Abstract
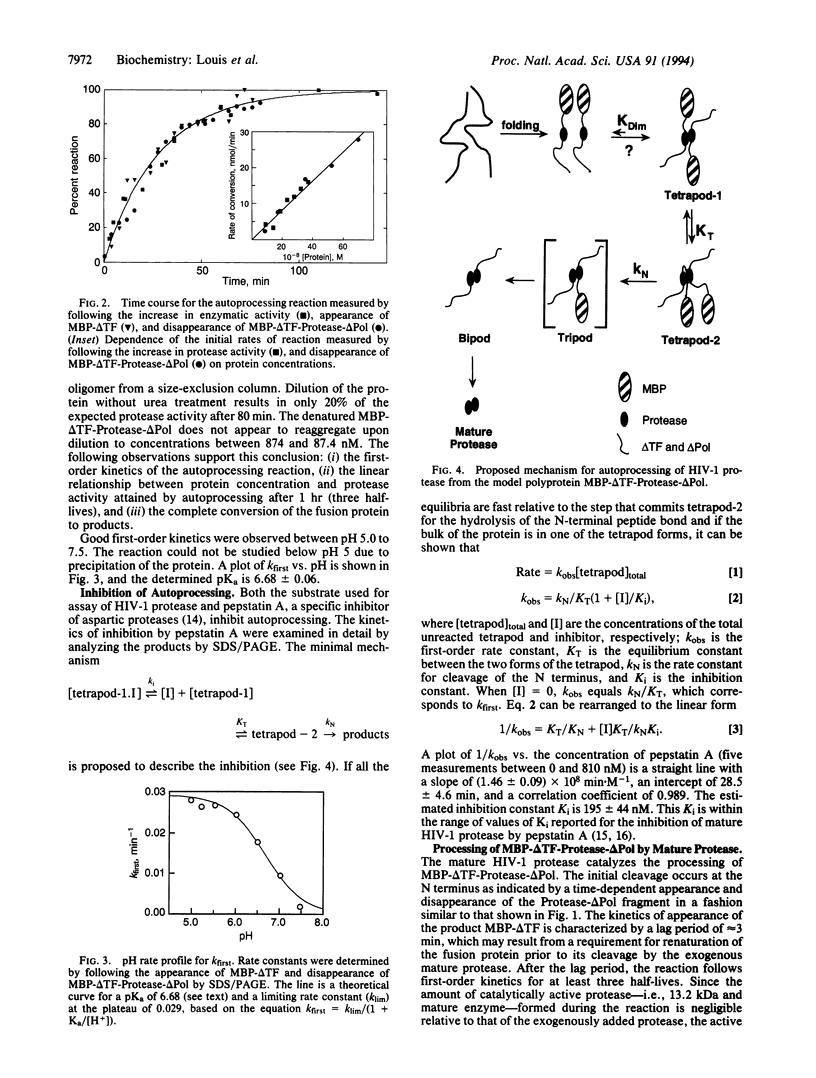
Upon renaturation, the polyprotein MBP-delta TF-Protease-delta Pol, consisting of HIV-1 protease and short native sequences from the trans-frame protein (delta TF) and the polymerase (delta Pol) fused to the maltose-binding protein (MBP) of Escherichia coli, undergoes autoprocessing to produce the mature protease in two steps. The initial step corresponds to cleavage of the N-terminal sequence to release the protein intermediate Protease-delta Pol, which has enzymatic activity comparable to that of the mature enzyme. Subsequently, the mature enzyme is formed by a slower cleavage at the C terminus. The rate of increase in enzymatic activity is identical to that of the appearance of MBP-delta TF and the disappearance of the MBP-delta TF-Protease-delta Pol. Initial rates are linearly dependent on the protein concentration, indicating that the N-terminal cleavage is first-order in protein concentration. The reaction is competitively inhibited by pepstatin A and has a pH rate profile similar to that of the mature enzyme. These results and molecular modeling studies are discussed in terms of a mechanism in which a dimeric full-length fusion protein must form prior to rate-limiting intramolecular cleavage of the N-terminal sequence that leads to an increase in enzymatic activity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Billich A., Hammerschmid F., Winkler G. Purification, assay and kinetic features of HIV-1 proteinase. Biol Chem Hoppe Seyler. 1990 Mar;371(3):265–272. [PubMed] [Google Scholar]
- Broadhurst A. V., Roberts N. A., Ritchie A. J., Handa B. K., Kay C. Assay of HIV-1 proteinase: a colorimetric method using small peptide substrates. Anal Biochem. 1991 Mar 2;193(2):280–286. doi: 10.1016/0003-2697(91)90022-l. [DOI] [PubMed] [Google Scholar]
- Burstein H., Bizub D., Kotler M., Schatz G., Vogt V. M., Skalka A. M. Processing of avian retroviral gag polyprotein precursors is blocked by a mutation at the NC-PR cleavage site. J Virol. 1992 Mar;66(3):1781–1785. doi: 10.1128/jvi.66.3.1781-1785.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein H., Bizub D., Skalka A. M. Assembly and processing of avian retroviral gag polyproteins containing linked protease dimers. J Virol. 1991 Nov;65(11):6165–6172. doi: 10.1128/jvi.65.11.6165-6172.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darke P. L., Nutt R. F., Brady S. F., Garsky V. M., Ciccarone T. M., Leu C. T., Lumma P. K., Freidinger R. M., Veber D. F., Sigal I. S. HIV-1 protease specificity of peptide cleavage is sufficient for processing of gag and pol polyproteins. Biochem Biophys Res Commun. 1988 Oct 14;156(1):297–303. doi: 10.1016/s0006-291x(88)80839-8. [DOI] [PubMed] [Google Scholar]
- Duplay P., Bedouelle H., Fowler A., Zabin I., Saurin W., Hofnung M. Sequences of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J Biol Chem. 1984 Aug 25;259(16):10606–10613. [PubMed] [Google Scholar]
- Fitzgerald P. M., McKeever B. M., VanMiddlesworth J. F., Springer J. P., Heimbach J. C., Leu C. T., Herber W. K., Dixon R. A., Darke P. L. Crystallographic analysis of a complex between human immunodeficiency virus type 1 protease and acetyl-pepstatin at 2.0-A resolution. J Biol Chem. 1990 Aug 25;265(24):14209–14219. [PubMed] [Google Scholar]
- Fitzgerald P. M., Springer J. P. Structure and function of retroviral proteases. Annu Rev Biophys Biophys Chem. 1991;20:299–320. doi: 10.1146/annurev.bb.20.060191.001503. [DOI] [PubMed] [Google Scholar]
- Hartsuck J. A., Koelsch G., Remington S. J. The high-resolution crystal structure of porcine pepsinogen. Proteins. 1992 May;13(1):1–25. doi: 10.1002/prot.340130102. [DOI] [PubMed] [Google Scholar]
- Hyland L. J., Tomaszek T. A., Jr, Meek T. D. Human immunodeficiency virus-1 protease. 2. Use of pH rate studies and solvent kinetic isotope effects to elucidate details of chemical mechanism. Biochemistry. 1991 Aug 27;30(34):8454–8463. doi: 10.1021/bi00098a024. [DOI] [PubMed] [Google Scholar]
- Jackson S. E., Fersht A. R. Folding of chymotrypsin inhibitor 2. 2. Influence of proline isomerization on the folding kinetics and thermodynamic characterization of the transition state of folding. Biochemistry. 1991 Oct 29;30(43):10436–10443. doi: 10.1021/bi00107a011. [DOI] [PubMed] [Google Scholar]
- Kay J., Dunn B. M. Viral proteinases: weakness in strength. Biochim Biophys Acta. 1990 Jan 30;1048(1):1–18. doi: 10.1016/0167-4781(90)90015-t. [DOI] [PubMed] [Google Scholar]
- Kohl N. E., Emini E. A., Schleif W. A., Davis L. J., Heimbach J. C., Dixon R. A., Scolnick E. M., Sigal I. S. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräusslich H. G. Human immunodeficiency virus proteinase dimer as component of the viral polyprotein prevents particle assembly and viral infectivity. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3213–3217. doi: 10.1073/pnas.88.8.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. L., Lin Y. Z., Koelsch G., Gustchina A., Wlodawer A., Tang J. Enzymic activities of two-chain pepsinogen, two-chain pepsin, and the amino-terminal lobe of pepsinogen. J Biol Chem. 1992 Aug 25;267(24):17257–17263. [PubMed] [Google Scholar]
- Louis J. M., McDonald R. A., Nashed N. T., Wondrak E. M., Jerina D. M., Oroszlan S., Mora P. T. Autoprocessing of the HIV-1 protease using purified wild-type and mutated fusion proteins expressed at high levels in Escherichia coli. Eur J Biochem. 1991 Jul 15;199(2):361–369. doi: 10.1111/j.1432-1033.1991.tb16132.x. [DOI] [PubMed] [Google Scholar]
- Marciniszyn J., Jr, Huang J. S., Hartsuck J. A., Tang J. Mechanism of intramolecular activation of pepsinogen. Evidence for an intermediate delta and the involvement of the active site of pepsin in the intramolecular activation of pepsinogen. J Biol Chem. 1976 Nov 25;251(22):7095–7102. [PubMed] [Google Scholar]
- Nashed N. T., Louis J. M., Sayer J. M., Wondrak E. M., Mora P. T., Oroszlan S., Jerina D. M. Continuous spectrophotometric assay for retroviral proteases of HIV-1 and AMV. Biochem Biophys Res Commun. 1989 Sep 15;163(2):1079–1085. doi: 10.1016/0006-291x(89)92331-0. [DOI] [PubMed] [Google Scholar]
- Navia M. A., Fitzgerald P. M., McKeever B. M., Leu C. T., Heimbach J. C., Herber W. K., Sigal I. S., Darke P. L., Springer J. P. Three-dimensional structure of aspartyl protease from human immunodeficiency virus HIV-1. Nature. 1989 Feb 16;337(6208):615–620. doi: 10.1038/337615a0. [DOI] [PubMed] [Google Scholar]
- Rich D. H. Pepstatin-derived inhibitors of aspartic proteinases. A close look at an apparent transition-state analogue inhibitor. J Med Chem. 1985 Mar;28(3):263–273. doi: 10.1021/jm00381a001. [DOI] [PubMed] [Google Scholar]
- Richards A. D., Phylip L. H., Farmerie W. G., Scarborough P. E., Alvarez A., Dunn B. M., Hirel P. H., Konvalinka J., Strop P., Pavlickova L. Sensitive, soluble chromogenic substrates for HIV-1 proteinase. J Biol Chem. 1990 May 15;265(14):7733–7736. [PubMed] [Google Scholar]
- Satow Y., Cohen G. H., Padlan E. A., Davies D. R. Phosphocholine binding immunoglobulin Fab McPC603. An X-ray diffraction study at 2.7 A. J Mol Biol. 1986 Aug 20;190(4):593–604. doi: 10.1016/0022-2836(86)90245-7. [DOI] [PubMed] [Google Scholar]
- Skalka A. M. Retroviral proteases: first glimpses at the anatomy of a processing machine. Cell. 1989 Mar 24;56(6):911–913. doi: 10.1016/0092-8674(89)90621-1. [DOI] [PubMed] [Google Scholar]
- Spurlino J. C., Lu G. Y., Quiocho F. A. The 2.3-A resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J Biol Chem. 1991 Mar 15;266(8):5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Miller M., Jaskólski M., Sathyanarayana B. K., Baldwin E., Weber I. T., Selk L. M., Clawson L., Schneider J., Kent S. B. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989 Aug 11;245(4918):616–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]





