Abstract
Kinase suppressor of Ras 1 (KSR1) has been implicated in tumorigenesis in multiple cancers, including skin, pancreatic and lung carcinomas. However, our recent study revealed a role of KSR1 as a tumour suppressor in breast cancer, the expression of which is potentially correlated with chemotherapy response. Here, we aimed to further elucidate the KSR1-regulated signalling in response to genotoxic agents in breast cancer. Stable isotope labelling by amino acids in cell culture (SILAC) coupled to high-resolution mass spectrometry (MS) was implemented to globally characterise cellular protein levels induced by KSR1 in the presence of doxorubicin or etoposide. The acquired proteomic signature was compared and GO-STRING analysis was subsequently performed to illustrate the activated functional signalling networks. Furthermore, the clinical associations of KSR1 with identified targets and their relevance in chemotherapy response were examined in breast cancer patients. We reveal a comprehensive repertoire of thousands of proteins identified in each dataset and compare the unique proteomic profiles as well as functional connections modulated by KSR1 after doxorubicin (Doxo-KSR1) or etoposide (Etop-KSR1) stimulus. From the up-regulated top hits, several proteins, including STAT1, ISG15 and TAP1 are also found to be positively associated with KSR1 expression in patient samples. Moreover, high KSR1 expression, as well as high abundance of these proteins, is correlated with better survival in breast cancer patients who underwent chemotherapy. In aggregate, our data exemplify a broad functional network conferred by KSR1 with genotoxic agents and highlight its implication in predicting chemotherapy response in breast cancer.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-015-3443-y) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Genotoxicity, KSR1, Proteomics, SILAC
Introduction
KSR1 was originally identified in D. melanogaster and C. elegans as a novel protein kinase, operating between Ras and Raf in the Ras signalling pathway [1–3]. Intriguingly, in the murine and human KSR1, instead of a lysine required for its kinase activity at subdomain II, an arginine residue is invariantly present. Therefore, mammalian KSR1 is extensively referred to as a pseudokinase [4, 5]. The role of KSR1 as a scaffolding protein was initially demonstrated by the finding that murine KSR1 (mKSR1) can cooperate with activated Ras to enhance MEK and MAPK activation [6]. Moreover, KSR1 is able to translocate from the cytoplasm to the plasma membrane where it forms a complex with Raf-1, MEK1 and 14-3-3 in the presence of activated Ras, supporting its scaffolding function in the MAPKs pathway [7, 8]. At the same time, the catalytic activity of KSR1 was confirmed by the fact that tumour necrosis factor alpha (TNFα) and ceramide can induce KSR1 autophosphorylation and increase its capacity to phosphorylate and activate Raf-1 [9]. A body of evidence hence supports a dual function for KSR1 as an active kinase and a scaffold protein [4].
Given its essential role in the Ras-Raf-MAPKs signalling pathway, most work has focused on studying the function of KSR1 in Ras-dependent tumours. Indeed, KSR1 has been shown to contribute to oncogenesis in various forms of Ras-activated cancer, such as skin [10], pancreatic [11] and lung carcinomas [12]. However, Ras mutations are rare in breast cancer [13] and KSR1′s biologic functions have not been comprehensively studied in this context. Recently, our group assessed the clinical significance of KSR1 in breast cancer patients and revealed a positive association of KSR1 with overall and disease-free survival [14]. Of note, KSR1 overexpression inhibits tumour growth both in vitro and in vivo. Moreover, KSR1 enhances BRCA1 stabilisation and diminishes its ubiquitination by increasing both BARD1 abundance and BARD1-BRCA1 interaction. Additionally, a SILAC-based phosphoproteomics was employed to study KSR1-modulated phospho-proteins and revealed an inhibitory role of KSR1 in regulating p53 transcriptional activity via DBC1 phosphorylation [15]. Interestingly, previous studies indicated a trend in association of KSR1 expression with response to chemotherapy and that KSR1 overexpression in MCF7 cells increased cisplatin sensitivity and apoptosis [14, 16]. Therefore, a profile of global protein expression changes and KSR1-regulated signalling in response to genotoxic agents (doxorubicin and etoposide) is needed to further elucidate the biological functions of KSR1 in breast cancer.
In the present study, we performed SILAC-coupled mass spectrometry (MS) quantitative proteomics to identify and quantify the protein alterations upon KSR1 overexpression in the presence of doxorubicin or etoposide in MCF7 cells. We present a detailed repertoire of thousands of proteins identified in each dataset and compare the distinctive proteomic signature as well as functional networks regulated by KSR1 in response to doxorubicin (Doxo-KSR1) or etoposide (Etop-KSR1). From the most significantly differentiated proteins, we identify numerous molecules, including STAT1, ISG15 and TAP1 that are positively associated with KSR1 expression in patient samples. Furthermore, high KSR1 expression, as well as a high level of the characterised proteins, is correlated with better clinical outcome in breast cancer patients who received chemotherapy. Our data illustrate a comprehensive functional network that KSR1 is involved in and highlight its significance in predicting chemotherapy response in patients.
Experimental procedures
Cell lines, reagents and plasmids
MCF7 cells were obtained from ATCC. The cells were authenticated (STR profiling) and tested for mycoplasma contamination every four weeks. MCF7 cells were maintained in SILAC medium as described below for at least seven passages and then seeded into 100 mm dishes. All cells were incubated at 37 °C in humidified 5 % CO2. FuGENE® HD transfection reagent was obtained from Promega. Doxorubicin and etoposide were purchased from Sigma Aldrich. Plasmids containing human wild type KSR1 (pCMV6-KSR1) and empty vector (pCMV6-vector) were obtained from OriGene.
SILAC cell culture
To generate SILAC conditions, normal DMEM medium deficient in arginine (R) and lysine (K) was complemented with stable isotope-encoded arginine and lysine as described previously [15]. Briefly, for “medium” labelling we used L-[13C6] arginine (R6) and L-[2H4] lysine (K4), and for “heavy” labelling we used L-[13C6, 15N4] arginine (R10) and L-[13C6, 15N2] lysine (K8). For the “light” condition, or unlabelled, L-[12C6,14N4] arginine (R0) and L-[12C6,14N2] lysine (K0) were used. The tissue culture media were supplemented with 10 % dialyzed fetal bovine serum with 10 kDa MWCO, 1 % (10 mg/ml) streptomycin/(10,000 units/ml) penicillin, 2 mM glutamine and 1 mM sodium pyruvate. The SILAC media were obtained from Dundee Cell Products Ltd (Dundee, UK). MCF7 cells were grown in these custom DMEM mediums along with 10 % dialysed FCS.
Protein digestion and peptide fractionation
Prior to protein digestion, equal amounts of protein (80 µg) from unlabelled and labelled samples were combined. Samples were reduced in 10 mM DTT and alkylated in 50 mM Iodoacetamide prior to boiling in loading buffer, and then separated by one-dimensional SDS-PAGE (4–12 % Bis–Tris Novex mini-gel, Invitrogen) and visualised by colloidal Coomassie staining (Novex, Invitrogen). The entire protein gel lanes were excised and cut into 10 slices each. Every gel slice was subjected to in-gel digestion with trypsin overnight at 37 °C. The resulting tryptic peptides were extracted by formic acid (1 %) and acetonitrile (CH3CN), lyophilized in a speedvac and resuspended in 1 % formic acid.
Mass Spectrometry methods
Trypsin-digested peptides were separated using an Ultimate 3000 RSLC (Thermo Scientific) nanoflow LC system. On average 0.5 µg was loaded with a constant flow of 5 µl/min onto an Acclaim PepMap100 nanoViper C18 trap column (100 µm inner-diameter, 2 cm; Thermo Scientific). After trap enrichment, peptides were eluted onto an Acclaim PepMap RSLC nanoViper, C18 column (75 µm, 15 cm; Thermo Scientific) with a linear gradient of 2–40 % solvent B (80 % acetonitrile with 0.08 % formic acid) over 65 min with a constant flow of 300 nl/min. The HPLC system was coupled to a linear ion trap Orbitrap hybrid mass spectrometer (LTQ-Orbitrap Velos, Thermo Scientific) via a nano electrospray ion source (Thermo Scientific). The spray voltage was set to 1.2 kV, and the temperature of the heated capillary was set to 250 °C. Full-scan MS survey spectra (m/z 335–1800) in profile mode were acquired in the Orbitrap with a resolution of 60,000 after accumulation of 1000,000 ions. The fifteen most intense peptide ions from the preview scan in the Orbitrap were fragmented by collision-induced dissociation (normalised collision energy, 35 %; activation Q, 0.250; and activation time, 10 ms) in the LTQ Orbitrap after the accumulation of 10,000 ions. Maximal filling times were 1000 ms for the full scans and 150 ms for the MS/MS scans. Precursor ion charge state screening was enabled, and all unassigned charge states as well as singly charged species were rejected. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum retention period of 180 s and a relative mass window of 15 ppm. The lock mass option was enabled for survey scans to improve mass accuracy [17]. Data were acquired using the Xcalibur software.
Proteome quantification
The raw mass spectrometric data files obtained for each experiment were collated into a single quantitated dataset using MaxQuant (1.3.0.5) [18] and the Andromeda search engine software (1.3.0.5) [19]. The sequence database used was from ipi.HUMAN v.3.68. Enzyme specificity was set to that of trypsin, allowing for cleavage N-terminal to proline residues and between aspartic acid and proline residues. Other parameters used were: (i) variable modifications, methionine oxidation, protein N-acetylation, gln → pyro-glu; (ii) fixed modifications, cysteine carbamidomethylation; (iii) database: target-decoy human MaxQuant; (iv) heavy labels: R6K4 and R10K8; (v) MS/MS tolerance: FTMS- 10 ppm, ITMS- 0.6 Da; (vi) minimum peptide length, 6; (vii) maximum missed cleavages, 2; (viii) maximum of labelled amino acids, 3; and (ix) false discovery rate, 1 %. Peptide ratios were calculated for each arginine- and/or lysine-containing peptide as the peak area of labelled arginine/lysine divided by the peak area of non-labelled arginine/lysine for each single-scan mass spectrum. Peptide ratios for all arginine- and lysine-containing peptides sequenced for each protein were averaged. Data are normalised using 1/median ratio value for each identified protein group per labelled sample.
KM Plotter analysis
KM Plotter online survival analysis, which collates a massive dataset of gene expression data and survival information of more than 3000 patients, was used [20]. The expression levels of KSR1, ISG15, STAT1, TAP1, MX1 and HLA-C were selected. The relevance on relapse free survival (RFS) in breast cancer patients who received systematic chemotherapy was assessed. A survival curve with the hazard ratio (HR) with 95 % confidence intervals and logrank P value was displayed. The generated P value does not include correction for multiple hypothesis testing.
Bioinformatics and statistical analyses
All the bioinformatic analysis was performed in R [21] and SWI-Prolog [22], using Real [23] for connecting the two systems. To characterise the top hits in different datasets, Significant B test was performed and P < 0.05 was considered significant [18]. To visualise the modulated proteomics, the heatmap of quantified values showing the overall pattern of regulation was displayed. GO analysis was performed at the level 2 of the three GO domains: biologic process (BP), cellular component (CC) and molecular function (MF) [24]. Furthermore, the hyper-geometric test from GOstats package was used to identify GO terms that are enriched in the de-regulated genes in each condition [25]. For every overrepresented GO term, a network connecting de-regulated genes in this GO term was drawn showing the networks amongst these genes in the STRING protein–protein interactions database [26]. Only edges with a confidence value greater than 500 were included (range: 0–999). Prolog was used for the overall control of our pipeline and the graph operations on the STRING database. The correlation analysis of gene expressions from TCGA dataset was performed using GraphPad Prism V5.0 (GraphPad Software, San Diego, CA).
Results
Establishing a quantitative proteomics approach to identify global protein changes induced by KSR1 in response to doxorubicin or etoposide
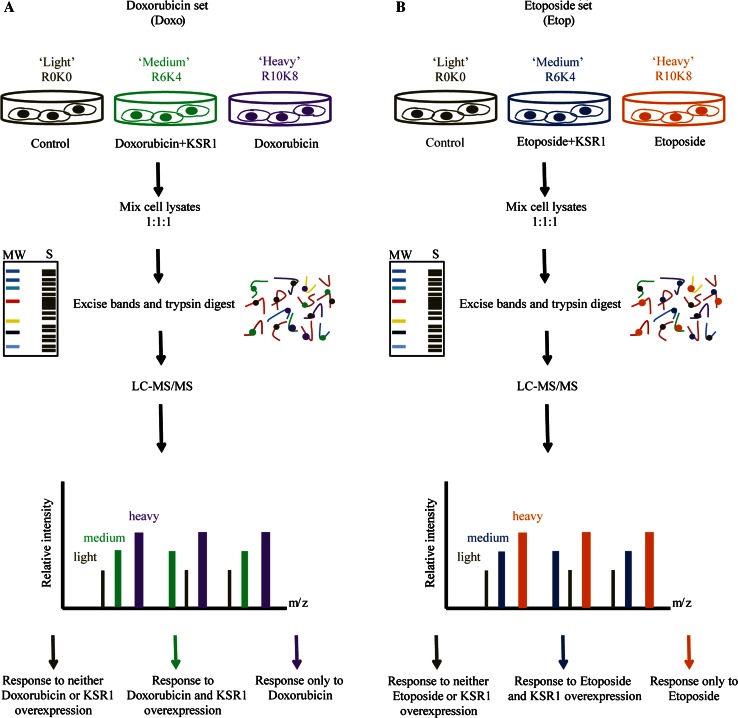
To characterise and compare the global proteomic alterations prompted by KSR1 in the presence of doxorubicin or etoposide, we conducted a triple-labelling SILAC strategy coupled to high-resolution quantitative MS as readily validated in our two recent studies [15, 27]. Following cell labelling, in the doxorubicin-KSR1 (Doxo-KSR1) set, R0K0 (Light (L)), R6K4 (Medium (M)) and R10K8 (Heavy (H)) labelled cells were treated with control (DMSO), overexpression of KSR1 (24 h) in the presence of doxorubicin (1 µm, 2 h) or doxorubicin only, respectively (Fig. 1a). In the etoposide-KSR1 (Etop-KSR1) set, R0K0, R6K4 and R10K8 labelled cells were treated with control (DMSO), overexpression of KSR1 (24 h) in the presence of etoposide (40 µm, 3 h) or etoposide alone, respectively (Fig. 1b).
Fig. 1.
Strategy to identify global protein changes induced by KSR1 in response to doxorubicin or etoposide. a Three SILAC labeled cell populations were subjected to DMSO (Control), treatment with KSR1 overexpression in the presence of doxorubicin, and treatment with doxorubicin alone. Subsequently, total protein lysates were prepared, and light, medium, and heavy cell populations were mixed 1:1:1, digested and fractionated to peptides. SILAC labelled peptides were then processed through LC–MS/MS. The peptides were then identified and quantified using quantitation software MaxQuant. b Similarly, cells were treated with DMSO (Control), treatment with KSR1 overexpression in the presence of etoposide, and treatment with etoposide alone. The acquired lysates were subject to same protocol as described above
For each analysis, from more than 20,000 non-redundant peptide sequences, a proteome comprising more than 2000 distinguishable and unambiguously identified proteins was assembled with a minimum of two peptides with a false discovery rate (FDR) of 1 % (minimal peptide length 7 aa). Detailed information for each identified protein, including protein IDs, number of peptides used for identification and their sequence and % coverage, normalised fold changes and log2 ratios are shown in Supplementary Table S1. The log2 ratios’ distribution shows the excellent quantitative precision of the experiment (Supplementary Fig. 1). In total, approximately 2000 proteins were identified in Doxo-KSR1 and Etop-KSR1 sets and their distribution according to fold changes was determined. Around 85 % of the proteome identified in Doxo-KSR1 and Etop-KSR1 was identical, highlighting a very high degree of overlap and consistency (Fig. 2a). We then used Significant B test to distinguish the most differentiated proteins in each dataset (P < 0.05 according to Significance B test) [18].
Fig. 2.
Global proteomic alterations affected by KSR1 in the presence of doxorubicin or etoposide. a Venn diagram showing a very high degree of overlap and consistency of the proteome identified in Doxo-KSR1 and Etop-KSR1 datasets. b Venn diagram showing the counts of proteins significantly modulated upon doxorubicin or etoposide stimulus (H/L), and proteins markedly differentiated by KSR1 in the presence of doxorubicin or etoposide (M/L), according to Significant B test (P < 0.05). c Venn diagram showing the numbers of proteins significantly up- or down-regulated in each dataset. d Heatmaps showing the commonly and exclusively regulated proteins upon doxorubicin or etoposide stimulus (upper panel), and proteins significantly affected by KSR1 in the presence of doxorubicin or etoposide (lower panel)
Global proteomic alterations induced by doxorubicin or etoposide in MCF7
Firstly, we sought to assess the global proteomic changes induced by doxorubicin or etoposide alone, two major genotoxic agents in cancer treatment. The H/L ratios representing Doxo/Control or Etop/Control from two acquired datasets were analysed and compared to delineate a comprehensive proteomic profile contributed by different genotoxic drugs. A total of 164 proteins were significantly modulated upon doxorubicin stimulus, whereas 162 molecules were markedly differentiated in response to etoposide (Fig. 2b). Specifically, 93 proteins showed significant up-regulation and 71 were down-regulated upon doxorubicin treatment. There was a pronounced increase in expression of 96 molecules in the presence of etoposide, while a decrease was evident in the levels of 66 proteins (Fig. 2c). Interestingly, there were 70 mutual proteins greatly affected by doxorubicin and etoposide, among which 45 were up-regulated and 25 were down-regulated (Fig. 2b, c).
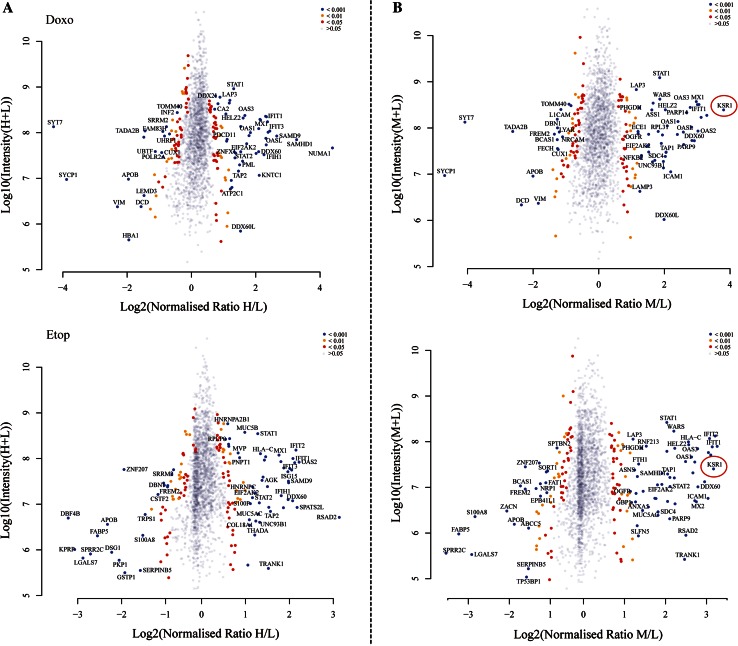
Heatmaps showing the commonly regulated proteins upon doxorubicin and etoposide stimulus were generated to highlight the similarity in their proteomic changes, whereas the ones belonging specifically to either doxorubicin or etoposide response were compared and presented (Fig. 2d). Furthermore, to distinguish the top hits upon doxorubicin or etoposide stimulus, the Log2 ‘normalized ratios’ (H/L) were plotted against Log10 ‘intensity’ (H/L) to demonstrate the significantly differentiated proteins, which were catalogued according to P values from Significant B test (Fig. 3a). A list of top 30 proteins that are significantly regulated in response to either doxorubicin or etoposide is presented in Supplementary Table S2.
Fig. 3.
Distribution of protein ratios from identified proteome in two datasets. Volcano plots showing the Log2 ‘normalized ratios’ (M/L) against Log10 ‘intensity’ (M/L) for each characterised protein in response to doxorubicin or etoposide stimulus (a), and affected by KSR1 in the presence of doxorubicin or etoposide (b). Proteins are displayed based on P values from Significant B test. Blue circles are proteins with a P value of <0.001; yellow are proteins with a P value between 0.001 and 0.01; red represent P values between 0.01 and 0.05; grey are proteins whose fold change is not significant (P > 0.05). KSR1 is highlighted showing a pronounced increase in its protein abundance, supporting a good reproducibility and accuracy of the technique (b)
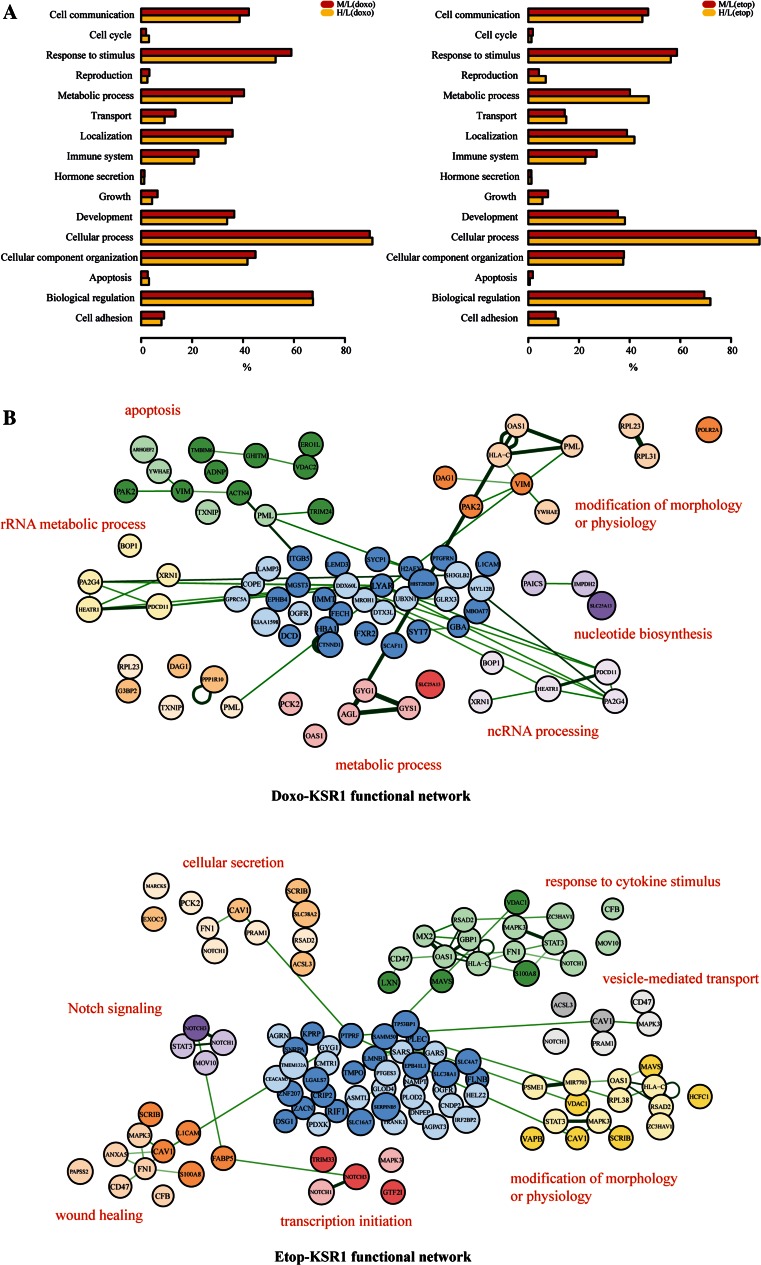
In the characterised proteome, a profile of differentially modulated proteins and associated pathways induced by either doxorubicin or etoposide alone was revealed. To gain insights into the distinctively affected cellular activities, the top GO categories to which the differential proteins belong were uncovered. GO analysis on cellular components (CC), biological processes (BP) and molecular function (MF) was subsequently performed. In terms of the localisation of the identified proteins, upon doxorubicin or etoposide stimulus, a similar distribution between membrane/cytoplasmic and nuclear cell components was observed (Supplementary Fig. 2). Molecular functions, such as transporter activity, structural molecule activity, enzyme regulator activity and binding activity, were amongst the most affected ones in response to doxorubicin or etoposide (Supplementary Fig. 2). Moreover, a number of associated biological processes were unveiled, including cell cycle, growth, apoptosis, transport, immune system and cell adhesion, highlighting a broad effect of both doxorubicin and etoposide on cell fate (Fig. 4a).
Fig. 4.
Characterization of the functional portrait conferred by KSR1 in the presence of doxorubicin or etoposide. a Functional profiles of top GO biologic processes affected by doxorubicin or etoposide stimulus (H/L), or by KSR1 in the presence of doxorubicin or etoposide (M/L) are presented. x-axis shows the percentage of hits that belong to a GO biologic process term. b The functional protein–protein interaction networks belonging exclusively to Doxo-KSR1 or Etop-KSR1 are shown. Protein nodes with lighter colour are up-regulated, whereas darker colour represents down-regulation
We then incorporated the STRING database with the GO analyses to define the functional protein–protein interaction networks induced by doxorubicin or etoposide [26]. Not surprisingly, a large number of functional connections were affected, such as mRNA splicing, cell killing and immune system process. A comparison of the signalling networks affected by either doxorubicin or etoposide showed a good overlap, supporting a similar mechanism of their genotoxic action. Conversely, differences were also observed. Doxorubicin showed a greater effect on metabolic process, whereas etoposide had a more pronounced impact on translational initiation, elongation, termination and cell surface receptor signalling. Representatives of the most enriched functional networks are displayed (Supplementary Fig. 3).
Global proteomic alterations induced by KSR1 overexpression in response to doxorubicin or etoposide
Although KSR1 has been indicated to play a potential role in chemotherapy response in breast cancer, a detailed proteomic picture and related signalling networks induced by genotoxic agents, which would further improve our understanding of its function, are still lacking [14, 16]. Therefore, we next determined the global proteome alterations upon KSR1 overexpression in the presence of doxorubicin or etoposide.
The quantifications of corresponding M/L ratios showing (Doxo+KSR1)/Control or (Etop+KSR1)/Control from two datasets were evaluated and then compared to provide a complete proteomic repertoire. In summary, upon KSR1 overexpression, there were 157 and 171 proteins greatly modulated in the presence of doxorubicin or etoposide, respectively (Fig. 2b). Moreover, 88 proteins showed substantial up-regulation and 69 were down-regulated in the Doxo+KSR1 dataset, whereas 98 and 73 were significantly up or down modulated in the Etop+KSR1 dataset (Fig. 2c). Among these, 92 were commonly affected with 57 showing an increase and 35 a decrease in their protein levels (Fig. 2b, c).
Similarly, to visualise the proteomic changes, we created heatmaps highlighting the mutual targets induced by KSR1 in response to doxorubicin or etoposide, as well as the ones exclusively belonging to either Doxo-KSR1 or Etop-KSR1 (Fig. 2d). In addition, to characterise the evident hits upon KSR1 overexpression in the presence of doxorubicin or etoposide, the Log2 ‘normalized ratios’ (M/L) were plotted against Log10 ‘intensity’ (M/L) to exhibit the significantly differentiated proteins based on P values from Significant B test (Fig. 3b). A marked increase in protein abundance of KSR1 itself was also recorded, confirming a good reproducibility and accuracy of SILAC-based proteomics (Fig. 3b). Lists of the top 30 proteins that are significantly regulated by Doxo-KSR1 (Table 1) or Etop-KSR1 (Table 2) are presented. We then investigated the proteomic changes in more detail to unveil the biological processes and molecular functions conferred by KSR1 overexpression in response to doxorubicin or etoposide. GO localisation analysis revealed that the identified proteins from KSR1-Doxo and KSR1-Etop datasets are disseminated in numerous cellular components, including extracellular matrix, cell membrane, cytoplasm, nuclei, synapse and other organelles (Supplementary Fig. 2). Likewise, they are implicated in a broad range of biological processes, including cell communication, apoptosis, response to stimulus, growth, transport, development and cell adhesion (Fig. 4a).
Table 1.
A list of top 30 proteins that are significantly regulated by KSR1 overexpression in the presence of doxorubicin
| Symbols | GeneIDs | Full descriptions | P value |
|---|---|---|---|
| SYT7 | 9066 | Synaptotagmin VII | 7.16E−39 |
| IFIT2 | 3433 | Interferon-induced protein with tetratricopeptide repeats 2 | 4.63E−32 |
| IFIT1 | 3434 | Interferon-induced protein with tetratricopeptide repeats 1 | 4.20E−31 |
| MX1 | 4599 | MX dynamin-like GTPase 1 | 2.15E−30 |
| KSR1 | 8844 | Kinase suppressor of ras 1 | 2.28E−27 |
| DDX58 | 23586 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | 8.26E−27 |
| SYCP1 | 6847 | Synaptonemal complex protein 1 | 1.59E−25 |
| OAS3 | 4940 | 2′-5′-oligoadenylate synthetase 3, 100 kDa | 1.85E−21 |
| IFIT3 | 3437 | Interferon-induced protein with tetratricopeptide repeats 3 | 7.28E−21 |
| ISG15 | 9636 | ISG15 ubiquitin-like modifier | 7.04E−19 |
| OAS2 | 4939 | 2′-5′-oligoadenylate synthetase 2, 69/71 kDa | 8.13E−18 |
| IFIH1 | 64135 | Interferon induced with helicase C domain 1 | 2.52E−17 |
| TADA2B | 93624 | Transcriptional adaptor 2B | 3.89E−17 |
| DDX60 | 55601 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 | 1.57E−16 |
| SAMD9 | 54809 | Sterile alpha motif domain containing 9 | 8.87E−16 |
| PARP14 | 54625 | Poly (ADP-ribose) polymerase family, member 14 | 2.63E−14 |
| PARP9 | 83666 | Poly (ADP-ribose) polymerase family, member 9 | 5.30E−14 |
| OASL | 8638 | 2′-5′-oligoadenylate synthetase-like | 2.53E−13 |
| WARS | 7453 | Tryptophanyl-tRNA synthetase | 1.93E−12 |
| STAT1 | 6772 | Signal transducer and activator of transcription 1, 91 kDa | 2.11E−12 |
| LGALS3BP | 3959 | Lectin, galactoside-binding, soluble, 3 binding protein | 3.27E−12 |
| HLA-C | 3107 | Major histocompatibility complex, class I, C | 7.35E−12 |
| RNF213 | 57674 | Ring finger protein 213 | 5.16E−10 |
| SAMHD1 | 25939 | SAM domain and HD domain 1 | 2.44E−09 |
| TAP1 | 6890 | Transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) | 2.51E−09 |
| OAS1 | 4938 | 2′-5′-oligoadenylate synthetase 1, 40/46 kDa | 3.38E−09 |
| ICAM1 | 3383 | Intercellular adhesion molecule 1 | 3.62E−09 |
| PML | 5371 | Promyelocytic leukaemia | 3.66E−09 |
| HELZ2 | 85441 | Helicase with zinc finger 2, transcriptional coactivator | 9.35E−09 |
| HLA-A | 3105 | Major histocompatibility complex, class I, A | 1.65E−08 |
Table 2.
A list of top 30 proteins that are significantly regulated by KSR1 overexpression in the presence of etoposide
| Symbols | GeneIDs | Full descriptions | P value |
|---|---|---|---|
| OAS2 | 4939 | 2′-5′-oligoadenylate synthetase 2, 69/71 kDa | 3.18E−33 |
| MX1 | 4599 | MX dynamin-like GTPase 1 | 2.97E−32 |
| IFIT1 | 3434 | Interferon-induced protein with tetratricopeptide repeats 1 | 7.00E−31 |
| IFIT2 | 3433 | Interferon-induced protein with tetratricopeptide repeats 2 | 1.24E−29 |
| DDX58 | 23586 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | 1.92E−24 |
| OAS3 | 4940 | 2′-5′-oligoadenylate synthetase 3, 100 kDa | 6.84E−21 |
| HLA-C | 3107 | Major histocompatibility complex, class I, C | 9.21E−21 |
| KSR1 | 8844 | Kinase suppressor of ras 1 | 1.92E−19 |
| CFB | 629 | Complement factor B | 5.48E−19 |
| ISG15 | 9636 | ISG15 ubiquitin-like modifier | 1.10E−18 |
| IFIH1 | 64135 | Interferon induced with helicase C domain 1 | 4.79E−18 |
| IFIT3 | 3437 | Interferon-induced protein with tetratricopeptide repeats 3 | 4.91E−18 |
| SPRR2C | 6702 | Small proline-rich protein 2C (pseudogene) | 3.18E−17 |
| DDX60 | 55601 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 | 2.97E−16 |
| HELZ2 | 85441 | Helicase with zinc finger 2, transcriptional coactivator | 6.21E−16 |
| MX2 | 4600 | MX dynamin-like GTPase 2 | 7.51E−16 |
| WARS | 7453 | Tryptophanyl-tRNA synthetase | 1.21E−15 |
| ICAM1 | 3383 | Intercellular adhesion molecule 1 | 1.91E−15 |
| PARP14 | 54625 | Poly (ADP-ribose) polymerase family, member 14 | 1.33E−14 |
| FABP5 | 2171 | Fatty acid binding protein 5 (psoriasis-associated) | 1.60E−14 |
| SAMD9 | 54809 | Sterile alpha motif domain containing 9 | 3.67E−14 |
| OAS1 | 4938 | 2′-5′-oligoadenylate synthetase 1, 40/46 kDa | 7.40E−14 |
| STAT1 | 6772 | Signal transducer and activator of transcription 1, 91 kDa | 1.96E−13 |
| LGALS3BP | 3959 | Lectin, galactoside-binding, soluble, 3 binding protein | 1.71E−12 |
| LGALS7 | 3963 | Lectin, galactoside-binding, soluble, 7 | 5.08E−12 |
| S100A8 | 6279 | S100 calcium binding protein A8 | 1.97E−11 |
| RSAD2 | 91543 | Radical S-adenosyl methionine domain containing 2 | 8.80E−11 |
| TRANK1 | 9881 | Tetratricopeptide repeat and ankyrin repeat containing 1 | 1.47E−10 |
| OASL | 8638 | 2′-5′-oligoadenylate synthetase-like | 3.64E−10 |
| STAT2 | 6773 | Signal transducer and activator of transcription 2, 113 kDa | 7.00E−10 |
We next sought to define the exclusive as well as shared modulated functional protein–protein interaction networks in the two datasets to further address the action of KSR1 in response to different genotoxic agents. From the significantly differentiated proteins, we selected candidates according to their appearance only in the Doxo-KSR1 (65 proteins, Fig. 2d) or the Etop-KSR1 (79 proteins, Fig. 2d) dataset or in both (92 proteins, Fig. 2d) for follow-up analysis. We first determined GO terms that were enriched in these three different groups of proteins and then portrayed the STRING network for each group with an overrepresented GO term. As expected, broadly integrated functional networks were uncovered. Of note, a unique profile of functional connections stimulated by KSR1 with doxorubicin or etoposide was delineated, highlighting the discrepancies of KSR1-regulated signalling in different contexts. For Doxo-KSR1, the most overrepresented exclusive functional networks were related to apoptosis, rRNA metabolic processes, ncRNA processing and nucleotide biosynthesis (Fig. 4b). Conversely, for Etop-KSR1, the most enriched functional connections were found to be cellular secretion, response to cytokine stimulus, Notch signalling, wound healing, transcription initiation and vesicle-mediated transport (Fig. 4b).
Clinical significance of SILAC-based proteomics identified targets in breast cancer
From the top hits modulated by KSR1 in the presence of genotoxic agents, some proteins, which were up-regulated upon KSR1 overexpression in the presence of genotoxic agents, have been previously indicated to play a role in doxorubicin or etoposide response, including ISG15 [28], STAT1 [29–32], TAP1 [33], MX1 [34] and HLA-C [35] (Tables 1, 2). Of note, recent work has shown that doxorubicin increased levels of the posttranslational modifier ISG15, which resulted in ISGylation of the p53 family protein. As a result, ISGylation inhibited the capability of ΔNp63α to stimulate anchorage-independent cell growth and tumour formation in vivo as well to repress the activity of proapoptotic p53 family members. These findings indicate a tumour suppressive role for ISG15 and highlight a molecular rationale for therapeutic use of doxorubicin [28]. Moreover, chemotherapeutic drugs, such as doxorubicin, can induce the activation of STAT1 and enhance the cooperation of p53 and STAT1 in the induction of cell death [30]. In breast cancer, the dual EGFR/HER2 tyrosine kinase inhibitor lapatinib and doxorubicin enhance the STAT1-dependent antitumour immune response [36, 37]. Consistently, our proteomic data also showed significant increase levels of ISG15 and STAT1 upon chemotherapeutic drugs and supports their role as responsive markers.
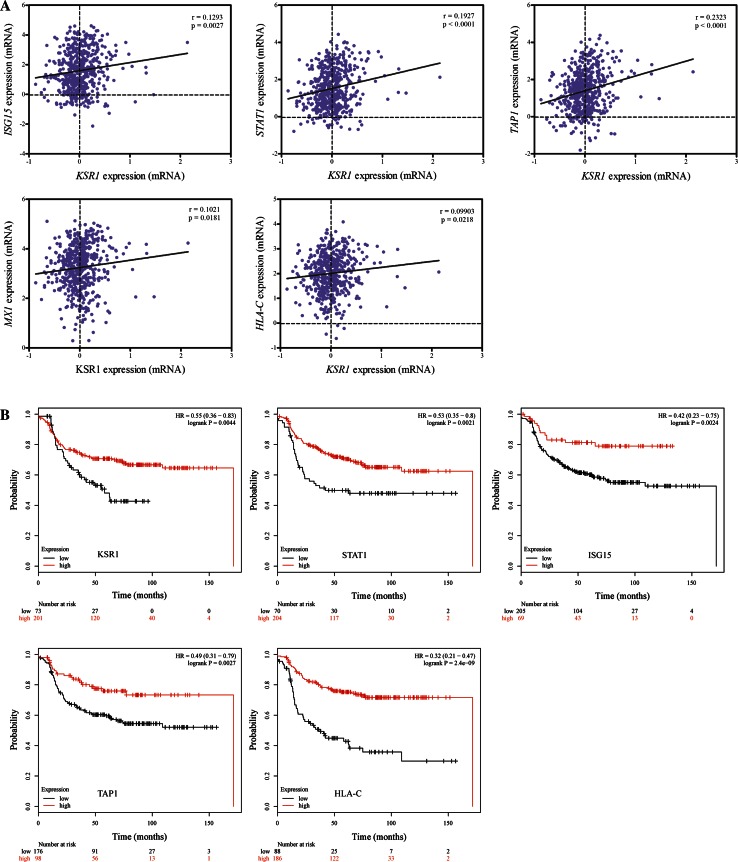
To further examine the clinical associations of KSR1 with ISG15, STAT1, TAP1, MX1 and HLA-C and their relevance in chemotherapy response, we carried out a series of analyses using extensive data from breast cancer patients. First, we took advantage of gene expression data from the well-established The Cancer Genome Atlas (TCGA) dataset [38] and analysed the mRNA expression correlation of KSR1 with ISG15, STAT1, TAP1, MX1 and HLA-C individually in approximately 600 breast cancer patients. A trend of potential positive correlations of KSR1 mRNA with ISG15, STAT1, TAP1, MX1 and HLA-C was observed, supporting our proteomic data (Fig. 5a).
Fig. 5.
Clinical significance of SILAC-based proteomics identified targets in breast cancer. a The correlation analysis of KSR1 mRNA with ISG15, STAT1, TAP1, MX1 and HLA-C gene expressions from TCGA dataset. b Kaplan–Meier curves show associations of expression levels of KSR1, STAT1, ISG15, TAP1 and HLA-C with relapse free survival (RFS) in breast cancer patients who underwent systematic chemotherapy
Next, we utilised the survival analysis tool KM Plotter, which incorporates comprehensive gene expression data and survival information from more than 3000 patients [20]. The relevance of the levels of KSR1, ISG15, STAT1, TAP1, MX1 and HLA-C on relapse free survival (RFS) in breast cancer patients who received chemotherapy was analysed. Interestingly, high KSR1 expressions are significantly associated with better survival in this subgroup of patients (Fig. 5b), supporting our previous finding of KSR1 acting as a tumor suppressor [14]. Likewise, KM Plotter analysis also demonstrated pronounced correlations between high levels of ISG15, STAT1, TAP1 and HLA-C, which were positively regulated upon KSR1 overexpression in the presence of genotoxic agents, with longer relapse free survival in patients who underwent chemotherapy (Fig. 5b).
Discussion
KSR1 was originally characterised as an evolutionary conserved protein kinase in the Ras signalling pathway in Drosophila and Caenorhabditis elegans [1–3], whereas mammalian KSR1 is broadly described as a pseudokinase due to the absence of a key lysine residue essential for its kinase activity [4, 5]. Recently, emerging evidence demonstrates dual function of mammalian KSR1 as an active kinase and a scaffold protein in the Ras-Raf-MAPK pathway [4]. Given its importance in this oncogenic signalling cascade, a number of studies attempted to address the role of KSR1 in Ras-dependent cancers. KSR1 has been shown to contribute to oncogenesis, while deficiency of KSR1 prevents Ras signalling and tumour development in various types of cancer, including skin and pancreatic [10–12]. However, in breast cancer where Ras mutations are rare [13], several lines of evidence suggest that KSR1 plays a different role. Firstly, KSR1 may not act as a positive regulator of the canonical Ras-RAF-MAPKs pathway, as no significant changes were observed in the phosphorylation of major members of this pathway, (i.e. ERK1/2), upon KSR1 overexpression [14]. Secondly, KSR1 up-regulation inhibited breast cancer growth in vitro and in vivo, whereas breast cancer patients with high KSR1 had better disease-free and overall survival [14]. Moreover, KSR1 overexpression in MCF7 cells enhanced cisplatin sensitivity and apoptosis and a trend correlation of KSR1 expression with patients’ response to chemotherapy was indicated [14, 16]. Therefore, a detailed profile of KSR1-regulated signalling in response to genotoxic agents is needed to improve our understanding of its role in breast cancer.
Here, we implemented SILAC-based high-resolution MS quantitative proteomics to identify and quantify global protein changes upon KSR1 overexpression in the presence of either doxorubicin or etoposide in MCF7 cells. Doxorubicin and etoposide are widely used anticancer agents that target human type IIA topoisomerases (Top2) resulting in the inhibition of DNA replication [39, 40]. Although both drugs are Top2 poisons, not only do the kinetics of formation of Top2 cleavage complexes (Top2 cc) differ (slow in the case of doxorubicin to very rapid in the case of etoposide), but also the numbers of Top2-mediated DNA double-strand breaks (DSB) and single-strand breaks (SSB) vary [40]. Moreover, the cellular response to Top2 inhibitors is complex and a broad range of DNA processes are implicated. Thanks to the SILAC-based quantitative proteomics, we were able to quantify and compare the affected proteome in response to doxorubicin and etoposide. As expected, a large number of proteins were significantly modulated upon doxorubicin and etoposide stimulus, whereas approximately 70 of them were mutually identified in both treatments. GO-STRING analysis revealed multiple signalling networks activated by doxorubicin and etoposide and supports the complexity of the cellular response to Top2 poisons. Of note, a good overlap of biological processes perturbed by either doxorubicin or etoposide was observed, highlighting the similarity in their mechanism of action in cytotoxicity. Consistently, discrepancies also exist in downstream effects, as doxorubicin and etoposide surely have additional targets besides Top2 [40]. For instance, metabolic processes were more enriched upon doxorubicin stimulus, whereas translational initiation, elongation and termination were more pronounced when after etoposide treatment. Further elucidation of doxorubicin- or etoposide- exclusive targets will shed new light on the mechanism of action of these Top2 drugs.
Clearly, the biological function of KSR1 in breast cancer is different from its role in other Ras-dependent tumours. Previously, we reported a tumour suppressive action of KSR1 in breast cancer by stabilising BRCA1 through a reduction of its ubiquitination and by promoting the BRCA1-BARD1 interaction. Moreover, a trend in correlation of KSR1 expression with response to chemotherapy was indicated and KSR1 abundance in MCF7 cells was positively associated with cisplatin sensitivity and apoptosis [14, 16]. These data support KSR1 in a continuum with clinical relevance and position KSR1 in the major oncoprotein pathways in breast tumorigenesis. Importantly, it led us to further investigate the KSR1-regulated proteome and associated signalling in response to genotoxic agents. To our knowledge, this is the first global analysis comparing the proteomic profiles induced by KSR1 in the presence of doxorubicin or etoposide. Approximately two hundred proteins were significantly affected upon KSR1 overexpression after either doxorubicin or etoposide treatment. Of note, a large number of these identified proteins were commonly affected in both treatments, indicating a high degree of similarity in KSR1-regulated proteome induced by genotoxic agents.
In-depth GO analysis confirmed a resemblance in both cellular component distributions and biological processes revealed in KSR1-Doxo and KSR1-Etop datasets. Moreover, we delineated a distinctive signature of functional signalling networks regulated by proteins in the KSR1-Doxo or KSR1-Etop dataset, indicating a certain degree of discrepancies in the KSR1-regulated proteome in response to different drugs. Apoptosis, rRNA metabolic process, ncRNA processing and nucleotide biosynthesis were amongst the most enriched functional connections for Doxo-KSR1, whereas cellular secretion, response to cytokine stimulus, Notch signalling, wound healing, transcription initiation and vesicle-mediated transport were characterised for Etop-KSR1. In concordance with our work, previous studies have documented an important role of KSR1 in cytokine-induced apoptosis [41, 42], a protective factor against bacterial infection [43], as well as an essential player in regulating glucose tolerance and insulin sensitivity [44]. These data suggest diverse effects conferred by KSR1 and add complexity to the KSR1-regulated signalling, which requires future investigation.
Furthermore, from the modulated proteomic profile, we identified a number of KSR1-regulated genes that are implicated in chemotherapy response in breast cancer. The expression levels of these proteins, including ISG15, STAT1, TAP1, MX1 and HLA-C, were up-regulated upon KSR1 overexpression in the presence of genotoxic agents. Likewise, this similar trend of positive correlations of KSR1 mRNA levels with ISG15, STAT1, TAP1, MX1 and HLA-C was also seen in the tumour samples from the TCGA database. Of note, high expression levels of KSR1, as well as the identified proteins ISG15, STAT1, TAP1 and HLA-C, are correlated with better outcome in patients who underwent chemotherapy. These findings which are consistent with previous results support their role in doxorubicin or etoposide response, highlighting the tumour suppressive action of KSR1 [14], ISG15 [28] and STAT1 [36].
Additional work is required to examine the KSR1-regulated proteome in detail, such as investigation of the identified signalling pathways in MCF7 and other models. Moreover, it would also be interesting to address the underlying mechanism by which KSR1 facilitates the expression of ISG15, STAT1, TAP1, MX1 and HLA-C in the case of genotoxic agents.
Collectively, we present a comprehensive profile of KSR1-regulated proteome in the presence of genotoxic agents, as well as distinct functional networks belonging exclusively to Doxo-KSR1 and Etop-KSR1. Notably, our results also underscore the clinical relevance of KSR1 expression in chemotherapy response in breast cancer patients.
Electronic supplementary material
Supplementary Fig. 1 The log2 ratios’ distribution of protein ratios from identified proteome in two datasets. Dot plots showing the Log2 ‘normalized ratios’ (y-axis) for each characterized protein in response to doxorubicin or etoposide stimulus (a), and affected by KSR1 in the presence of doxorubicin or etoposide (b) are displayed. Supplementary material 1 (TIFF 1140 kb)
Supplementary Fig. 2 Characterization of the cellular profile affected by identified proteome. GO analyses of cellular components (upper panel) and molecular functions (upper panel) in two datasets are presented. x-axis shows the percentage of hits that belong to a GO term. Supplementary material 2 (TIFF 1644 kb)
Supplementary Fig. 3 Characterization of functional networks conferred by doxorubicin or etoposide treatment. The functional protein-protein interaction networks induced by doxorubicin (a) or etoposide (b) are presented. Protein nodes with lighter colour are up-regulated, whereas darker colour represents down-regulation. Supplementary material 3 (TIFF 2554 kb)
Acknowledgments
We would like to thank Darren Patten for helpful discussion. This work was supported by Action Against Cancer and Hilary Craft in particular, the Pink Ribbon Foundation and the Sally Roter PhD Studentship.
Conflict of interest
The authors declare no competing financial interests.
Footnotes
Hua Zhang and Nicos Angelopoulos have contributed equally to this work.
Contributor Information
Hua Zhang, Email: h.zhang10@imperial.ac.uk.
Georgios Giamas, Email: g.giamas@imperial.ac.uk.
References
- 1.Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83(6):879–888. doi: 10.1016/0092-8674(95)90204-X. [DOI] [PubMed] [Google Scholar]
- 2.Kornfeld K, Hom DB, Horvitz HR. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell. 1995;83(6):903–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- 3.Sundaram M, Han M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 1995;83(6):889–901. doi: 10.1016/0092-8674(95)90205-8. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, Koo CY, Stebbing J, Giamas G. The dual function of KSR1: a pseudokinase and beyond. Biochem Soc Trans. 2013;41(4):1078–1082. doi: 10.1042/BST20130042. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H, Photiou A, Grothey A, Stebbing J, Giamas G. The role of pseudokinases in cancer. Cell Signal. 2012;24(6):1173–1184. doi: 10.1016/j.cellsig.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 6.Therrien M, Michaud NR, Rubin GM, Morrison DK. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 1996;10(21):2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- 7.Xing H, Kornfeld K, Muslin AJ. The protein kinase KSR interacts with 14-3-3 protein and Raf. Curr Biol. 1997;7(5):294–300. doi: 10.1016/S0960-9822(06)00152-7. [DOI] [PubMed] [Google Scholar]
- 8.Michaud NR, Therrien M, Cacace A, Edsall LC, Spiegel S, Rubin GM, Morrison DK. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc Natl Acad Sci U S A. 1997;94(24):12792–12796. doi: 10.1073/pnas.94.24.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Yao B, Delikat S, Bayoumy S, Lin XH, Basu S, McGinley M, Chan-Hui PY, Lichenstein H, Kolesnick R. Kinase suppressor of Ras is ceramide-activated protein kinase. Cell. 1997;89(1):63–72. doi: 10.1016/S0092-8674(00)80183-X. [DOI] [PubMed] [Google Scholar]
- 10.Lozano J, Xing R, Cai Z, Jensen HL, Trempus C, Mark W, Cannon R, Kolesnick R. Deficiency of kinase suppressor of Ras1 prevents oncogenic ras signaling in mice. Cancer Res. 2003;63(14):4232–4238. [PubMed] [Google Scholar]
- 11.Xing HR, Cordon-Cardo C, Deng X, Tong W, Campodonico L, Fuks Z, Kolesnick R. Pharmacologic inactivation of kinase suppressor of ras-1 abrogates Ras-mediated pancreatic cancer. Nat Med. 2003;9(10):1266–1268. doi: 10.1038/nm927. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Zafrullah M, Yang X, Yin X, Zhang Z, Fuks Z, Kolesnick R. Downregulation of KSR1 in pancreatic cancer xenografts by antisense oligonucleotide correlates with tumor drug uptake. Cancer Biol Ther. 2008;7(9):1490–1495. doi: 10.4161/cbt.7.9.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adjei AA. Blocking oncogenic Ras signaling for cancer therapy. J Natl Cancer Inst. 2001;93(14):1062–1074. doi: 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- 14.Stebbing J, Zhang H, Xu Y, Lit LC, Green AR, Grothey A, Lombardo Y, Periyasamy M, Blighe K, Zhang W, et al. KSR1 regulates BRCA1 degradation and inhibits breast cancer growth. Oncogene. 2014;34(16):2103. doi: 10.1038/onc.2014.129. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Xu Y, Filipovic A, Lit LC, Koo CY, Stebbing J, Giamas G. SILAC-based phosphoproteomics reveals an inhibitory role of KSR1 in p53 transcriptional activity via modulation of DBC1. Br J Cancer. 2013;109(10):2675–2684. doi: 10.1038/bjc.2013.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim M, Yan Y, Kortum RL, Stoeger SM, Sgagias MK, Lee K, Lewis RE, Cowan KH. Expression of kinase suppressor of Ras1 enhances cisplatin-induced extracellular signal-regulated kinase activation and cisplatin sensitivity. Cancer Res. 2005;65(10):3986–3992. doi: 10.1158/0008-5472.CAN-03-2334. [DOI] [PubMed] [Google Scholar]
- 17.Olsen JV, Mann M. Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc Natl Acad Sci U S A. 2004;101(37):13417–13422. doi: 10.1073/pnas.0405549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 19.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 20.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 21.R Core Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
- 22.Wielemaker J, Schrijvers T, Triska M. Lager to: SWI-Prolog. Theory and Practice of Logic Programming. 2012;12(1–2):67–96. doi: 10.1017/S1471068411000494. [DOI] [Google Scholar]
- 23.Angelopoulos N, Costa VS, Azevedo J, Wielemaker J, Camacho R, Wessels L (2013) Integrative functional statistics in logic programming. In: Proceedings of practical aspects of declarative languages. vol 7752. Rome, pp 190–205
- 24.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23(2):257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 26.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y, Zhang H, Lit LC, Grothey A, Athanasiadou M, Kiritsi M, Lombardo Y, Frampton AE, Green AR, Ellis IO, et al. The kinase LMTK3 promotes invasion in breast cancer through GRB2-mediated induction of integrin beta1. Sci Signal. 2014;7(330):ra58. doi: 10.1126/scisignal.2005170. [DOI] [PubMed] [Google Scholar]
- 28.Jeon YJ, Jo MG, Yoo HM, Hong SH, Park JM, Ka SH, Oh KH, Seol JH, Jung YK, Chung CH. Chemosensitivity is controlled by p63 modification with ubiquitin-like protein ISG15. J Clin Invest. 2012;122(7):2622–2636. doi: 10.1172/JCI61762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu S, Waguespack M, Barker SA, Li S. Doxorubicin directs the accumulation of interleukin-12 induced IFN gamma into tumors for enhancing STAT1 dependent antitumor effect. Clin Cancer Res. 2007;13(14):4252–4260. doi: 10.1158/1078-0432.CCR-06-2894. [DOI] [PubMed] [Google Scholar]
- 30.Youlyouz-Marfak I, Gachard N, Le Clorennec C, Najjar I, Baran-Marszak F, Reminieras L, May E, Bornkamm GW, Fagard R, Feuillard J. Identification of a novel p53-dependent activation pathway of STAT1 by antitumour genotoxic agents. Cell Death Differ. 2008;15(2):376–385. doi: 10.1038/sj.cdd.4402270. [DOI] [PubMed] [Google Scholar]
- 31.Cao Y, Eble JM, Moon E, Yuan H, Weitzel DH, Landon CD, Nien CY, Hanna G, Rich JN, Provenzale JM, et al. Tumor cells upregulate normoxic HIF-1alpha in response to doxorubicin. Cancer Res. 2013;73(20):6230–6242. doi: 10.1158/0008-5472.CAN-12-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Datta S, Parajuli N, Tymoszuk P, Ottina E, Parson W, Sgonc R, Villunger A, Doppler W. Replenishment of the B cell compartment after doxorubicin-induced hematopoietic toxicity is facilitated by STAT1. J Leukoc Biol. 2014;95(6):853–866. doi: 10.1189/jlb.0113053. [DOI] [PubMed] [Google Scholar]
- 33.Kaneno R, Shurin GV, Kaneno FM, Naiditch H, Luo J, Shurin MR. Chemotherapeutic agents in low noncytotoxic concentrations increase immunogenicity of human colon cancer cells. Cell Oncol (Dordr) 2011;34(2):97–106. doi: 10.1007/s13402-010-0005-5. [DOI] [PubMed] [Google Scholar]
- 34.Xu J, Gao M, Fan S, Meng Q, Goldberg ID, Abounader R, Ressom H, Laterra JJ, Rosen EM. Effect of Akt inhibition on scatter factor-regulated gene expression in DU-145 human prostate cancer cells. Oncogene. 2007;26(20):2925–2938. doi: 10.1038/sj.onc.1210088. [DOI] [PubMed] [Google Scholar]
- 35.Alcoceba M, Sebastian E, Marin L, Balanzategui A, Sarasquete ME, Chillon MC, Jimenez C, Puig N, Corral R, Pardal E, et al. HLA specificities are related to development and prognosis of diffuse large B-cell lymphoma. Blood. 2013;122(8):1448–1454. doi: 10.1182/blood-2013-02-483420. [DOI] [PubMed] [Google Scholar]
- 36.Hannesdottir L, Tymoszuk P, Parajuli N, Wasmer MH, Philipp S, Daschil N, Datta S, Koller JB, Tripp CH, Stoitzner P, et al. Lapatinib and doxorubicin enhance the Stat1-dependent antitumor immune response. Eur J Immunol. 2013;43(10):2718–2729. doi: 10.1002/eji.201242505. [DOI] [PubMed] [Google Scholar]
- 37.Tymoszuk P, Doppler W. Impact of STAT1 and CD8 T cells on the antineoplastic activity of lapatinib and doxorubicin against spontaneous mammary tumors. Oncoimmunology. 2013;2(12):e26689. doi: 10.4161/onci.26689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tacar O, Sriamornsak P, Dass CR. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J Pharm Pharmacol. 2013;65(2):157–170. doi: 10.1111/j.2042-7158.2012.01567.x. [DOI] [PubMed] [Google Scholar]
- 40.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17(5):421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan F, John SK, Polk DB. Kinase suppressor of Ras determines survival of intestinal epithelial cells exposed to tumor necrosis factor. Cancer Res. 2001;61(24):8668–8675. [PubMed] [Google Scholar]
- 42.Yan F, John SK, Wilson G, Jones DS, Washington MK, Polk DB. Kinase suppressor of Ras-1 protects intestinal epithelium from cytokine-mediated apoptosis during inflammation. J Clin Invest. 2004;114(9):1272–1280. doi: 10.1172/JCI200421022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Li X, Carpinteiro A, Goettel JA, Soddemann M, Gulbins E. Kinase suppressor of Ras-1 protects against pulmonary Pseudomonas aeruginosa infections. Nat Med. 2011;17(3):341–346. doi: 10.1038/nm.2296. [DOI] [PubMed] [Google Scholar]
- 44.Klutho PJ, Costanzo-Garvey DL, Lewis RE. Regulation of glucose homeostasis by KSR1 and MARK2. PLoS One. 2011;6(12):e29304. doi: 10.1371/journal.pone.0029304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 The log2 ratios’ distribution of protein ratios from identified proteome in two datasets. Dot plots showing the Log2 ‘normalized ratios’ (y-axis) for each characterized protein in response to doxorubicin or etoposide stimulus (a), and affected by KSR1 in the presence of doxorubicin or etoposide (b) are displayed. Supplementary material 1 (TIFF 1140 kb)
Supplementary Fig. 2 Characterization of the cellular profile affected by identified proteome. GO analyses of cellular components (upper panel) and molecular functions (upper panel) in two datasets are presented. x-axis shows the percentage of hits that belong to a GO term. Supplementary material 2 (TIFF 1644 kb)
Supplementary Fig. 3 Characterization of functional networks conferred by doxorubicin or etoposide treatment. The functional protein-protein interaction networks induced by doxorubicin (a) or etoposide (b) are presented. Protein nodes with lighter colour are up-regulated, whereas darker colour represents down-regulation. Supplementary material 3 (TIFF 2554 kb)