Pal pathway (15) (Fig. 4 and 5). (At the 30-min time point, processing of PacC53 to PacC27 also is visible, but this second proteolysis step is pH independent [14, 41, 42]).
FIG 4.
Proteolytic processing activation of PacC in null endocytic mutants. Wild-type and mutant cells expressing wild-type Myc-PacC72 from the gene replacement allele pacC900 were precultured under acidic conditions and shifted to alkaline conditions for the indicated times. Cells were collected and analyzed by Western blotting with anti-Myc antibody. Anti-hexokinase (hxk) was used as a loading control. (A) Wild-type versus myoAS371A cells. (B) Wild-type versus sagAΔ cells. (C) Wild-type versus fimAΔ cells. (D) Wild-type versus arfBΔ cells.
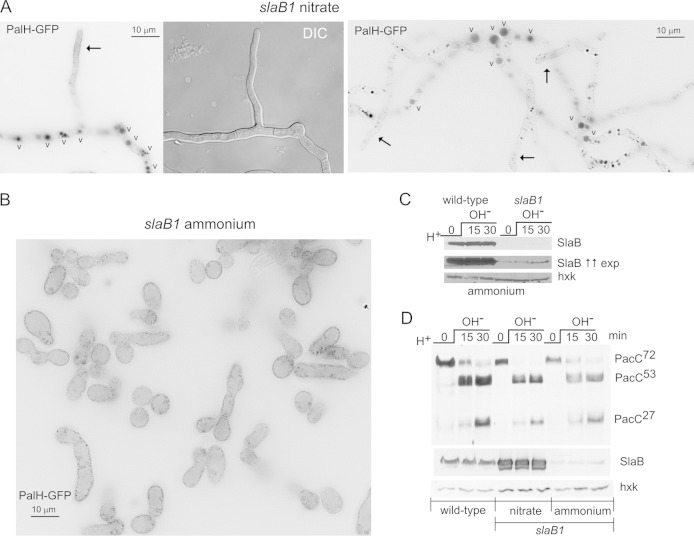
FIG 5.
On ammonium, slaB1 completely prevents PalH endocytosis but does not alter the proteolytic processing activation of PacC. (A) slaB1 hyphae cultured on nitrate, expressing PalH-GFP as described in the legend to Fig. 3. (Left) Hyphae are localized to the plasma membrane at the nascent branch and predominate in the vacuoles (indicated by the letter “v”). (Right) Hyphal tip cells showing weak plasma membrane staining (arrows) and basal conidiospores showing large and conspicuously fluorescent vacuoles. (B) A large field at the same magnification as those used for panel A showing the homogenous population of yeast-like cells resulting from cultivating the slaB1 mutant on ammonium. The PalH-GFP puncta noticeable at the plasma membrane are large pits (not shown). (C) Western blot analysis of SlaB in slaB1 cells cultured on ammonium and shifted from acidic to alkaline pH. The anti-SlaB Western blots in the top and middle panels represent two different exposures (exp) for the same experiment. The lower panel is an anti-hexokinase (hxk) loading control. The middle panel was deliberately overexposed to reveal traces of SlaB. (D) Normal proteolytic processing activation of PacC in slaB1 cells, cultured on ammonium or nitrate, compared to the wild type. As for panel C, the anti-SlaB blot was deliberately overexposed to illustrate the extent of downregulation. Nitrate conditions result in marked overexpression of SlaB.
The highly reproducible proteolytic processing activation pattern of PacC72 to PacC53 and PacC27 in wild-type cells shifted to alkaline conditions is displayed on the left in Fig. 4. PacC processing patterns of the different mutants are shown on the right. Accelerated endocytosis due to myoAS371E did not promote pH signaling (Fig. 4A). Neither the weak endocytosis-impairing sagAΔ strain nor the stronger endocytosis-impairing arfBΔ and fimAΔ strains substantially altered proteolytic PacC activation either (Fig. 4B, C, and D). These data, although strongly suggesting that endocytosis is not required for signaling, left open the possibility that residual endocytosis that, in all likelihood, takes place in the arfBΔ or fimAΔ strain might suffice to enable pH signaling.
To eliminate this possibility, we used slaB1. In slaB1 cells cultured on nitrate, the distribution of (overexpressed) PalH-GFP was essentially indistinguishable from that of the wild type, predominating very strongly in the endosomal system and localizing weakly to hyphal tips, a localization that was more conspicuous in young branches (Fig. 5A). As noted above, slaB1 cells on ammonium showed a uniform phenotype, with the population entirely consisting of yeast-like giant cells. These cells contained PalH localizing exclusively to the plasma membrane and associated endocytic pits (Fig. 5B). Strong SlaB downregulation in slaB1 cells cultured in ammonium conditions, both at acidic pH and after shifting cells to alkaline pH, was confirmed by Western blotting (Fig. 5C). In these, SlaB was almost undetectable compared to the level in the wild-type control (note that SlaB was detected only when Western blots were deliberately overexposed) (Fig. 5C, middle). Thus, we proceeded to analyze pH signaling in slaB1 cells cultured on ammonium (Fig. 5D). Despite strong SlaB downregulation, PacC processing in response to alkaline pH was essentially normal (Fig. 5D). Cells cultured in nitrate-containing medium, which results in strong SlaB upregulation, showed no effect either (Fig. 5D).
In summary, none of the four different alleles impairing endocytosis to different degrees, including slaB1, apparently resulting in a complete endocytic block, affect the PacC proteolytic cascade to any detectable extent, strongly indicating that endocytosis is not required at all for pH signaling.
PalB foci at the plasma membrane.
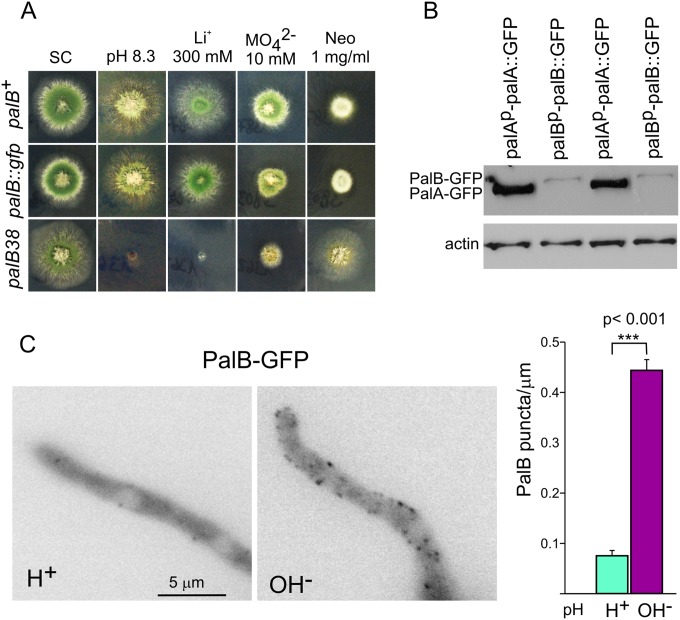
A prediction derived from the fact that recruitment of the signaling protease PalB to ESCRTs is required for pH signaling (9), made in the context of ESCRT recruitment to plasma membrane-associated puncta, is that at alkaline pH PalB also should localize to cortical structures if endocytosis was not required. Thus, we tagged palB by gene replacement. We confirmed that palB::gfp is functional by showing that strains carrying this allele behave as the wild type in diagnostic tests for pH regulation (Fig. 6A). However, the signal of endogenously tagged PalB-GFP was insufficient for microscopic analyses, and indeed, Western blotting demonstrated that levels of PalB-GFP were markedly lower than those of endogenously tagged PalA-GFP, which were used previously to visualize cortical pH signaling structures (10) (Fig. 6B). (This is consistent with the catalytic role attributed to PalB.) To circumvent this, we expressed PalB under the control of the alcohol dehydrogenase gene promoter (alcAp) and compared ethanol-grown cells shifted to acidic and alkaline conditions by epifluorescence microscopy. These experiments showed that PalB-GFP localized to cortical structures in an alkaline pH-dependent manner (Fig. 6C).
FIG 6.
PalB is recruited to cortical puncta in an alkaline-dependent manner. (A) Diagnostic tests of pH regulation for palB-GFP. The null palB38 mutation prevents growth at pH 8.3 or on 0.3 M LiCl-containing media, and it leads to hypersensitivity to 10 mM sodium molybdate and to resistance to 1 mg/ml of neomycin (Neo). A palB-GFP gene replacement strain grows like the wild type at pH 8.3 and on LiCl and sodium molybdate plates, and it is as sensitive as the wild type to neomycin, indicating that tagging does not impair function. (B) Western blot analysis of cells expressing endogenously tagged PalA-GFP or PalB-GFP. Actin is a loading control. (C) Cells expressing PalB-GFP under the control of alcAp were cultured on ethanol medium at acidic pH and shifted to acidic (H+; pH 5.2) or alkaline (OH−; pH 8.2) pH for 30 min before being photographed. The diagram on the right is a quantitation of the number of cortical structures per micron counted in 32 hyphal tip cells cultured under acidic or alkaline conditions. Error bars are standard errors. The two sets of measurements are significantly different (P < 0.001) as determined with the Mann-Whitney U test.
DISCUSSION
Previously, we proposed a comprehensive model for pH regulation in which signal transduction takes place in plasma membrane-associated foci scaffolded by ESCRT-III polymer (10). Strong supporting evidence came from the findings that, in cells exposed to alkaline environment, ESCRT subunits and late-acting pH signaling proteins PalC and PalA are recurrently recruited to plasma membrane-associated foci (10), with downregulation of ESCRT-disassembling Vps4 ATPase stabilizing PalC at these locales, indicating that recurrence at the plasma membrane reflects ESCRT-III polymerization/depolymerization cycles (10). Evidence strongly indicates that ubiquitination of the pH signaling arrestin-like protein PalF/Rim8 crucially regulates signal transduction: the alkaline pH-dependent ubiquitin attachment to the arrestin docks ESCRT-I Vps23 to receptor-arrestin complexes to organize subsequent ESCRT polymerizing steps at foci (2, 4, 10, 17). A recent report using S. cerevisiae essentially concurred with the above-described conclusions (43).
Two important predictions derive from the above-described model. One is the cortical localization of pH signaling proteolysis. Indisputable evidence showed that PalB (9) and Rim13 (44) associate with ESCRT-III, and that this association is required for signaling. Thus, PalB, like ESCRTs, should be recruited to cortical foci in an alkaline pH-dependent manner to mediate PacC proteolysis. Indeed, the Mitchell laboratory (44) demonstrated that S. cerevisiae Rim13 is recruited to foci when cells encounter alkaline pH, whereas others showed that these foci actually are cortical (43). Here, we report that PalB, like its yeast orthologue, also is recruited to cortical foci, further supporting the contention that the plasma membrane model applies to distant ascomycete clades, in spite of the existence of minor differences in mechanistic detail between A. nidulans and S. cerevisiae (for example, PalB contains an MIT domain that contributes to its recruitment to ESCRT-III [9] that is absent from Rim13).
If so, endocytosis should be dispensable for signaling. In yeast, endocytosis involves >30 proteins that are recruited in an orderly fashion to endocytic sites to mediate sequentially clathrin coat assembly, cargo selection, membrane bending, actin polymerization, and membrane scission (38, 45). It has been reported that ablation of the early endocytic factor End3 (46) or downregulation of Las17 (47) does not prevent pH signaling in yeast (43), which was taken as evidence against endocytosis involvement. However, the extent to which these genetic manipulations impair endocytosis was determined by FM4-64 uptake (thus, bulk membrane internalization), and this only qualitatively, leaving open the possibility that they impair, yet do not block, all endocytosis. End3 acts with clathrin, Sla1 (48), Sla2 (49), and Pan1 (50) in the coat assembly step (38, 45). Indeed, in a quantitative analysis of 400 viable null endocytosis-impairing mutations (51), end3Δ ranked 315th (the greatest impairment being first), whereas sla1Δ ranked at position 12 and pan1Δ and sla2Δ could not be ranked, as they are lethal in the tested genetic backgrounds. Similarly, downregulation of Las17, a regulator of actin polymerization, may be insufficient to block all endocytosis, leaving open the possibility that basal levels suffice to sustain pH signaling, should this involve endocytosis.
To address this issue, we exploited the limited set of endocytosis mutations available in A. nidulans (23), taking advantage of two phenotypic features to estimate their degree of endocytic impairment. One was the extent to which any given mutation affects colony growth, a very sensitive criterion, because endocytosis is essential for hyphal growth (21, 22). Second, similar to the study by Burston et al. (51) in yeast, we took advantage of the fact that the synaptobrevin SynA, like its yeast Snc1 orthologue (32, 33), is efficiently taken up by endocytosis and rapidly recycled to the apex exocytically (25, 28, 29). This enables the use of uniform plasma membrane SynA distribution as a diagnostic of endocytic impairment. According to both criteria, the sagAΔ mutant impaired endocytosis very weakly; arfBΔ and fimAΔ mutants, which are phenotypically similar to each other, prevented endocytosis substantially, even though they gave rise to heterogeneous populations; finally, we used the regulatable slaB1 allele and ammonium growth, which completely prevented hyphal growth and SynA or PalH endocytosis. The absence of any detectable effect in sensitive PacC proteolytic processing assays by any mutation included in this panel represents strong evidence that pH signaling does not require endocytosis.
The alkaline pH signal received by PalH is decoded by the PalF arrestin-like protein (Rim8 in S. cerevisiae). Fungal arrestins generally act as ubiquitin ligase adaptors that mediate ubiquitination of their cognate plasma membrane protein partners, promoting their endocytic- and MVB pathway-mediated downregulation (52–56). In contrast, rather than promoting PalH internalization, PalF stabilizes PalH at the plasma membrane (4) and PalF/Rim8 ubiquitination plays a crucial, positive-acting role in pH signaling by recruiting ESCRTs to cortical sites (4, 10, 17). If this ESCRT recruitment occurred on endosomes, ubiquitinated PalF/Rim8 inevitably would be sorted for MVB degradation. Thus, a highly speculative interpretation is that PalF-mediated recruitment of ESCRTs to the plasma membrane evolved as a mechanism by which pH signaling proteins exploit the scaffolding potential of ESCRT-III polymers without facing the threat of destruction.
ACKNOWLEDGMENTS
We are very grateful to Greg May for myoAS371E, to Brian Shaw for arfBΔ and fimAΔ mutants, to Giorgos Diallinas for the sagAΔ mutant, and to José M. Rodríguez for constructing, in the initial stages of this project, the p1398 plasmid and the transformed strain expressing PalB-GFP. Elena Reoyo provided excellent technical assistance.
This work was supported by Ministerio de Economía y Competitividad (Spain) grants BIO2009-07281 and BIO2012-30965 and by the Comunidad de Madrid (grant S2010/BMD2414). Daniel Lucena-Agell was the holder of an FPI fellowship.
REFERENCES
- 1.Negrete-Urtasun S, Reiter W, Díez E, Denison SH, Tilburn J, Espeso EA, Peñalva MA, Arst HN Jr. 1999. Ambient pH signal transduction in Aspergillus: completion of gene characterization. Mol Microbiol 33:994–1003. doi: 10.1046/j.1365-2958.1999.01540.x. [DOI] [PubMed] [Google Scholar]
- 2.Herranz S, Rodríguez JM, Bussink HJ, Sánchez-Ferrero JC, Arst HN Jr, Peñalva MA, Vincent O. 2005. Arrestin-related proteins mediate pH signaling in fungi. Proc Natl Acad Sci U S A 102:12141–12146. doi: 10.1073/pnas.0504776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calcagno-Pizarelli AM, Negrete-Urtasun S, Denison SH, Rudnicka JD, Bussink HJ, Munera-Huertas T, Stanton L, Hervás-Aguilar A, Espeso EA, Tilburn J, Arst HN Jr, Peñalva MA. 2007. Establishment of the ambient pH signaling complex in Aspergillus nidulans: PalI assists plasma membrane localization of PalH. Eukaryot Cell 6:2365–2375. doi: 10.1128/EC.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hervás-Aguilar A, Galindo A, Peñalva MA. 2010. Receptor-independent ambient pH signaling by ubiquitin attachment to fungal arrestin-like PalF. J Biol Chem 285:18095–18102. doi: 10.1074/jbc.M110.114371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peñalva MA, Lucena-Agell D, Arst HN Jr. 2014. Liaison alcaline: Pals entice non-endosomal ESCRTs to the plasma membrane for pH signaling. Curr Opin Microbiol 22:49–59. doi: 10.1016/j.mib.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Negrete-Urtasun S, Denison SH, Arst HN Jr. 1997. Characterization of the pH signal transduction pathway gene palA of Aspergillus nidulans and identification of possible homologs. J Bacteriol 179:1832–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vincent O, Rainbow L, Tilburn J, Arst HN Jr, Peñalva MA. 2003. YPXL/I is a protein interaction motif recognised by Aspergillus PalA and its human homologue AIP1/Alix. Mol Cell Biol 23:1647–1655. doi: 10.1128/MCB.23.5.1647-1655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galindo A, Hervás-Aguilar A, Rodríguez-Galán O, Vincent O, Arst HN Jr, Tilburn J, Peñalva MA. 2007. PalC, one of two Bro1 domain proteins in the fungal pH signaling pathway, localizes to cortical structures and binds Vps32. Traffic 8:1346–1364. doi: 10.1111/j.1600-0854.2007.00620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodríguez-Galán O, Galindo A, Hervás-Aguilar A, Arst HN Jr, Peñalva MA. 2009. Physiological involvement in pH signalling of Vps24-mediated recruitment of Aspergillus PalB cysteine protease to ESCRT-III. J Biol Chem 284:4404–4412. doi: 10.1074/jbc.M808645200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galindo A, Calcagno-Pizarelli AM, Arst HN Jr, Peñalva MA. 2012. An ordered pathway for the assembly of ESCRT-containing fungal ambient pH signalling complexes at the plasma membrane. J Cell Sci 125:1784–1795. doi: 10.1242/jcs.098897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denison SH, Orejas M, Arst HN Jr. 1995. Signaling of ambient pH in Aspergillus involves a cysteine protease. J Biol Chem 270:28519–28522. doi: 10.1074/jbc.270.48.28519. [DOI] [PubMed] [Google Scholar]
- 12.Calcagno-Pizarelli AM, Hervás-Aguilar A, Galindo A, Abenza JF, Peñalva MA, Arst HN Jr. 2011. Rescue of Aspergillus nidulans severely debilitating null mutations in ESCRT-0, I, II and III genes by inactivation of a salt-tolerance pathway allows examination of ESCRT gene roles in pH signalling. J Cell Sci 124:4064–4076. doi: 10.1242/jcs.088344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espeso EA, Roncal T, Díez E, Rainbow L, Bignell E, Álvaro J, Suárez T, Denison SH, Tilburn J, Arst HN Jr, Peñalva MA. 2000. On how a transcription factor can avoid its proteolytic activation in the absence of signal transduction. EMBO J 19:719–728. doi: 10.1093/emboj/19.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Díez E, Álvaro J, Espeso EA, Rainbow L, Suárez T, Tilburn J, Arst HN Jr, Peñalva MA. 2002. Activation of the Aspergillus PacC zinc-finger transcription factor requires two proteolytic steps. EMBO J 21:1350–1359. doi: 10.1093/emboj/21.6.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peñas MM, Hervás-Aguilar A, Múnera-Huertas T, Reoyo E, Peñalva MA, Arst HN Jr, Tilburn J. 2007. Further characterization of the signaling proteolysis step in the Aspergillus nidulans pH signal transduction pathway. Eukaryot Cell 6:960–970. doi: 10.1128/EC.00047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peñalva MA, Tilburn J, Bignell E, Arst HN Jr. 2008. Ambient pH gene regulation in fungi: making connections. Trends Microbiol 16:291–300. doi: 10.1016/j.tim.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Herrador A, Herranz S, Lara D, Vincent O. 2010. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol Cell Biol 30:897–907. doi: 10.1128/MCB.00132-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefkowitz RJ, Shenoy SK. 2005. Transduction of receptor signals by beta-arrestins. Science 308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 19.Upadhyay S, Shaw BD. 2008. The role of actin, fimbrin and endocytosis in growth of hyphae in Aspergillus nidulans. Mol Microbiol 68:690–705. doi: 10.1111/j.1365-2958.2008.06178.x. [DOI] [PubMed] [Google Scholar]
- 20.Lee SC, Schmidtke SN, Dangott LJ, Shaw BD. 2008. Aspergillus nidulans ArfB plays a role in endocytosis and polarized growth. Eukaryot Cell 7:1278–1288. doi: 10.1128/EC.00039-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Araujo-Bazán L, Peñalva MA, Espeso EA. 2008. Preferential localization of the endocytic internalization machinery to hyphal tips underlies polarization of the actin cytoskeleton in Aspergillus nidulans. Mol Microbiol 67:891–905. doi: 10.1111/j.1365-2958.2007.06102.x. [DOI] [PubMed] [Google Scholar]
- 22.Hervás-Aguilar A, Peñalva MA. 2010. Endocytic machinery protein SlaB is dispensable for polarity establishment but necessary for polarity maintenance in hyphal tip cells of Aspergillus nidulans. Eukaryot Cell 9:1504–1518. doi: 10.1128/EC.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peñalva MA. 2010. Endocytosis in filamentous fungi: Cinderella gets her reward. Curr Opin Microbiol 13:684–689. doi: 10.1016/j.mib.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Cove DJ. 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta 113:51–56. doi: 10.1016/S0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 25.Pantazopoulou A, Peñalva MA. 2011. Characterization of Aspergillus nidulans RabCRab6. Traffic 12:386–406. doi: 10.1111/j.1600-0854.2011.01164.x. [DOI] [PubMed] [Google Scholar]
- 26.Hernández-González M, Peñalva MA, Pantazopoulou A. 2015. Conditional inactivation of Aspergillus nidulans sarA uncovers the morphogenetic potential of regulating endoplasmic reticulum (ER) exit. Mol Microbiol 95:491–508. doi: 10.1111/mmi.12880. [DOI] [PubMed] [Google Scholar]
- 27.Peñalva MA. 2005. Tracing the endocytic pathway of Aspergillus nidulans with FM4-64. Fungal Genet Biol 42:963–975. doi: 10.1016/j.fgb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Taheri-Talesh N, Horio T, Araujo-Bazán LDX, Espeso EA, Peñalva MA, Osmani SA, Oakley BR. 2008. The tip growth apparatus of Aspergillus nidulans. Mol Biol Cell 19:1439–1449. doi: 10.1091/mbc.E07-05-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abenza JF, Pantazopoulou A, Rodríguez JM, Galindo A, Peñalva MA. 2009. Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic 10:57–75. doi: 10.1111/j.1600-0854.2008.00848.x. [DOI] [PubMed] [Google Scholar]
- 30.Pantazopoulou A, Peñalva MA. 2009. Organization and dynamics of the Aspergillus nidulans Golgi during apical extension and mitosis. Mol Biol Cell 20:4335–4347. doi: 10.1091/mbc.E09-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taheri-Talesh N, Xiong Y, Oakley BR. 2012. The functions of myosin II and myosin V homologs in tip growth and septation in Aspergillus nidulans. PLoS One 7:e31218. doi: 10.1371/journal.pone.0031218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valdez-Taubas J, Pelham HR. 2003. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr Biol 13:1636–1640. doi: 10.1016/j.cub.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HR. 2000. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell 11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita RA, May GS. 1998. Constitutive activation of endocytosis by mutation of myoA, the myosin I gene of Aspergillus nidulans. J Biol Chem 273:14644–14648. doi: 10.1074/jbc.273.23.14644. [DOI] [PubMed] [Google Scholar]
- 35.Jones GW, Hooley P, Farrington SM, Shawcross SG, Iwanejko LA, Strike P. 1999. Cloning and characterisation of the sagA gene of Aspergillus nidulans: a gene which affects sensitivity to DNA-damaging agents. Mol Gen Genet 261:251–258. doi: 10.1007/s004380050964. [DOI] [PubMed] [Google Scholar]
- 36.Gillingham AK, Munro S. 2007. Identification of a guanine nucleotide exchange factor for Arf3, the yeast orthologue of mammalian Arf6. PLoS One 2:e842. doi: 10.1371/journal.pone.0000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams AE, Botstein D, Drubin DG. 1991. Requirement of yeast fimbrin for actin organization and morphogenesis in vivo. Nature 354:404–408. doi: 10.1038/354404a0. [DOI] [PubMed] [Google Scholar]
- 38.Kaksonen M, Toret CP, Drubin DG. 2005. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 39.Kaksonen M, Sun Y, Drubin DG. 2003. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115:475–487. doi: 10.1016/S0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 40.Kaksonen M, Toret CP, Drubin DG. 2006. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 41.Peñalva MA, Arst HN Jr. 2004. Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu Rev Microbiol 58:425–451. doi: 10.1146/annurev.micro.58.030603.123715. [DOI] [PubMed] [Google Scholar]
- 42.Hervás-Aguilar A, Rodríguez JM, Tilburn J, Arst HN Jr, Peñalva MA. 2007. Evidence for the direct involvement of the proteasome in the proteolytic processing of the Aspergillus nidulans zinc finger transcription factor PacC. J Biol Chem 282:34735–34747. doi: 10.1074/jbc.M706723200. [DOI] [PubMed] [Google Scholar]
- 43.Obara K, Kihara A. 2014. Signaling events of the Rim101 pathway occur at the plasma membrane in a ubiquitination-dependent manner. Mol Cell Biol 34:3525–3534. doi: 10.1128/MCB.00408-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian S, Woolford CA, Desai JV, Lanni F, Mitchell AP. 2012. cis- and trans-acting localization determinants of pH response regulator Rim13 in Saccharomyces cerevisiae. Eukaryot Cell 11:1201–1209. doi: 10.1128/EC.00158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Idrissi FZ, Blasco A, Espinal A, Geli MI. 2012. Ultrastructural dynamics of proteins involved in endocytic budding. Proc Natl Acad Sci U S A 109:E2587–E2594. doi: 10.1073/pnas.1202789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benedetti H, Raths S, Crausaz F, Riezman H. 1994. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell 5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Y, Martin AC, Drubin DG. 2006. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell 11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 48.Howard JP, Hutton JL, Olson JM, Payne GS. 2002. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J Cell Biol 157:315–326. doi: 10.1083/jcb.200110027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wesp A, Hicke L, Palecek J, Lombardi R, Aust T, Munn AL, Riezman H. 1997. End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol Biol Cell 8:2291–2306. doi: 10.1091/mbc.8.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wendland B, Emr SD. 1998. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol 141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burston HE, Maldonado-Baez L, Davey M, Montpetit B, Schluter C, Wendland B, Conibear E. 2009. Regulators of yeast endocytosis identified by systematic quantitative analysis. J Cell Biol 185:1097–1110. doi: 10.1083/jcb.200811116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 53.Nikko E, Sullivan JA, Pelham HR. 2008. Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep 9:1216–1221. doi: 10.1038/embor.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becuwe M, Vieira N, Lara D, Gomes-Rezende J, Soares-Cunha C, Casal M, Haguenauer-Tsapis R, Vincent O, Paiva S, Leon S. 2012. A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. J Cell Biol 196:247–259. doi: 10.1083/jcb.201109113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karachaliou M, Amillis S, Evangelinos M, Kokotos AC, Yalelis V, Diallinas G. 2013. The arrestin-like protein ArtA is essential for ubiquitination and endocytosis of the UapA transporter in response to both broad-range and specific signals. Mol Microbiol 88:301–317. doi: 10.1111/mmi.12184. [DOI] [PubMed] [Google Scholar]
- 56.Becuwe M, Leon S. 7 November 2014. Integrated control of transporter endocytosis and recycling by the arrestin-related protein Rod1 and the ubiquitin ligase Rsp5. eLife doi: 10.7554/eLife.03307. [DOI] [PMC free article] [PubMed] [Google Scholar]