Abstract
Metformin is the most widely prescribed medication for the treatment of type 2 diabetes (T2D). However ~25% of patients treated with metformin develop gastrointestinal (GI) side-effects leading to discontinuation of therapy in approximately 5% cases. We hypothesised that reduced transport of metformin via Organic Cation Transporter 1 (OCT1) could increase metformin concentration in the intestine, leading to increased risk of severe GI side-effects and drug discontinuation. We compared the phenotype, carriage of reduced-function OCT1 variants, and concomitant prescribing of drugs known to inhibit OCT1 transport in 251 intolerant and 1915 fully metformin tolerant T2D patients. We showed that women and older people were more likely to be intolerant to metformin. Concomitant use of medications, known to inhibit OCT1 activity, was associated with intolerance (odds ratio OR=1.63, 95% CI 1.22-2.17, p=0.001) as was carriage of two reduced-function OCT1 alleles compared to carriage of one or no deficient allele (OR=2.41, 95% CI 1.48-3.93, p < 0.001). Individuals with two reduced function OCT1 alleles who were treated with OCT1 inhibitors were over four times more likely to develop intolerance (OR=4.13, 95% CI 2.09-8.16, p < 0.001). Our results suggest that reduced OCT1 transport is an important determinant of metformin intolerance.
Metformin is recommended as first-line therapy for type 2 diabetes (T2D) (1), and currently is used by over 120 million patients worldwide. It ameliorates hyperglycemia by inhibiting hepatic gluconeogenesis, and increasing peripheral glucose uptake (2). It may also increase gut glucose utilisation (3). At a molecular level it has been suggested that metformin interferes with glucagon signalling (4), and more recently, that it inhibits mitochondrial glycerol-3-phosphate dehydrogenase, leading to reduction of hepatic gluconeogenesis (5). Activation of AMP-activated protein kinase may mediate metformin effects on lipid metabolism and insulin sensitivity (6). Metformin is recommended as first-line therapy for T2D because of its efficacy, safety (lack of weight gain, low risk of hypoglycemia), relatively low cost, and potential cardiovascular benefit (7).
Metformin treatment is, however, frequently associated with gastrointestinal (GI) side-effects (20-30% of patients) (2) and this can negatively affect quality of life and adherence in T2D patients (8). Approximately 5% of patients develop severe GI symptoms and discontinue the treatment with metformin, which could deprive them of the beneficial effects of the drug. Common metformin GI symptoms include nausea, diarrhea, vomiting, bloating and abdominal pain (9). The pathophysiology of metformin induced GI intolerance is unclear, although different hypotheses have been proposed, including stimulation of intestinal serotonin secretion, changes in incretin and glucose metabolism, and bile-salt malabsorption (9). It is hypothesised that GI intolerance is related to high concentration of metformin in the intestine after oral administration of the drug (10, 11).
Metformin is an organic cation, and carrier proteins mediate its oral absorption, hepatic uptake and renal elimination. Several solute carrier (SLC) transporters, expressed in the membranes of the enterocytes, could be involved in the absorption of metformin from the intestinal lumen, including organic cation transporter 1 (OCT1), plasma membrane monoamine transporter (PMAT), carnitine/cation transporter 1 (OCTN1) and organic cation transporter 3 (OCT3) (12-15). While there are no established common loss-of-functions variants of other metformin gut transporters, the human OCT1 gene (SLC22A1) is highly polymorphic, and four OCT1 variants: R61C (rs12208357), M420del (rs72552763), G401S (rs34130495) and G465R (rs34059508), showed reduced metformin transport in vitro (16). In addition to genetic variation, a number of commonly prescribed drugs have been shown to inhibit transport via OCT1 in vitro (e.g. tricyclic antidepressants (TCAs), proton pump inhibitors (PPIs), alpha-adrenoreceptor antagonists, calcium-channel blockers (verapamil and diltiazem)) (17).
We hypothesised that reduced transport of metformin by OCT1 could increase metformin concentration in the intestine, resulting in increased risk of GI intolerance and drug discontinuation. Therefore we assessed the role of five reduced-function variants in OCT1 (R61C, C88R (rs55918055), G401S, M420del, and G465R), and concomitant use of OCT1 inhibiting drugs in metformin intolerance, in a large cohort of metformin treated T2D patients from Tayside, Scotland.
RESEARCH DESIGN AND METHODS
Study Population
In this observational cohort study, we identified patients with T2D who were receiving treatment with metformin, using data from the Genetics of Diabetes Audit and Research Tayside Study (GoDARTS) database. The GoDARTS resource includes nearly 10,000 patients with T2D. Since October 1997, DNA was collected from the patients for genetic studies. Retrospective and prospective longitudinal data is collected on each individual with T2D from diagnosis of diabetes, including prescribing, biochemistry and clinical data, which can be obtained in an anonymised form. The GoDARTS study was approved by the Tayside Medical Ethics Committee. Informed consent was obtained for all participants. The use of the GoDARTS bioresource for the study of metformin pharmacogenetics was approved by the Tayside Tissue Bank.
The study included all GoDARTS patients with T2D, who were incident users of metformin in the period from 1st January 1994 to 1st June 2011.
Definition of Intolerance
We established a proxy phenotype of metformin intolerance based upon prescribing patterns. Patients who stopped metformin within the first 6 months of treatment (immediate release form, IR), and switched to another oral hypoglycemic agent, including metformin slow release forms, within 6 months of the last metformin IR prescription, were identified as intolerant. We excluded patients who switched to insulin within 6 months of the last metformin prescription, as well as patients who ever received daily dose of metformin immediate or slow release formulations, of 2000 mg or more.
Patients who were prescribed ≥ 2000 mg of metformin IR form for more than 6 months were defined as tolerant. Patients in both groups with serum creatinine levels above 120μmol/L were excluded from the analysis.
From a total of 6,265 patients, incident users of metformin with T2D, based on our definition, we classified 251 patients as intolerant, and 1,915 patients as tolerant (2,166 patients totally).
Anthropometric and Biochemical Variables
Values closest to metformin index date were taken for weight, BMI, and serum creatinine levels (within one year either side of metformin start), and for HbA1c (within 6 months prior to metformin start). Creatinine clearance was estimated using the Cockcroft-Gault formula (18).
Metformin Dose
Daily dose of metformin was defined as the last prescribed dose for intolerant patients, and as an average dose in the first 6 months of metformin treatment for tolerant patients.
OCT1 Inhibiting Medications
We identified all patients who were prescribed, concomitantly with metformin, medications shown to inhibit OCT1 activity in vitro. This included prescriptions for tricyclic antidepressants (TCAs) (19, 20), citalopram (17, 19), proton pump inhibitors (PPIs) (21), verapamil (19, 20), diltiazem (19), doxazosin (19, 20), spironolactone (19, 20), clopidogrel (22), rosiglitazone (23), quinine (17, 19), tramadol (19, 24), and codeine (25). There were only few or no prescriptions for other OCT1 inhibiting drugs (including prazosin, disopyramide, quinidine, repaglinide, propafenone, ketoconazole, morphine, tropisetron, ondansetron, antipsychotics, tyrosine kinase inhibitors) (17, 19, 20, 23, 25-27).
Genotyping
M420del and R61C variants were genotyped previously in the whole of GoDARTS using TaqMan genotyping assays (Applied Biosystems) (28). Genotypes of the other three variants were imputed from existing genome-wide data on 7319 GoDARTS patients using the 1000 genome reference panel and software IMPUTE2 (29, 30). The imputation quality information values were 0.932, 0.918 and 0.876 for C88R, G410S, and G465 respectively. The imputed genotype data were further supplemented by exomechip data on 4760 individuals (695 patients with definable phenotype in this study). High concordance rate between imputed genotypes called at a threshold of 0.9 and exomechip data were observed, as only 0.2%, 0.4%, and 0.5% variant carriers of C88R, G410S, and G465, respectively, were misclassified by imputed data.
A total of 1940 patients (90% out of 2166 patients in the study) had available genotype data for the five OCT1 variants. All variants were in line with Hardy-Weinberg equilibrium (p > 0.05). The OCT1 diplotypes were estimated using PLINK software (http://pngu.mgh.harvard.edu/purcell/plink/) (31).
Statistical Analysis
The differences between quantitative variables were estimated using a t-test (variables with normal distribution), or Mann-Whitney U test (variables with non-normal distribution). Comparisons between categorical variables were performed with χ2-test or Fisher’s exact test (in the case where expected frequencies were less or equal to 5). R61C, C88R, G401S, M420del, and G465R variants were analysed together, according to the number of haplotypes carrying reduced-function alleles: 0, 1, or 2 (OCT1 combined genotype). The combined genotype frequencies between the two groups were compared using the χ2-test (recessive and dominant model), and Cochran-Armitage trend test (additive model). Logistic regression analysis was used to assess the effects of OCT1 combined genotype, and co-prescribed medications known to inhibit OCT1 activity in vitro, on the outcome phenotype of metformin intolerance as defined above. Based on previous studies, showing significant effect of OCT1 combined genotype in individuals with two dysfunctional alleles (32, 33), a recessive model was assumed. Age, sex, and weight were included as covariates. In a sensitivity analysis, patients were matched for age and sex. Conditional logistic regression was used for the analysis of matched groups. The statistical analyses were performed with SAS software v 9.3 (SAS Institute Inc., Cary, North Carolina, USA). Statistical significance was defined as p < 0.05.
RESULTS
Metformin intolerance phenotype
The characteristics of metformin intolerant and tolerant patients are shown in Table 1. Intolerant patients were on average 10 years older (p < 0.001), were more commonly women (p < 0.001), had lower weight and BMI (p < 0.001), lower creatinine clearance (p < 0.001), lower HbA1c (p =0.003), and were treated with lower metformin dose (p < 0.001).
Table 1. Baseline characteristics of metformin intolerant and tolerant patients.
| Intolerant group (n=251) | Tolerant group (n=1915) | p | |
|---|---|---|---|
| Age (years) | 67.8 ±10.5 | 58.0±10.8 | <0.001 |
| Age at diagnosis (years) | 62.8±10.5 | 54.8±10.5 | <0.001 |
| Females/Males (Females %) | 141/110 (56.2%) | 767/1148 (40.1%) | <0.001 |
| Weight (kg) | 82.8±17.1 | 92.2±18.5 | <0.001 |
| BMI (kg/m2) | 30.6±5.8 | 32.6±6.2 | <0.001 |
| HbA1c (%) (mmol/mol) |
8.3 (7.7-9.5) 67 (61-80) |
8.7 (7.8-9.9) 72 (62-85) |
0.003 |
| Creatinine (μmol/l) | 87.2±13.9 | 87.0±14.5 | 0.808 |
| Creatinine clearance (ml/min) | 75.4 (58.2-92.8) | 97.4 (77.6-121.6) | <0.001 |
| Antidiabetic drug-naive | 136 (54.2%) | 1173 (61.3%) | 0.031 |
| Use of OCT1 inhibiting drugs | 120 (47.8%) | 627 (32.7%) | <0.001 |
| Metformin daily dose (mg) | 1000 (1000-1000) | 1000 (1000-1500) | <0.001 |
Data are presented as means±SD, medians (interquartile range), or numbers (percentages).
To assess whether we had identified a group who developed gastrointestinal (GI) side-effects after starting metformin, we explored the use of GI drugs that could be prescribed for GI side-effects (compound alginates, antispasmodics, antidiarrhoeals and antiemetics), between the two groups before and after metformin commencement (Supplementary Table 1). Consistent with a prescribing pattern suggesting GI intolerance, more patients were treated with anti-diarrhoeal medications after metformin initiation in the intolerant group than the tolerant group. Although not statistically significant, there was also a similar trend for higher use of antispasmodics and other drugs altering gut motility, as well as antiemetics, after metformin initiation in the intolerant compared to the tolerant group (Supplementary Table 1).
OCT1 interacting drugs and metformin intolerance
Almost half of the intolerant patients (47.8%) were taking OCT1 inhibiting drugs compared to 32.7% tolerant patients (p < 0.001) (Table 1). In a logistic regression analysis, adjusted for age, sex, and weight, the concomitant use of OCT1 inhibiting medications was significantly associated with intolerance (odds ratio OR=1.63, 95% CI 1.22-2.17, p=0.001) (data for other covariates are not shown).
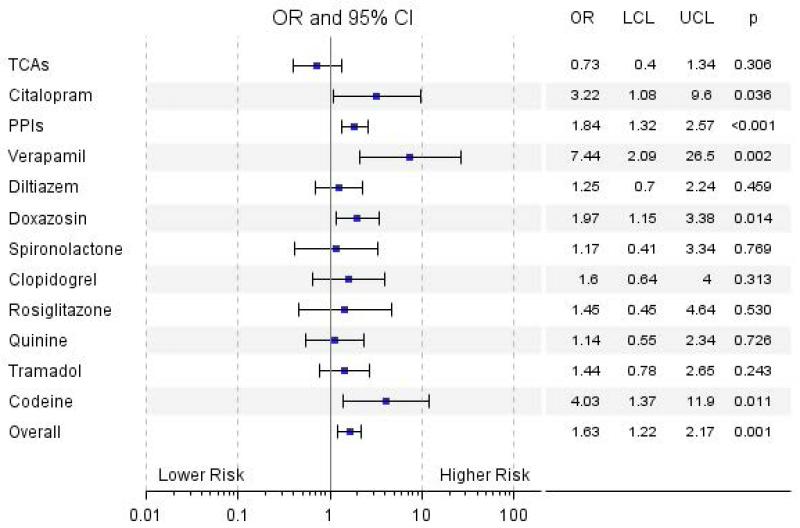
Next, we explored differences in concomitant treatment with individual OCT1 inhibiting drug/drug class between intolerant and tolerant patients. The numbers of patients taking different OCT1 inhibiting medications are shown in Supplementary Table 2. Odds ratios for the association of the individual drugs/drug classes with intolerance are shown in a forest plot in Figure 1. The concomitant use of citalopram, PPIs, verapamil, doxazosin, and codeine, was significantly associated with metformin intolerance. Interestingly, patients treated with verapamil had seven times higher odds of developing intolerance (OR=7.44, 95% CI 2.09-26.5, p=0.002). A high proportion of intolerant patients were using PPIs (28.3%). As PPIs are used to treat indigestion and reflux, and more intolerant patients were taking PPIs before metformin initiation than tolerant patients (56 (22.3%) intolerant vs. 232 (12.1%) tolerant patients, p < 0.001), it is possible that the result for PPIs is confounded by prior GI symptoms. Therefore we studied the use of histamine H2-receptor antagonists (H2RAs) between intolerant and tolerant patients, as these are used for the same indication as PPIs yet do not inhibit OCT1 (19). There were no significant differences in percentage of patients treated with H2RAs (19 (7.6%) intolerant vs. 117 (6.1%) tolerant patients, p = 0.370) in the intolerant group and the tolerant group, suggesting that the result seen for PPIs does reflect OCT1 inhibition.
Figure 1.
Association of the individual drugs/drug classes with metformin intolerance. The analysis was adjusted for age, sex, weight, and co-treatment with other OCT1 inhibitors. TCAs, tricyclic antidepressants; PPIs, proton pump inhibitors; OR, odds ratio; LCL, lower confidence limit; UCL, higher confidence limit.
OCT1 genotypes and metformin intolerance
We explored the linkage disequilibrium between the five OCT1 variants by haplotype analysis using directly genotyped exomechip data. As shown in Supplementary table 3, C88R and G465R substitutions only occurred with M420del variant, while R61C and G401S occurred only on the wild-type background of other polymorphisms, in line with previous studies in Caucasians (16, 27, 33). Further diplotype data in Supplementary Table 4 showed that the number of reduced-function haplotypes in each patient could be simply characterized by the total number of variant alleles in R61C, M420del and G401S (OCT1 combined genotype).
The numbers of patients in the intolerant and tolerant group according to the number of deficient OCT1 alleles are shown in the Supplementary Table 5. There was a significant difference in the combined genotype frequency between the two groups in the recessive model (p < 0.001).
The combined genotype was added to the logistic regression model, adjusted for age, sex, weight, and the overall use of OCT1 inhibiting drugs (Table 2). In addition to the concomitant treatment with OCT1 inhibiting medications, the presence of two reduced-function alleles was independently associated with intolerance to metformin (OR=2.41, 95% CI 1.48-3.93, p < 0.001). When patients were grouped according to combination of OCT1 genotype and the use of OCT1 inhibiting drugs, carriers of two low-activity alleles who were also treated with OCT1 interacting drugs, had four-fold higher odds of intolerance compared to patients with one or no deficient alleles who were not taking OCT1 inhibitors (OR=4.13, 95% CI 2.09-8.16, p < 0.001) (Table 3).
Table 2. Logistic regression model of metformin intolerance.
| OR (95% CI) | p | |
|---|---|---|
| Age | 1.10 (1.08-1.12) | <0.001 |
| Sex (Females vs Males) | 1.85 (1.33-2.57) | <0.001 |
| Weight | 0.99 (0.98-1.00) | 0.064 |
| Use of OCT1 inhibiting drugs | 1.64 (1.20-2.25) | 0.002 |
| Two reduced-function OCT1 alleles | 2.41 (1.48-3.93) | <0.001 |
OR, odds ratio for intolerance. Logistic regression analysis included 205 intolerant and 1650 tolerant patients.
Table 3. Joint effects of OCT1 genotype and OCT1 interacting drugs on intolerance.
| OR (95% CI) | p | |
|---|---|---|
| One or no reduced-function allele carriers not treated with OCT1 inhibiting drugs* | 1.00 | |
| One or no reduced-function allele carriers treated with OCT1 inhibiting drugs† | 1.62 (1.16-2.26) | 0.005 |
| Two reduced-function alleles carriers not treated with OCT1 inhibiting drugs ‡ | 2.27 (1.13-4.58) | 0.022 |
| Two reduced-function alleles carriers treated with OCT1 inhibiting drugs§ | 4.13 (2.09-8.16) | <0.001 |
OR, odds ratio for intolerance. Analysis was adjusted for age, sex, and weight.
93 intolerant and 1030 tolerant patients;
84 intolerant and 516 tolerant patients;
12 intolerant and 70 tolerant patients;
16 intolerant and 34 tolerant patients.
Sensitivity analysis
Since there were large differences in age and sex between cases and controls, we carried out a sensitivity analysis by comparing the intolerant group (n=231) with age- and sex-matched subgroup of tolerant patients (n=709). In this sensitivity analysis we confirmed the main findings obtained with the larger tolerant group (Supplementary Figure 1, Supplementary Tables 6-7).
To guard against potential bias originating from the imputed data, we performed another sensitivity analysis in a subset of 660 patients using only directly genotyped data by TaqMan and exomechip. Carriage of two dysfunctional OCT1 alleles showed significant association with intolerance even in this much smaller cohort (OR=2.99, 95% CI 1.40-6.38, p=0.005, Supplementary Table 8).
To test our definition of intolerance, we carried out additional analyses excluding intolerant patients who had more than 2 metformin prescriptions, and excluding patients who transitioned to metformin slow release forms. Exclusion of these two small subgroups did not alter the results (data not shown).
DISCUSSION
A number of studies have investigated effects of genetic variants on metformin efficacy (34), including the first genome-wide association study of metformin response performed by our group (35). However, so far, only one small study has examined the pharmacogenetics of metformin GI side-effects in T2D (36) and no studies have investigated the use of co-prescribing on risk of metformin intolerance. To our knowledge, this is the first study that addressed the genetics of metformin intolerance, and the role of OCT1, in a large cohort of T2D patients. We found that concomitant use of drugs that inhibit OCT1 increases metformin intolerance, with some drugs such as verapamil increasing odds of intolerance seven fold. In addition, we showed a significant effect of OCT1 genotype on intolerance: patients carrying two OCT1 reduced-function alleles had more than twice the odds of intolerance compared to one or no deficient allele carriers. Together these results point to a key role of metformin transport by OCT1 in mediating GI intolerance with metformin treatment.
Although our results provide strong evidence that inhibition of OCT1 transport increases the risk of GI intolerance, the precise mechanism for this remains uncertain. Results from earlier studies suggested basolateral localization of OCT1 in enterocytes (14, 37, 38), however a recent study demonstrated apical localization of OCT1 in Caco-2 cell monolayers and mouse and human enterocytes (15). If OCT1 has a role in efflux of metformin (either via the basolateral or apical route), then inhibition could increase metformin concentration in the enterocytes. Alternatively, if OCT1 has a role in transport of metformin from the lumen into the enterocytes, then OCT1 inhibition could increase luminal metformin concentration. Although proposed hypotheses of metformin GI intolerance are inconclusive, and it is unclear whether adverse effects could be attributed to the drug present in the mucosa or in the lumen, increased metformin concentrations in the gut may affect intestinal serotonin concentration (39), bile salt absorption (40) or potentially alter the microbiome (41).
An alternative mechanism for the increased metformin intolerance with reduced OCT1 transport is that OCT1 inhibition is altering systemic concentrations of metformin, and it is the higher circulating metformin concentrations that result in metformin intolerance. However, the role of OCT1 in metformin pharmacokinetics has been extensively studied and the data are unclear. There were no differences in the pharmacokinetic properties of metformin between Oct1 (-/-) and Oct1 (+/+) mice after oral application (42). On the other hand, results of the studies on OCT1 reduced-function variants and metformin pharmacokinetics in humans have been contradictory (32, 33, 42). In a study by Christensen et al, the number of reduced-function alleles was associated with lower trough metformin levels (33). It has been suggested that decrease in metformin levels could be a combined result of reduced intestinal absorption, an increased renal clearance (32), and decreased distribution (33). In contrast, in a study in 20 healthy volunteers, patients carrying one or more OCT1 reduced-function alleles had slightly higher metformin plasma concentrations than patients with wild-type OCT1 (42). However, in this study, only two patients had two inactive alleles, and it has been suggested that major changes may be seen only in subjects carrying two OCT1 low-activity alleles (recessive model) (33, 43). This is in line with our results, as a significant effect of OCT1 genotype on metformin intolerance was only observed in the recessive model. Additional studies are needed to clarify contradictory findings on the effect of OCT1 variants on metformin pharmacokinetics, as well as response (16, 28, 33).
We show that use of concomitant drugs that inhibit OCT1 transport in vitro increases risk of metformin intolerance. Many drugs used clinically have been identified as OCT1 inhibitors in vitro (17). We included ten drugs and two drug classes in our analysis, based on their frequency of use in the study cohort and their reported half maximal inhibitory concentrations (IC50). Strong inhibitory effects on OCT1-mediated metformin transport in vitro were demonstrated for verapamil (20), rosiglitazone (23) , PPIs (21) and clopidogrel (22). For other included drugs/drugs classes in our study, in vitro inhibition measurements with metformin as OCT1 substrate were not performed. However, based on the IC50 values for 4-(4-(dimethylamino)styryl)-N-methylpyridinium (ASP) transport, spironolactone, most of the TCAs, diltiazem, doxazosin and citalopram were classified as strong OCT1 inhibitors (19, 20), while quinine and tramadol were weaker inhibitors (17, 19, 24). A recent study showed that codeine could also significantly inhibit OCT1-mediated uptake (25). Metformin transport however may be more sensitive to inhibition compared to the model substrates because of its lower apparent affinity (20).
We see striking differences between drug classes, with large effects particularly seen for verapamil, PPIs, doxazosin, codeine and citalopram. We report the largest effect of co-prescribing on metformin intolerance with verapamil use, with an OR of 7.44 for intolerance. This is in keeping with the in vitro findings showing a very potent inhibitory effect of verapamil on OCT1 (20). Whilst it is possible that the use of PPIs may reflect GI problems prior to starting metformin, we show that the use of H2RAs, that are used for the same indication as PPIs, are not associated with intolerance. The effect of all OCT1 inhibiting drugs remains significant despite removal of PPI use from the analysis (OR=1.46, 95% CI 1.08-1.99, p=0.016), and there is no reason why patients selected to be treated with drugs such as verapamil, doxazosin, or citalopram should have increased GI symptoms, other than by interaction with metformin use.
It is possible that the drugs reported to inhibit OCT1 also inhibit other cation-selective transporters involved in metformin absorption, or distribution and clearance, thus contributing to metformin intolerance in addition to OCT1 inhibition alone. This may explain the additional impact of use of these drugs on intolerance risk, in patients already carrying two loss-of-function OCT1 variants. Although most of the studied inhibiting drugs have a higher affinity to OCT1 compared to OCT2 and OCT3 (17, 44), PPIs are shown to inhibit OCT1, OCT2, and OCT3 with similar IC50 values (21). There is also a large overlapping of substrates and inhibitors between PMAT and OCTs (45). Although extensive in vitro data for potential PMAT inhibitors are lacking, verapamil for instance, exhibited similar inhibitory potencies toward PMAT and OCTs (45).
We observed higher use of antiemetic drugs (drugs used in nausea and vertigo) in intolerant group prior to and after metformin initiation. It is unclear the reason for this and it may reflect the small numbers in these groups. However, adjustment for prior use of antiemetics, as well as exclusion of these patients, did not affect significantly our results. From 14 intolerant patients with prior use of these drugs, 8 were treated with prochlorperazine and 4 with domperidone. It is interesting, however, that prochlorperazine showed also weak inhibition of ASP transport by OCT1 (19), and domperidone has recently been identified as OCT2 inhibitor (46).
Other identified significant risk factors for metformin intolerance in our study were older age and female sex, with a trend towards lower weight. Estimated creatinine clearance was calculated based on an equation with these three variables, and thus was significantly different between the two groups, although creatinine levels were similar. Our results are in agreement with the findings of a small study that investigated genetics of common metformin GI side-effects (36). The presence of side-effects in this prospective study correlated positively with age, and 76% of 53 cases were women. Interestingly, the authors analysed 7 variants in OCT1, OCT2 and MATE1 genes, reporting that, of these, two common OCT1 polymorphisms in high linkage disequilibrium with each other were associated with increased prevalence of metformin side-effects (M408V and 8 bp insertion), while R61C, M420del, and G465R variants, or OCT1 haplotypes, showed no association (36). However, M408V showed normal uptake of metformin in vitro (16), and the function of 8 bp insertion is unknown. In contrast to our study, the phenotype used was that of mild intolerance, and no interacting drugs were studied. The small sample size of the previous study means the power to identify effects of the OCT1 reduced-function variants was limited.
The main limitation of our study is the surrogate phenotype of metformin GI intolerance based on discontinuation of the drug in the first months of treatment. However, patients were switched to another oral hypoglycemic agent, so they did not stop metformin because of sufficient glycaemic control; and their HbA1c values were lower than in tolerant patients, suggesting they did not stop metformin because of insufficient glycaemic control in the first months either. However, we cannot rule out other possible reasons for stopping the drug, mainly other non-gastrointestinal side-effects, e.g. rash or headache, or other reasons unrelated to metformin intolerance. Nevertheless, GI symptoms are the most common side-effects of metformin, and 5% of patients are not able to tolerate treatment because of their severity (2). This number is in line with our findings, as we identified 251 from a total of 6,265 incident metformin users as intolerant (4%). We also show that the intolerant group have increased use of anti-diarrheal drugs, and tendency of higher use of other GI drugs, after metformin initiation, supporting that they are having GI problems. Using a similar approach, we successfully developed proxy phenotype for statin intolerance previously (47).
Our study has potential clinical impact. Our data suggest that concomitant therapy with OCT1 inhibiting drugs, like PPIs and verapamil, is a risk factor for developing intolerance, and this could be avoided by prescribing alternative medications, that do not interact with OCT1, to patients suffering from side-effects. This is particularly true for the 8% of the population with two inactive OCT1 alleles, who are over four fold more likely to develop intolerance with co-prescribed OCT1 inhibiting drugs. Avoiding OCT1 inhibiting drugs in these individuals would prevent reduced efficacy associated with suboptimal metformin dosage, and potential cessation of metformin treatment and its substitution with second-line therapies. This needs to be established in a clinical trial before such recommendation can be implemented into practice.
In conclusion, we have identified age, female sex, reduced-function alleles of OCT1 and the concomitant use of OCT1 inhibiting medications, as risk factors for metformin intolerance. Future prospectively designed studies are needed to substantiate our findings and to identify other possible predictive biomarkers of metformin intolerance.
Supplementary Material
Supplementary Table 1 Use of gastrointestinal medications in metformin intolerant and tolerant patients
Supplementary Table 2 Number of patients treated concomitantly with different OCT1 inhibiting drugs in metformin intolerant and tolerant group.
Supplementary Table 3 Haplotype analysis of the five functional variants in the OCT1 gene for directly genotyped individuals in the GoDARTS (n=3023)
Supplementary Table 4 Classification of individuals directly genotyped for the five OCT1 SNPs according to the number of deficient OCT1 haplotypes
Supplementary Table 5 OCT1 combined genotype frequencies in metformin tolerant and intolerant patients
Supplementary Table 6 Conditional logistic regression model of metformin intolerance after matching patients for age and sex
Supplementary Table 7 Joint effects of OCT1 genotype and OCT1 interacting drugs on intolerance after matching patients for age and sex
Supplementary Table 8 Sensitivity analysis - logistic regression model of metformin intolerance for a subset of directly genotyped patients
Supplementary Figure 1 Effects of individual OCT1 inhibiting drugs/drug classes on metformin intolerance risk after matching patients for age and sex. Analysis was adjusted for weight and co-treatment with other OCT1 inhibitors. TCAs, tricyclic antidepressants; PPIs, proton pump inhibitors; OR, odds ratio; LCL, lower confidence limit; UCL, higher confidence limit.
Acknowledgements
We are grateful to all the participants who took part in this study, to the general practitioners, to the Scottish School of Primary Care for their help in recruiting the participants, and to the whole team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses.
Funding. The Wellcome Trust United Kingdom Type 2 Diabetes Case Control Collection (GoDARTS) cohort collection was funded by The Wellcome Trust and informatics support is provided by the Chief Scientist Office, Scotland. This work was specifically supported by an European Foundation for the Study of Diabetes Albert Renold Travel Fellowship to T.D. E.R.P. holds a Wellcome Trust New Investigator Award (102820/Z/13/Z).
Footnotes
Duality of Interest. We declare no conflict of interest.
REFERENCES
- 1.Inzucchi SE, Bergenstal RM, Buse JB, Diamant M, Ferrannini E, Nauck M, Peters AL, Tsapas A, Wender R, Matthews DR. Management of hyperglycemia in type 2 diabetes: a patient-centered approach: position statement of the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD) Diabetes Care. 2012;35:1364–1379. doi: 10.2337/dc12-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirpichnikov D, McFarlane SI, Sowers JR. Metformin: an update. Ann Intern Med. 2002;137:25–33. doi: 10.7326/0003-4819-137-1-200207020-00009. [DOI] [PubMed] [Google Scholar]
- 3.Mithieux G, Rajas F, Zitoun C. Glucose utilization is suppressed in the gut of insulin-resistant high fat-fed rats and is restored by metformin. Biochem Pharmacol. 2006;72:1757–1762. doi: 10.1016/j.bcp.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 4.Miller RA, Chu Q, Xie J, Foretz M, Viollet B, Birnbaum MJ. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature. 2013;494:256–260. doi: 10.1038/nature11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madiraju AK, Erion DM, Rahimi Y, Zhang XM, Braddock DT, Albright RA, Prigaro BJ, Wood JL, Bhanot S, MacDonald MJ, Jurczak MJ, Camporez JP, Lee HY, Cline GW, Samuel VT, Kibbey RG, Shulman GI. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature. 2014;510:542–546. doi: 10.1038/nature13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rena G, Pearson ER, Sakamoto K. Molecular mechanism of action of metformin: old or new insights? Diabetologia. 2013;56:1898–1906. doi: 10.1007/s00125-013-2991-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scarpello JH, Howlett HC. Metformin therapy and clinical uses. Diab Vasc Dis Res. 2008;5:157–167. doi: 10.3132/dvdr.2008.027. [DOI] [PubMed] [Google Scholar]
- 8.Florez H, Luo J, Castillo-Florez S, Mitsi G, Hanna J, Tamariz L, Palacio A, Nagendran S, Hagan M. Impact of metformin-induced gastrointestinal symptoms on quality of life and adherence in patients with type 2 diabetes. Postgrad Med. 2010;122:112–120. doi: 10.3810/pgm.2010.03.2128. [DOI] [PubMed] [Google Scholar]
- 9.Bouchoucha M, Uzzan B, Cohen R. Metformin and digestive disorders. Diabetes Metab. 2011;37:90–96. doi: 10.1016/j.diabet.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Wilcock C, Bailey CJ. Accumulation of metformin by tissues of the normal and diabetic mouse. Xenobiotica. 1994;24:49–57. doi: 10.3109/00498259409043220. [DOI] [PubMed] [Google Scholar]
- 11.Bailey CJ, Wilcock C, Scarpello JH. Metformin and the intestine. Diabetologia. 2008;51:1552–1553. doi: 10.1007/s00125-008-1053-5. [DOI] [PubMed] [Google Scholar]
- 12.Zhou M, Xia L, Wang J. Metformin transport by a newly cloned proton-stimulated organic cation transporter (plasma membrane monoamine transporter) expressed in human intestine. Drug Metab Dispos. 2007;35:1956–1962. doi: 10.1124/dmd.107.015495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamichi N, Shima H, Asano S, Ishimoto T, Sugiura T, Matsubara K, Kusuhara H, Sugiyama Y, Sai Y, Miyamoto K, Tsuji A, Kato Y. Involvement of carnitine/organic cation transporter OCTN1/SLC22A4 in gastrointestinal absorption of metformin. J Pharm Sci. 2013;102:3407–3417. doi: 10.1002/jps.23595. [DOI] [PubMed] [Google Scholar]
- 14.Muller J, Lips KS, Metzner L, Neubert RH, Koepsell H, Brandsch M. Drug specificity and intestinal membrane localization of human organic cation transporters (OCT) Biochem Pharmacol. 2005;70:1851–1860. doi: 10.1016/j.bcp.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Han TK, Everett RS, Proctor WR, Ng CM, Costales CL, Brouwer KL, Thakker DR. Organic cation transporter 1 (OCT1/mOct1) is localized in the apical membrane of Caco-2 cell monolayers and enterocytes. Mol Pharmacol. 2013;84:182–189. doi: 10.1124/mol.112.084517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, Castro RA, Ianculescu AG, Yue L, Lo JC, Burchard EG, Brett CM, Giacomini KM. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest. 2007;117:1422–1431. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nies AT, Koepsell H, Damme K, Schwab M. Organic cation transporters (OCTs, MATEs), in vitro and in vivo evidence for the importance in drug therapy. Handb Exp Pharmacol. 2011:105–167. doi: 10.1007/978-3-642-14541-4_3. [DOI] [PubMed] [Google Scholar]
- 18.Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron. 1976;16:31–41. doi: 10.1159/000180580. [DOI] [PubMed] [Google Scholar]
- 19.Ahlin G, Karlsson J, Pedersen JM, Gustavsson L, Larsson R, Matsson P, Norinder U, Bergstrom CA, Artursson P. Structural requirements for drug inhibition of the liver specific human organic cation transport protein 1. J Med Chem. 2008;51:5932–5942. doi: 10.1021/jm8003152. [DOI] [PubMed] [Google Scholar]
- 20.Ahlin G, Chen L, Lazorova L, Chen Y, Ianculescu AG, Davis RL, Giacomini KM, Artursson P. Genotype-dependent effects of inhibitors of the organic cation transporter, OCT1: predictions of metformin interactions. Pharmacogenomics J. 2011;11:400–411. doi: 10.1038/tpj.2010.54. [DOI] [PubMed] [Google Scholar]
- 21.Nies AT, Hofmann U, Resch C, Schaeffeler E, Rius M, Schwab M. Proton pump inhibitors inhibit metformin uptake by organic cation transporters (OCTs) PLoS One. 2011;6:e22163. doi: 10.1371/journal.pone.0022163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Song F, Tu M, Wang K, Zhao L, Wu X, Zhou H, Xia Z, Jiang H. In vitro interaction of clopidogrel and its hydrolysate with OCT1, OCT2 and OAT1. Int J Pharm. 2014;465:5–10. doi: 10.1016/j.ijpharm.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Bachmakov I, Glaeser H, Fromm MF, Konig J. Interaction of oral antidiabetic drugs with hepatic uptake transporters: focus on organic anion transporting polypeptides and organic cation transporter 1. Diabetes. 2008;57:1463–1469. doi: 10.2337/db07-1515. [DOI] [PubMed] [Google Scholar]
- 24.Tzvetkov MV, Saadatmand AR, Lotsch J, Tegeder I, Stingl JC, Brockmoller J. Genetically polymorphic OCT1: another piece in the puzzle of the variable pharmacokinetics and pharmacodynamics of the opioidergic drug tramadol. Clin Pharmacol Ther. 2011;90:143–150. doi: 10.1038/clpt.2011.56. [DOI] [PubMed] [Google Scholar]
- 25.Tzvetkov MV, dos Santos Pereira JN, Meineke I, Saadatmand AR, Stingl JC, Brockmoller J. Morphine is a substrate of the organic cation transporter OCT1 and polymorphisms in OCT1 gene affect morphine pharmacokinetics after codeine administration. Biochem Pharmacol. 2013;86:666–678. doi: 10.1016/j.bcp.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 26.Minematsu T, Giacomini KM. Interactions of tyrosine kinase inhibitors with organic cation transporters and multidrug and toxic compound extrusion proteins. Mol Cancer Ther. 2011;10:531–539. doi: 10.1158/1535-7163.MCT-10-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzvetkov MV, Saadatmand AR, Bokelmann K, Meineke I, Kaiser R, Brockmoller J. Effects of OCT1 polymorphisms on the cellular uptake, plasma concentrations and efficacy of the 5-HT(3) antagonists tropisetron and ondansetron. Pharmacogenomics J. 2012;12:22–29. doi: 10.1038/tpj.2010.75. [DOI] [PubMed] [Google Scholar]
- 28.Zhou K, Donnelly LA, Kimber CH, Donnan PT, Doney AS, Leese G, Hattersley AT, McCarthy MI, Morris AD, Palmer CN, Pearson ER. Reduced-function SLC22A1 polymorphisms encoding organic cation transporter 1 and glycemic response to metformin: a GoDARTS study. Diabetes. 2009;58:1434–1439. doi: 10.2337/db08-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzvetkov MV, Vormfelde SV, Balen D, Meineke I, Schmidt T, Sehrt D, Sabolic I, Koepsell H, Brockmoller J. The effects of genetic polymorphisms in the organic cation transporters OCT1, OCT2, and OCT3 on the renal clearance of metformin. Clin Pharmacol Ther. 2009;86:299–306. doi: 10.1038/clpt.2009.92. [DOI] [PubMed] [Google Scholar]
- 33.Christensen MM, Brasch-Andersen C, Green H, Nielsen F, Damkier P, Beck-Nielsen H, Brosen K. The pharmacogenetics of metformin and its impact on plasma metformin steady-state levels and glycosylated hemoglobin A1c. Pharmacogenet Genomics. 2011;21:837–850. doi: 10.1097/FPC.0b013e32834c0010. [DOI] [PubMed] [Google Scholar]
- 34.Todd JN, Florez JC. An update on the pharmacogenomics of metformin: progress, problems and potential. Pharmacogenomics. 2014;15:529–539. doi: 10.2217/pgs.14.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou K, Bellenguez C, Spencer CC, Bennett AJ, Coleman RL, Tavendale R, Hawley SA, Donnelly LA, Schofield C, Groves CJ, Burch L, Carr F, Strange A, Freeman C, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin A, Craddock N, Deloukas P, Dronov S, Duncanson A, Edkins S, Gray E, Hunt S, Jankowski J, Langford C, Markus HS, Mathew CG, Plomin R, Rautanen A, Sawcer SJ, Samani NJ, Trembath R, Viswanathan AC, Wood NW, Harries LW, Hattersley AT, Doney AS, Colhoun H, Morris AD, Sutherland C, Hardie DG, Peltonen L, McCarthy MI, Holman RR, Palmer CN, Donnelly P, Pearson ER. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet. 2011;43:117–120. doi: 10.1038/ng.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarasova L, Kalnina I, Geldnere K, Bumbure A, Ritenberga R, Nikitina-Zake L, Fridmanis D, Vaivade I, Pirags V, Klovins J. Association of genetic variation in the organic cation transporters OCT1, OCT2 and multidrug and toxin extrusion 1 transporter protein genes with the gastrointestinal side effects and lower BMI in metformin-treated type 2 diabetes patients. Pharmacogenet Genomics. 2012;22:659–666. doi: 10.1097/FPC.0b013e3283561666. [DOI] [PubMed] [Google Scholar]
- 37.Jonker JW, Wagenaar E, Mol CA, Buitelaar M, Koepsell H, Smit JW, Schinkel AH. Reduced hepatic uptake and intestinal excretion of organic cations in mice with a targeted disruption of the organic cation transporter 1 (Oct1 [Slc22a1]) gene. Mol Cell Biol. 2001;21:5471–5477. doi: 10.1128/MCB.21.16.5471-5477.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang DS, Jonker JW, Kato Y, Kusuhara H, Schinkel AH, Sugiyama Y. Involvement of organic cation transporter 1 in hepatic and intestinal distribution of metformin. J Pharmacol Exp Ther. 2002;302:510–515. doi: 10.1124/jpet.102.034140. [DOI] [PubMed] [Google Scholar]
- 39.Cubeddu LX, Bonisch H, Gothert M, Molderings G, Racke K, Ramadori G, Miller KJ, Schworer H. Effects of metformin on intestinal 5-hydroxytryptamine (5-HT) release and on 5-HT3 receptors. Naunyn Schmiedebergs Arch Pharmacol. 2000;361:85–91. doi: 10.1007/s002109900152. [DOI] [PubMed] [Google Scholar]
- 40.Carter D, Howlett HC, Wiernsperger NF, Bailey CJ. Differential effects of metformin on bile salt absorption from the jejunum and ileum. Diabetes Obes Metab. 2003;5:120–125. doi: 10.1046/j.1463-1326.2003.00252.x. [DOI] [PubMed] [Google Scholar]
- 41.Napolitano A, Miller S, Nicholls AW, Baker D, Van Horn S, Thomas E, Rajpal D, Spivak A, Brown JR, Nunez DJ. Novel gut-based pharmacology of metformin in patients with type 2 diabetes mellitus. PLoS One. 2014;9:e100778. doi: 10.1371/journal.pone.0100778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shu Y, Brown C, Castro RA, Shi RJ, Lin ET, Owen RP, Sheardown SA, Yue L, Burchard EG, Brett CM, Giacomini KM. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin Pharmacol Ther. 2008;83:273–280. doi: 10.1038/sj.clpt.6100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zolk O. Current understanding of the pharmacogenomics of metformin. Clin Pharmacol Ther. 2009;86:595–598. doi: 10.1038/clpt.2009.144. [DOI] [PubMed] [Google Scholar]
- 44.Koepsell H, Lips K, Volk C. Polyspecific organic cation transporters: structure, function, physiological roles, and biopharmaceutical implications. Pharm Res. 2007;24:1227–1251. doi: 10.1007/s11095-007-9254-z. [DOI] [PubMed] [Google Scholar]
- 45.Engel K, Wang J. Interaction of organic cations with a newly identified plasma membrane monoamine transporter. Mol Pharmacol. 2005;68:1397–1407. doi: 10.1124/mol.105.016832. [DOI] [PubMed] [Google Scholar]
- 46.Wittwer MB, Zur AA, Khuri N, Kido Y, Kosaka A, Zhang X, Morrissey KM, Sali A, Huang Y, Giacomini KM. Discovery of potent, selective multidrug and toxin extrusion transporter 1 (MATE1, SLC47A1) inhibitors through prescription drug profiling and computational modeling. J Med Chem. 2013;56:781–795. doi: 10.1021/jm301302s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donnelly LA, Doney AS, Tavendale R, Lang CC, Pearson ER, Colhoun HM, McCarthy MI, Hattersley AT, Morris AD, Palmer CN. Common nonsynonymous substitutions in SLCO1B1 predispose to statin intolerance in routinely treated individuals with type 2 diabetes: a go-DARTS study. Clin Pharmacol Ther. 2011;89:210–216. doi: 10.1038/clpt.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 Use of gastrointestinal medications in metformin intolerant and tolerant patients
Supplementary Table 2 Number of patients treated concomitantly with different OCT1 inhibiting drugs in metformin intolerant and tolerant group.
Supplementary Table 3 Haplotype analysis of the five functional variants in the OCT1 gene for directly genotyped individuals in the GoDARTS (n=3023)
Supplementary Table 4 Classification of individuals directly genotyped for the five OCT1 SNPs according to the number of deficient OCT1 haplotypes
Supplementary Table 5 OCT1 combined genotype frequencies in metformin tolerant and intolerant patients
Supplementary Table 6 Conditional logistic regression model of metformin intolerance after matching patients for age and sex
Supplementary Table 7 Joint effects of OCT1 genotype and OCT1 interacting drugs on intolerance after matching patients for age and sex
Supplementary Table 8 Sensitivity analysis - logistic regression model of metformin intolerance for a subset of directly genotyped patients
Supplementary Figure 1 Effects of individual OCT1 inhibiting drugs/drug classes on metformin intolerance risk after matching patients for age and sex. Analysis was adjusted for weight and co-treatment with other OCT1 inhibitors. TCAs, tricyclic antidepressants; PPIs, proton pump inhibitors; OR, odds ratio; LCL, lower confidence limit; UCL, higher confidence limit.