ABSTRACT
A rapid and global emergence of azole resistance has been observed in the pathogenic fungus Aspergillus fumigatus over the past decade. The dominant resistance mechanism appears to be of environmental origin and involves mutations in the cyp51A gene, which encodes a protein targeted by triazole antifungal drugs. Whole-genome sequencing (WGS) was performed for high-resolution single-nucleotide polymorphism (SNP) analysis of 24 A. fumigatus isolates, including azole-resistant and susceptible clinical and environmental strains obtained from India, the Netherlands, and the United Kingdom, in order to assess the utility of WGS for characterizing the alleles causing resistance. WGS analysis confirmed that TR34/L98H (a mutation comprising a tandem repeat [TR] of 34 bases in the promoter of the cyp51A gene and a leucine-to-histidine change at codon 98) is the sole mechanism of azole resistance among the isolates tested in this panel of isolates. We used population genomic analysis and showed that A. fumigatus was panmictic, with as much genetic diversity found within a country as is found between continents. A striking exception to this was shown in India, where isolates are highly related despite being isolated from both clinical and environmental sources across >1,000 km; this broad occurrence suggests a recent selective sweep of a highly fit genotype that is associated with the TR34/L98H allele. We found that these sequenced isolates are all recombining, showing that azole-resistant alleles are segregating into diverse genetic backgrounds. Our analysis delineates the fundamental population genetic parameters that are needed to enable the use of genome-wide association studies to identify the contribution of SNP diversity to the generation and spread of azole resistance in this medically important fungus.
IMPORTANCE
Resistance to azoles in the ubiquitous ascomycete fungus A. fumigatus was first reported from clinical isolates collected in the United States during the late 1980s. Over the last decade, an increasing number of A. fumigatus isolates from the clinic and from nature have been found to show resistance to azoles, suggesting that resistance is emerging through selection by the widespread usage of agricultural azole antifungal compounds. Aspergillosis is an emerging clinical problem, with high rates of treatment failures necessitating the development of new techniques for surveillance and for determining the genome-wide basis of azole resistance in A. fumigatus.
INTRODUCTION
Fungal infections are increasingly recognized as a threat to human (1, 2), animal, and plant (3) populations. Aspergillus species are ubiquitous, saprophytic fungi with airborne conidia that grow on organic matter. Aspergillus fumigatus is the principal causative agent of human aspergillosis, which can range from allergic syndromes to invasive aspergillosis (IA), a life-threatening infection in immunocompromised hosts. Profoundly neutropenic patients receiving chemotherapy or hematopoietic stem cell or solid organ transplantation are at high risk of IA (4). Oral triazole antifungal drugs (itraconazole, voriconazole, and posaconazole) are effective against A. fumigatus and remain the front-line therapy in the management and prophylaxis of IA (5). However, the emergence of azole resistance in A. fumigatus isolates from Europe, Asia, the Middle East, and recently, Africa (6–8) is a global and evolving public health problem (9), although surprisingly, the United States seems to be exempt from this increase in azole resistance (10). Treatment failure due to azole-resistant strains is a major clinical concern in the management of patients with IA, with high rates of patient mortality being observed (11–13).
The true prevalence of azole resistance in Aspergillus species is largely unknown; however, frequencies of resistance varying between 0 and 6% have been determined for different clinical centers (14). Azole-resistant A. fumigatus strains have been isolated from both azole-naive and -exposed patients, as well as broadly from the natural environment in several countries. This has led to the hypothesis that the widespread use of structurally similar azoles in agriculture has led to increases in resistance in the clinical context (9, 15, 16).
The molecular basis of resistance to triazoles in A. fumigatus mainly involves point mutations at several codons in the cyp51A gene, which encodes lanosterol 14-α-sterol demethylase, combined with tandem repeats in the promoter region (17, 18). This enzyme is required for the biosynthesis of ergosterol, an essential component of the fungal cell membrane. Single-nucleotide polymorphisms (SNPs) in cyp51A cause structural alterations due to amino acid substitutions. Numerous SNPs that confer increased resistance to triazoles in vitro have been reported in this gene in A. fumigatus (19–23). However, a predominant resistance mutation, TR34/L98H, consists of a tandem repeat (TR) of 34 bases in the promoter of the cyp51A gene and a leucine-to-histidine change at codon 98 (24); this mutation is globally widespread in environmental isolates of the fungus. Recently, a novel cyp51A-mediated resistance mutation that leads to high-level voriconazole resistance, TR46/Y121F/T289A, has been described as occurring in environmental and clinical isolates of A. fumigatus found in the Netherlands (25), Belgium (26), Denmark (27), Germany (28), Tanzania (7), and India (29).
Typing methods, such as multilocus sequence typing (MLST), have fallen out of favor due to their low discriminatory power and considerable cost (30); the Aspergillus fumigatus MLST database is no longer curated, and higher-resolution methods are becoming more widely accessible for the analysis of small eukaryotic genomes due to falling costs (31). Driven by rapid technological advances, the application of whole-genome sequencing (WGS) to type microbial genomes is increasingly used to improve patient care (32–36). WGS is now positioned to become an essential primary platform for the management of antimicrobial resistance, including detection and surveillance of emerging drug-resistant microorganisms (33). While WGS is still a novel tool for the growing challenge of infectious diseases, broad applications of this technology beyond bacterial and viral infections have been swiftly embraced, and WGS has recently been used to study separate outbreaks of human fungal infections using whole-genome SNP phylogenetic analysis (37–39). Therefore, WGS is now poised to overtake other typing methods due to the high resolution of the data generated and the relatively low labor and monetary costs.
In this study, we determined the genetic background upon which cyp51A mutations occur by sequencing a diverse panel of clinical and environmental isolates of this fungus, and by doing so, we have demonstrated the utility of WGS to describe the evolutionary processes that underpin the emergence of azole resistance in A. fumigatus.
RESULTS
Species identification.
Twenty-two isolates were evaluated in our study (Table 1). The isolates were identified as A. fumigatus sensu stricto based on either sequencing of the internal transcribed spacer (ITS) region and amplification of parts of the β-tubulin and calmodulin genes (for the eight isolates from India and eight isolates from the Netherlands) or matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (for the six isolates obtained from Leeds, United Kingdom). All twenty-two isolates and two control strains (NCPF 7097/AF65 and NCPF 7367/AF293) were further analyzed using whole-genome sequencing (WGS).
TABLE 1 .
Clinical and environmental isolates of A. fumigatus used in this study and details of alignments
| Country, city | Source | Yr of isolation |
Collectora | Culture reference |
Triazole resistanceb |
No. of reads aligned (millions) |
Depth of coverage (×) |
% of reference genome coveredc |
|---|---|---|---|---|---|---|---|---|
| UK, Leeds | Clinical | 2012 | R.C.B. | 12-7505446 | Resistant | 51.9 | 174.1 | 94.4 |
| UK, Leeds | Clinical | 2012 | R.C.B. | 12-7505220 | Resistant | 48.3 | 161.9 | 95.0 |
| UK, Leeds | Clinical | 2009 | R.C.B. | 09-7500806 | Susceptible | 43.6 | 146.7 | 95.2 |
| UK, Leeds | Clinical | 2012 | R.C.B. | 12-7504652 | Susceptible | 43.7 | 145.6 | 93.1 |
| UK, Leeds | Clinical | 2012 | R.C.B. | 12-7504462 | Susceptible | 47.4 | 158.8 | 95.4 |
| UK, Leeds | Clinical | 2012 | R.C.B. | 12-7505054 | Susceptible | 50.1 | 168.7 | 93.2 |
| Netherlands, Nijmegen | Clinical | 2003 | J.F.M. | 08-12-12-13 | Resistant | 34.9 | 117.5 | 93.2 |
| Netherlands, Nijmegen | Clinical | 2005 | J.F.M. | 08-36-03-25 | Resistant | 45.2 | 152.5 | 94.8 |
| Netherlands, Nijmegen | Clinical | 2004 | J.F.M. | 08-31-08-91 | Resistant | 45.8 | 153.6 | 94.0 |
| Netherlands, Berghem | Environmental | 2008 | J.F.M. | 08-19-02-61 | Resistant | 51.7 | 173.2 | 93.7 |
| Netherlands, Berghem | Environmental | 2008 | J.F.M. | 08-19-02-30 | Susceptible | 44.5 | 150.1 | 94.8 |
| Netherlands, Nijmegen | Clinical | 2010 | J.F.M. | 10-01-02-27 | Resistant | 46.5 | 155.9 | 94.5 |
| Netherlands, Nijmegen | Environmental | 2008 | J.F.M. | 08-19-02-46 | Resistant | 48.6 | 163.1 | 93.8 |
| Netherlands, Nijmegen | Environmental | 2008 | J.F.M. | 08-19-02-10 | Resistant | 51.8 | 173.6 | 93.5 |
| India, Delhi | Clinical | 2009 | A.C. | Afu 942/09 | Resistant | 44.2 | 148.1 | 93.6 |
| India, Delhi | Clinical | 2009 | A.C. | Afu 1042/09 | Resistant | 41.8 | 139.8 | 93.5 |
| India, Delhi | Clinical | 2011 | A.C. | Afu 343/P/11 | Resistant | 38.4 | 128.9 | 94.8 |
| India, Delhi | Clinical | 2012 | A.C. | Afu 591/12 | Resistant | 34.4 | 115.4 | 93.5 |
| India, Delhi | Environmental | 2011 | A.C. | Afu 124/E11 | Resistant | 50 | 167.1 | 93.6 |
| India, Bihar | Environmental | 2011 | A.C. | Afu 166/E11 | Resistant | 42.8 | 143.6 | 93.6 |
| India, Bihar | Environmental | 2011 | A.C. | Afu 257/E11 | Resistant | 33.8 | 113.6 | 93.5 |
| India, Delhi | Environmental | 2011 | A.C. | Afu 218/E11 | Resistant | 46.5 | 155.8 | 93.5 |
| UK | Clinical | 1997 | NCPF | AF65 (NCPF 7097) | Susceptible | 57.5 | 192.8 | 95.1 |
| UK | Clinical | 1993 | NCPF | AF293 (NCPF 7367) | Susceptible | 49.3 | 164.5 | 98.1 |
R.C.B., Richard C. Barton; J.F.M., Jacques F. Meis, A.C., Anuradha Chowdhary; NCPF, National Collection of Pathogenic Fungi.
Resistance to triazole antifungal drugs was defined as an itraconazole MIC of >2 mg/liter using CLSI broth microdilution methods.
The AF293 genome was the reference genome for the number of reads aligned, the corresponding depth of coverage, and the percentage of the reference genome covered by reads.
Antifungal susceptibility testing.
Antifungal susceptibility testing (Table 2) showed that 18 isolates had MICs above the established epidemiological cutoff value for itraconazole (≥2 mg/liter). All of the wild-type isolates revealed low MICs for azole antifungals.
TABLE 2 .
In vitro antifungal susceptibility profiles of A. fumigatus isolates and corresponding SNPs in cyp51A with nonsynonymous substitutions
| Culture reference |
MIC (mg/liter) ofa: |
cyp51A amino acid substitution at position: |
Resistance marker |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITC | VOR | POS | ISA | CAS | MFG | AFG | AMB | F46 | L98 | M172 | N248 | D255 | S297 | E427 | F495 | ||
| 12-7505446 | >16 | 1 | 0.5 | ND | 0.125 | <0.015 | <0.015 | 0.25 | Y | H | V | T | E | S | K | F | TR34/L98H |
| 12-7505220 | >16 | 1 | 0.5 | ND | 0.125 | <0.015 | <0.015 | 0.5 | Y | H | V | T | E | S | K | F | TR34/L98H |
| 09-7500806 | 1 | 0.25 | 0.06 | ND | 0.06 | <0.015 | <0.015 | 0.5 | Y | L | V | T | E | S | K | F | None |
| 12-7504652 | 1 | 0.5 | 0.25 | ND | 0.125 | <0.015 | <0.015 | 0.25 | Y | L | V | T | E | S | K | F | None |
| 12-7504462 | 0.5 | 0.125 | 0.06 | ND | 0.125 | <0.015 | <0.015 | 0.5 | Y | L | V | T | E | S | K | F | None |
| 12-7505054 | 0.5 | 0.5 | 0.06 | ND | 0.5 | 0.063 | 0.06 | 0.5 | Y | L | V | T | E | S | K | F | None |
| 08-12-12-13 | >16 | 1 | 1 | 16 | 0.5 | 0.063 | 0.031 | 0.25 | Y | H | V | T | E | T | K | I | TR34/L98H |
| 08-36-03-25 | >16 | 1 | 0.5 | 8 | 0.5 | 0.008 | 0.008 | 0.5 | Y | H | V | T | E | T | K | I | TR34/L98H |
| 08-31-08-91 | >16 | 4 | 1 | 8 | 0.25 | 0.031 | 0.016 | 0.5 | Y | H | V | T | E | S | K | F | TR34/L98H |
| 08-19-02-61 | >16 | 2 | 0.25 | 4 | 0.5 | 0.031 | 0.008 | 0.25 | Y | H | V | T | E | S | K | F | TR34/L98H |
| 08-19-02-30 | 0.25 | 0.5 | 0.06 | 0.5 | 0.5 | 0.016 | 0.016 | 0.5 | Y | L | V | T | E | S | K | F | None |
| 10-01-02-27 | >16 | 4 | 0.5 | 4 | 0.5 | 0.016 | 0.016 | 0.5 | Y | H | V | T | E | S | K | F | TR34/L98H |
| 08-19-02-46 | >16 | 4 | 0.5 | 4 | 0.5 | 0.016 | 0.016 | 0.5 | Y | H | V | T | E | S | K | F | TR34/L98H |
| 08-19-02-10 | >16 | 2 | 0.5 | 4 | 0.5 | 0.031 | 0.016 | 0.25 | Y | H | V | T | E | S | K | F | TR34/L98H |
| Afu 942/09 | >16 | 2 | 2 | 8 | 0.125 | <0.015 | <0.015 | 0.25 | Y | H | V | T | E | S | K | F | TR34/L98H |
| Afu 1042/09 | >16 | 2 | 2 | 8 | 0.06 | <0.015 | <0.015 | 0.25 | Y | H | V | T | E | S | K | F | TR34/L98H |
| Afu 343/P/11 | >16 | 8 | >8 | >8 | 0.125 | <0.015 | <0.015 | 0.125 | Y | H | V | T | E | S | K | F | TR34/L98H |
| Afu 591/12 | >16 | 8 | >8 | 2 | 0.25 | ≤0.015 | 0.06 | 0.5 | Y | H | V | T | E | S | K | F | TR34/L98H |
| Afu 124/E11 | >16 | 8 | 1 | 8 | 0.06 | <0.015 | <0.015 | 0.125 | Y | H | V | T | E | S | K | F | TR34/L98H |
| Afu 166/E11 | 16 | 16 | 2 | 8 | ND | ND | ND | ND | Y | H | V | T | E | S | K | F | TR34/L98H |
| Afu 257/E11 | >16 | 8 | 1 | >8 | 0.125 | <0.015 | <0.015 | 0.125 | Y | H | V | T | E | S | K | F | TR34/L98H |
| Afu 218/E11 | >16 | 8 | 1 | >8 | 0.125 | <0.015 | <0.015 | 0.125 | Y | H | V | T | E | S | K | F | TR34/L98H |
| AF65 | 0.5 | 0.125 | 0.06 | ND | 0.125 | <0.015 | <0.015 | 0.5 | Y | L | V | T | E | S | K | F | None |
| AF293 | 0.5 | 0.25 | 0.06 | ND | 0.125 | <0.015 | <0.015 | 0.25 | F | L | M | N | D | S | E | F | None |
ITC, itraconazole; VOR, voriconazole; POS, posaconazole; ISA, isavuconazole; CAS, caspofungin; MFG, micafungin; AFG, anidulafungin; AMB, amphotericin B; ND, not done.
Sequence analysis.
All 24 genome sequences mapped to >92% of the de novo AF293 reference genome with >100× coverage for all isolates (average of 152.7×) (Table 1). The raw sequence reads covered at least 93.1% of the AF293 reference genome for all isolates (Table 1); 98.1% of the AF293 reference genome was covered by sequence reads from the resequenced AF293 isolate. We found that 73 SNPs were also called in the resequenced AF293 isolate (data not shown), reflecting mutations due to repeated subculture (40) or sequencing errors.
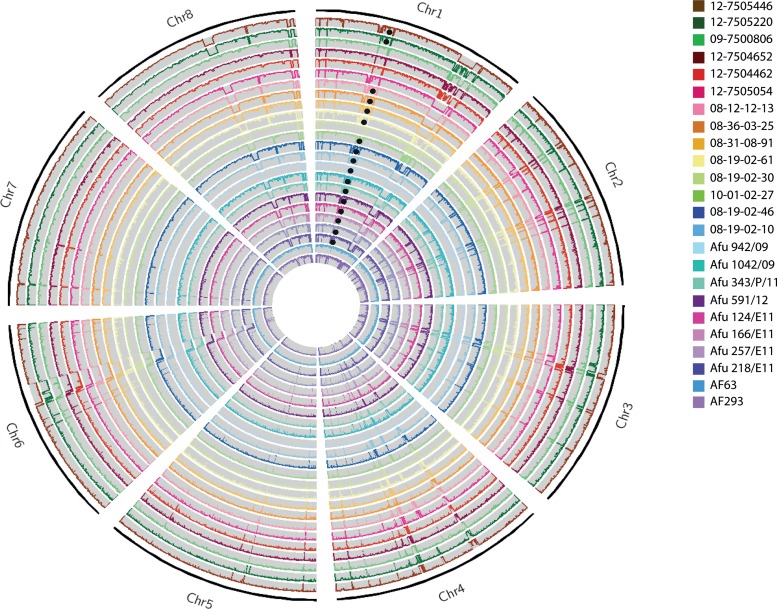
The normalized whole-genome depth of coverage was plotted to observe any possible ploidy events (Fig. 1). While none of the 24 A. fumigatus isolates sequenced in this study displayed copy number variation (CNV) or chromosomal CNVs, large-scale deletions in multiple chromosomes were observed. Surprisingly, deletions of over 300,000 bp were seen in all isolates except AF293 and AF65 in both chromosomes 1 and 6, accounting for the losses of 129 and 124 genes, respectively. No enrichment for a particular biological process or function was found for the genes deleted in either chromosome. However, three genes within these deletions have known functions associated with azole antifungals: Afu1g14330, an ABC transporter involved in fluconazole transport; Afu1g14050, an F-box protein whose transcript is induced by voriconazole; and Afu6g10130, an uncharacterized open reading frame (ORF) whose transcript is downregulated in response to voriconazole. A smaller deletion of 60,000 bp was observed in chromosome 8 in all isolates except 09-7500806, 12-7504652, 12-7504462, and AF65; this deletion covered a region of the genome encoding 16 genes. As for the other deletions, there was no enrichment for biological process or function in this set of deleted genes.
FIG 1 .
Circos (72) image of normalized whole-genome depth of coverage of all 24 A. fumigatus isolates (plotted as listed in the key), averaged over 10,000-bp bins. Black circles mark the presence of the TR34/L98H mutation. Chromosomes 1 and 6 show large deletions spanning >300 kbp in most isolates, except AF65 and AF293, while chromosome 8 displays a 60-kbp deletion in all isolates except AF65, 09-7500806, 12-7504652, and 12-7504462.
SNP analysis.
Among all 24 genomes, a total of 1,895,038 SNPs were identified, 217,498 of which were common to one or more isolates. All SNPs in cyp51A that caused a nonsynonymous amino acid substitution are summarized in Table 2. The genomes of 16 itraconazole (ITC)-resistant isolates showed the presence of the L98H substitution. The genomes of six azole-susceptible isolates (09-7500806, 12-7504652, 12-7504462, 12-7505054, 08-19-02-30, and AF65) and one control isolate, the AF293 reference genome, had no such mutations. With the exception of L98H, the SNPs detected in cyp51A, including F495I, K427E, S297T, E255D, T248N, V172M, and Y46F, have previously been observed among both azole-sensitive and -resistant A. fumigatus strains. As shown by the data in Table 2, all 17 isolates for which the itraconazole MICs were ≥4 mg/liter had the L98H amino acid substitution in addition to the 34-base-pair nucleotide tandem repeat in their promoter region that has been previously associated with itraconazole resistance. All of the isolates for which itraconazole MICs were ≤1 mg/liter had a wild-type (i.e., that seen in reference strain AF293) cyp51A amino acid sequence or had the Y46F, V172M, T248N, E255D, S297T, K427E, and F495I amino acid substitutions that were present in both fully susceptible and resistant strains.
Phylogenetic analysis.
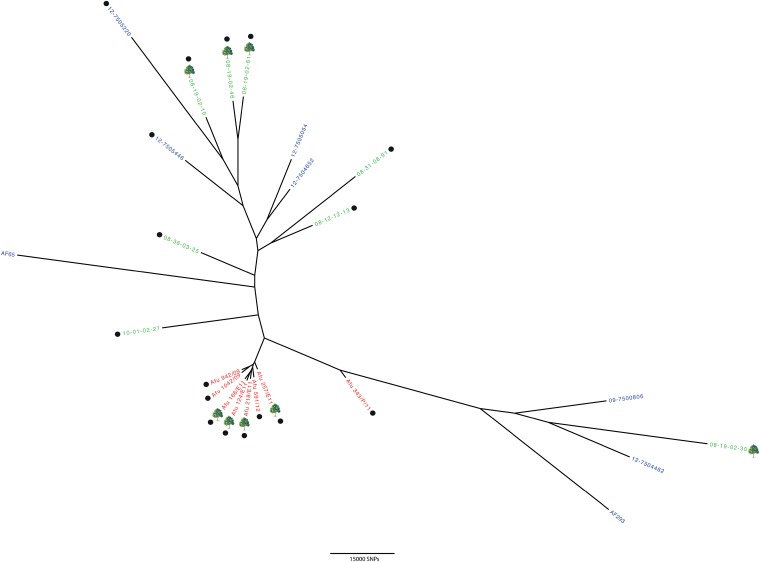
The 24 genomes were analyzed by whole-genome SNP phylogenetic analysis using RAxML (41). Phylogenetic analysis showed all Indian isolates to be very highly related to each other, much more so than isolates from the Netherlands or the United Kingdom (Fig. 2); this is with the exception of Afu 343/P/11, which is a clinical isolate and may have been acquired somewhere other than India (for instance through migrant working or tourism). Except for the tightly clustered isolates from India, there was no discernible relationship between the geographical location and the genotype of the cultured isolate. The phylogeny showed that clinical and environmental isolates were genotypically indistinguishable. Furthermore, phylogenetic analysis was unable to discriminate between azole-resistant and azole-susceptible strains, showing that the TR34/L98H allele occurs in diverse genetic backgrounds for both clinical and environmental isolates.
FIG 2 .
Phylogenetic analysis of A. fumigatus isolates representing azole-resistant and -susceptible genotypes from India, the Netherlands, and the United Kingdom. Bootstrap analysis was performed on WGST SNP data from 24 A. fumigatus genomes to generate an unrooted maximum-likelihood phylogeny, with all branches supported to 87% or higher. Isolates are color coded according to country of origin (red, India; green, the Netherlands; blue, United Kingdom), and environmental isolates are indicated by a tree symbol. Isolates marked with a black circle contain the TR34/L98H mutation in the cyp51A gene. Branch lengths represent the numbers of SNPs between taxa.
Genetic diversity.
When compared against the reference isolate AF293, isolates from the Netherlands and the United Kingdom had 155,728 and 148,979 high-confidence SNPs, respectively, whereas there were 83,447 high-confidence SNPs within the Indian isolates. The average pairwise SNP diversity was higher in isolates collected from the United Kingdom (27,914) and the Netherlands (23,623) than in those from India (5,100), showing that this population has less genetic diversity despite being collected across a wider geographic range. Interestingly, 72,065 SNPs were found to be common to all three countries, accounting for 86.36% of all Indian SNPs. However, as only ~2,500 SNPs (Table 3) separate the main cluster of Indian isolates (excluding the outlier Afu 343/P/11), it is clear that the Indian isolates are genetically depauperate, manifesting less than 10% of the diversity seen in Europe.
TABLE 3 .
SNP differences between each Indian A. fumigatus isolate, shown as unique SNPs between isolates
| Isolate | No. of SNP differences between isolate to left and: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Afu 942/09 | Afu 1042/09 | Afu 343/P/11 | Afu 591/12 | Afu 124/E11 | Afu 166/E11 | Afu 257/E11 | Afu 218/E11 | |
| Afu 942/09 | 2207 | 21,150 | 3067 | 1954 | 2406 | 2928 | 2353 | |
| Afu 1042/09 | 2233 | 21,261 | 3232 | 2148 | 2625 | 3100 | 2617 | |
| Afu 343/P/11 | 5136 | 5221 | 5479 | 4861 | 5017 | 5491 | 4941 | |
| Afu 591/12 | 1943 | 2082 | 20,369 | 1484 | 1704 | 2401 | 1615 | |
| Afu 124/E11 | 2564 | 2732 | 21,485 | 3218 | 2383 | 3359 | 2331 | |
| Afu 166/E11 | 2371 | 2564 | 20,996 | 2793 | 1738 | 2950 | 1991 | |
| Afu 257/E11 | 1645 | 1791 | 20,222 | 2242 | 1466 | 1702 | 1623 | |
| Afu 218/E11 | 2539 | 2777 | 21,141 | 2925 | 1907 | 2212 | 3092 | |
To evaluate the genetic divergence among populations, Wright’s fixation indexes (FST) were calculated for pairs of populations. The pairwise FST value for the United Kingdom and the Netherlands was zero, implying complete panmixis and interbreeding between these two populations. This finding of intermixed diversity among these two populations is supported by phylogenetic analysis (Fig. 2).
Pairwise FST values between Indian, United Kingdom, and Netherlands populations showed that there was a pronounced phylogeographic structure at the continental level, with an FST value of 0.314 for India versus the Netherlands and an FST value of 0.381 for India versus the United Kingdom.
Recombination analysis.
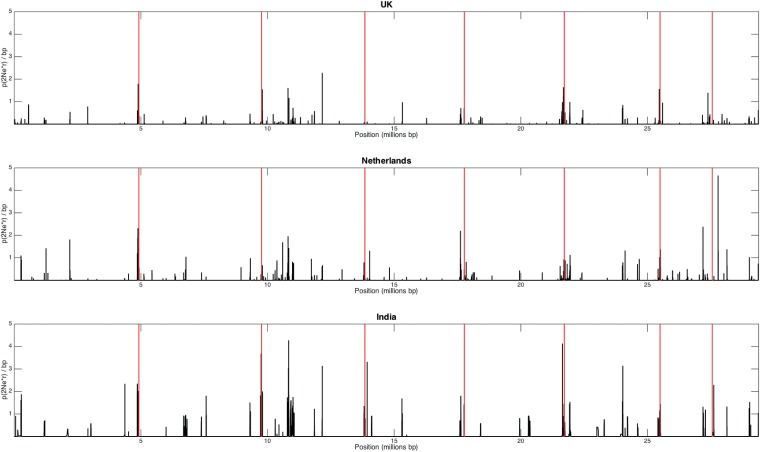
Population-wide recombination rates were estimated for each population (defined as a single country) using the program LDhat (42), which identifies patterns of linkage disequilibrium using Hudson’s composite likelihood method. The interval method used in this study specifically estimates a variable recombination rate using a Bayesian reversible-jump Markov chain Monte Carlo method under a crossing-over model. Evidence for the frequency of recombination in each population is presented in Fig. 3; the greater numbers of recombination hot spots in the United Kingdom and the Netherlands data reflect the high genetic diversity (Fig. 3). Given the lower genetic diversity observed in India (in terms of SNP numbers and also as seen in the phylogenetic analysis in Fig. 2), we anticipated this data set to be highly clonal, but on the contrary, the LDhat analysis (Fig. 3) detected similar numbers of recombination hot spots for the United Kingdom and the Netherlands, implying that the Indian data set is derived from isolates that are undergoing sexual or parasexual recombination. Overall, the rate of recombination in the Indian population (ρ = 0.0726/bp−1) was significantly higher (calculated using a t test) than the recombination rates observed in United Kingdom and Netherlands populations (ρ = 0.0070/bp−1 and ρ = 0.0104/bp−1, respectively).
FIG 3 .
Recombination analysis, using LDhat interval (42), of A. fumigatus isolates from the United Kingdom, the Netherlands, and India. The black peaks represent the recombination rate across the whole A. fumigatus genome, and the vertical red lines mark the chromosome boundaries.
In order to refute the existence of clonality in the Indian data set, we estimated the index of association (IA) and the modified statistic rBarD (73) to confirm the presence of recombination within each population of this data set. We assumed that the null hypothesis of no linkage disequilibrium and, therefore, no recombination would not be rejected if the resulting values of both statistics were not significantly different from the distribution of values obtained from 999 resamplings. For all three populations (India, United Kingdom, and the Netherlands), the null hypothesis could not be rejected, implying no significant linkage among loci, and we concluded, therefore, that all populations are recombining.
PCR and mining of de novo assembly of sequence reads has shown that both MAT1-1 and MAT1-2 loci are present in all populations investigated here (Table 4). While a chi-square test showed no significant difference in the frequency of either locus in the United Kingdom and Netherlands populations, a bias was observed in the Indian population, which contained significantly fewer MAT1-2 idiomorphs (P < 0.0304).
TABLE 4 .
Distribution of MAT1-1 and MAT1-2 idiomorphs (mating types) among isolates of A. fumigatus from three countries
| Country | % (no. of isolates) with mating type |
Chi-square value | |
|---|---|---|---|
| MAT1-1 | MAT1-2 | ||
| United Kingdom | 66.6 (4) | 33.4 (2) | 0.41 |
| The Netherlands | 62.5 (5) | 37.5 (3) | 0.47 |
| India | 87.5 (7) | 12.5 (1) | 0.034 |
DISCUSSION
To investigate the role whole-genome sequencing can play in the detection of azole resistance in A. fumigatus, we sequenced the genomes of 24 clinical and environmental isolates obtained from India, the Netherlands, and the United Kingdom. In the present study, we aimed to verify the applicability of using WGS to characterize known mutations in the cyp51A locus and to determine the genome-wide patterns of genetic diversity upon which these mutations reside.
We were able to characterize the genotype of cyp51A for all isolates in the study with high confidence. Of these, sixteen isolates with itraconazole MICs of ≥4 mg/liter had the L98H amino acid substitution accompanied by a characteristic 34-base-pair nucleotide tandem repeat in their promoter region. We did not detect any amino acid substitutions associated with resistance in cyp51A genes in the strains with itraconazole MICs of ≤1 mg/liter. Several mutations specific to the cyp51A gene, such as substitutions at codons G54, G138, P216, F219, M220, and G448, have also been described as responsible for resistance to azoles in clinical isolates of A. fumigatus (14–18), though none of these mutations were present in our analyzed sample collection. Interestingly, and in contrast with previous observations where all TR34/L98H mutations are accompanied by a pair of amino acid substitutions, S297T and F495I (44), only two of our 17 isolates with TR34/L98H showed S297T and F495I substitutions. This suggests that intragenic recombination in cyp51A may have occurred to unlink these mutations in this set of isolates. Whether or not this means that there have been multiple origins of the TR34/L98H allele will require broader WGS surveys; however, the observation that this pair of mutations has never been found unlinked to date suggests that the probability of de novo origination for this allele is low to nonexistent. In support of this interpretation, it is worth noting that a novel environmental cyp51A-mediated resistance mechanism (TR46/Y121F/T289A) was recently reported in the Netherlands (25), Belgium (26), Denmark (27), Germany (28), Tanzania (7), and India (29), superimposed on a more homogenous genetic background than TR34/L98H. This recently detected resistance mechanism has been observed to rapidly increase in frequency in Dutch hospitals and homes, where it is associated with voriconazole therapy failure (6, 25). Therefore, it appears that the contemporary rise in the frequency of TR34/L98H is not a unique event and that the emergence of A. fumigatus isolates in nature showing cyp51A-associated resistance alleles is an ongoing phenomenon.
Our demonstration of high levels of SNP diversity and a panmictic population structure within the United Kingdom and Netherlands is testament to the ability of this highly aerosolized (airborne) fungus to passively disperse. Our findings suggest that the TR34/L98H allele has existed long enough for the twin forces of recombination and gene flow to homogenize the genetic background upon which it occurs in Europe. The short branch lengths and high numbers of shared SNPs show that Indian isolates of A. fumigatus are broadly similar to those in Europe. However, and in sharp contrast, the Indian isolates show a far higher level of relatedness, manifesting less than 10% of the genetic diversity that is observed within the Netherlands and United Kingdom. While we have not sampled enough of the population space to delineate the full population structure and our collection is biased to isolates that are azole resistant, the A. fumigatus isolates from India that we sequenced were collected from locations separated by over 1,000 km. This suggests that there has been a recent expansion of this genotype within India and it is likely not a coincidence that these genotypes all contain the TR34/L98H allele. To further investigate the possibility that this allele has been introduced into the Indian genetic background recently, further sampling and sequencing of both azole-resistant and azole-sensitive isolates from this region are needed to clarify our finding.
There have been significant advances recently in our understanding of azole resistance mechanisms in other pathogenic fungi, such as Candida albicans (45) and Cryptococcus neoformans (46). In both species, aneuploidy for the chromosome bearing the ERG11 gene has been reported and found to be integral to antifungal resistance. We did not detect any chromosomal aneuploidy or copy number variation for cyp51A or cyp51B, the ortholog of ERG11, in any of the A. fumigatus isolates investigated here. However, we did detect large deletions in chromosomes 1, 6, and 8 in many of the isolates (Fig. 1). Analyzing the gene content of the deleted regions revealed two genes in chromosome 1 (Afu1g14330, an ABC transporter, and Afu1g14050, encoding an F-box protein) and one gene in chromosome 6 (Afu6g10130, uncharacterized but downregulated in response to voriconazole) that had a known function associated with azole antifungals. However, given the large-scale nature of the deletions (both over 300,000 bp in size), we do not believe that these deletions are part of an evolutionary process that is enabling A. fumigatus to evolve resistance to azole antifungals. Indeed, we see no deletions specific to either azole-sensitive or azole-resistant isolates, suggesting that these deletions occurred in response to a different process. Deletions on this scale have not previously been reported in A. fumigatus, representing an opportunity for further investigation.
While most mutations in azole-resistant A. fumigatus isolates were single nucleotide substitutions in the target gene (cyp51A), mutations at other genes have been reported as conferring resistance to azoles in this fungus. Furthermore, other resistance mechanisms could be responsible for azole resistance in A. fumigatus. The non-cyp51A-mediated resistance can potentially be explained by alternative resistance mechanisms that have been described previously, including (i) higher basal expression of the cdr1B efflux transporter (47), (ii) a mutation in the CCAAT-binding transcription factor complex subunit HapE (48), which was not seen in our data, and (iii) high cyp51B expression (49). A recent publication reported drug resistance due to RNA interference (RNAi)-dependent epimutations in Mucor circinelloides (50); we found five orthologs of genes involved in the RNAi pathway in Aspergillus nidulans, Cryptococcus neoformans, and Schizosaccharomyces pombe present in A. fumigatus isolate 09-7500806. These five genes (Afu3g11010, Afu8g05280, Afu5g11790, Afu4g02930, and Afu5g11440) all had protein domains associated with RNA silencing: PIWI and PAZ domains were present in both Afu3g11010 and Afu8g05280, Dicer domains were present in both Afu5g11790 and Afu4g02930, and a domain for Argonaute small interfering RNA (siRNA) chaperone complex subunit Arb1 was present in Afu5g11440. Transcriptome profiling has revealed that all five genes are expressed in A. fumigatus A1163 (51). While these five genes do not represent the full RNAi pathway, we believe that epigenetic regulation could be involved in mediating antifungal drug resistance within A. fumigatus.
The limited set of mutations in cyp51A conferring resistance makes them relatively simple targets for molecular detection because their roles in resistance are known a priori. However, the evolution of novel azole resistance mechanisms, such as has been seen for TR46/Y121F/T289A and cdr1B, makes the detection of these “known unknowns” much more challenging. As our method captures the entirety of SNP diversity for each isolate of A. fumigatus, it is theoretically possible to determine the contribution of each nucleotide in the genome to the generation of the resistance phenotype within the framework of a genome-wide association study (GWAS). This “hypothesis-free” approach will ultimately extend the WGS-enabled description of known mutations, such as those reported here, to describe the full suite of drug resistance alleles and their epistatic interactions that occur in A. fumigatus. However, before this ambition is realized, significant hurdles need to be addressed. Most fundamentally, a description of the genome-wide global population structure of A. fumigatus needs to be undertaken in order to delineate the essential population parameters that underpin a GWAS; namely, the extent of population genetic structure and genome-wide rates of linkage disequilibrium. From the limited set of isolates that we sequenced, it is already evident that genome-wide recombination occurs and that SNPs are shared across the United Kingdom, the Netherlands, and India, as well as within these countries. This is an encouraging finding, especially as mixed-model GWAS frameworks are now being used and shown to perform well when handling structured populations by estimating the phenotypic covariance due to genetic relatedness (43). Although GWAS has not yet been broadly applied to microbial populations, a recent study by Sheppard et al. (53) illustrated the requirement for novel techniques in order to take these population genetic variables into account, and similar approaches are likely amenable to analyzing fungal genome-wide SNP datasets such as we have described here.
We have shown the population of A. fumigatus to be broadly panmictic and recombinogenic, a phenomenon that has been previously observed and confirmed by independent groups (54–57). The process of recombination is expected to accelerate adaptation to novel environmental conditions, as has previously been described in the related nonpathogenic fungus Aspergillus nidulans (52). Furthermore, while there were fewer recombination hot spots observed within the Indian population, the genome-wide population recombination rate was higher than that seen in the United Kingdom and the Netherlands. Seven of eight Indian isolates were typed as being MAT1-1, with the chi-square test showing that this bias is statistically significant. Together, these findings provide tentative evidence that an outcrossing event may have resulted in the origin of the Indian genotypic cluster, and it is worth speculating that the resulting MAT1-1 TR34/L98 progeny are undergoing a rapid selective sweep by virtue of being highly fit. Clustered sexually recombining neighborhoods have previously been described in a related pathogenic fungus, Talaromyces (Penicillium) marneffei (58), suggesting that this type of fungal population structure may be more common than previously recognized. Whether our findings reflect the process of natural selection in A. fumigatus populations in response to the widespread use of agricultural azoles in Europe and India remains to be seen; however, the genomic approaches that we have detailed here will clearly underpin future research to decipher the population structure of A. fumigatus and map the future evolutionary trajectory of clinically relevant phenotypes.
MATERIALS AND METHODS
Isolates.
Twenty-four A. fumigatus isolates representing three geographical locations were included in the analysis (Table 1). Six clinical isolates, 12-7505446, 12-7505220, 09-7500806, 12-7504652, 12-7504462, and 12-7505054, were obtained from the Mycology Reference Centre, Leeds Teaching Hospitals National Health Service Trust, Leeds, United Kingdom. The eight isolates from the Netherlands included four clinical isolates (08-12-12-13, 08-36-03-25, 08-31-08-91, and 10-01-02-27) and four environmental soil isolates (08-19-02-61, 08-19-02-30, 08-19-02-46, and 08-19-02-10). Of the eight Indian isolates included in this study, four were clinical (Afu 942/09, Afu 1042/09, Afu 343/P/11, and Afu 591/12) and the remaining four (Afu 124/E11, Afu 166/E11, Afu 257/E11, and Afu 218/E11) originated from environmental soil sources. Additionally, the following two control isolates were included for comparison. (i) AF293 (NCPF 7367; National Collection of Pathogenic Fungi at Mycology Reference Laboratory, Public Health England, Bristol, United Kingdom) is a clinical isolate initially cultured in 1993 from lung tissue of a neutropenic patient with invasive aspergillosis in the United Kingdom; the whole-genome sequence of this strain was published in 2005 by Nierman et al. (59). (ii) AF65 is a clinical isolate originally cultured from a lung biopsy specimen from a patient with acute leukemia; this isolate was deposited at the NCPF in 1994 under accession number NCPF 7097 (60).
Aspergillus fumigatus identification
All 24 isolates were identified by macroscopic and microscopic morphological characteristics as A. fumigatus species complex; growth at 50°C differentiated A. fumigatus from Aspergillus lentulus. Six strains from Leeds, United Kingdom, were further identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), using a Bruker Daltonics BioTyper (Bruker, Bremen, Germany). All of the A. fumigatus isolates obtained from the Netherlands (n = 8) and India (n = 8) were previously described as A. fumigatus by sequencing of the internal transcribed spacer (ITS) region. In addition, the presence of any cryptic species within Aspergillus section Fumigati among these isolates was previously excluded by amplification of parts of the β-tubulin and calmodulin genes (61).
Antifungal susceptibility testing.
The in vitro antifungal susceptibility profiles of triazole antifungal drugs, determined according to the CLSI M38-A2 broth microdilution method (62), were previously described and published for isolates from India and the Netherlands (61). MICs were interpreted based on proposed breakpoints (63). For six isolates, the MICs of itraconazole were primarily determined at Mycology Reference Centre, Leeds, United Kingdom, using E test strips (bioMérieux, Marcy l’Etoile, France) on RPMI 1640 agar supplemented with 2% glucose, according to the manufacturer’s instructions, and the results were interpreted as described by Pfaller et al. (64). To ensure consistency of results, MICs for all 24 isolates were further confirmed using the CLSI M38-A2 broth microdilution method.
Conidial harvest.
All fungal isolates were subcultured on potato dextrose agar (PDA) plates and incubated at 35°C for 5 days until sporulation. Stock conidial suspensions were prepared by washing the surface of the PDA plates with 10 ml of sterile water containing 0.05% Tween 20. The conidial suspensions were filtered using Miracloth (EMD Chemicals, San Diego, CA, United States) to remove fungal hyphae, transferred to 50-ml sterile conical tubes, and centrifuged at maximum speed (10,000 × g) for 10 min. The supernatants were discarded, and the pellets were resuspended in 5 ml of sterile distilled water. The concentrations of the suspended conidial stocks were determined by counting the conidia using a hemocytometer chamber at ×400 magnification. Harvested conidia at concentrations of 2 × 108/ml were subjected to DNA extraction.
DNA extraction and quality assessment.
High-molecular-weight DNA was extracted with an optimized MasterPure yeast DNA purification kit (Epicentre Biotechnologies, Cambridge, United Kingdom) with an additional bead-beating step included. Harvested conidia were homogenized using 1.0-mm-diameter zirconia/silica beads (BioSpec Products, Bartlesville, OK) in a FastPrep-24 system (MP Biomedicals, Solon, OH) at 4.5 m/s for 45 s. Genomic DNA (gDNA) was quantified using a Qubit 2.0 fluorometer and dsDNA BR (double-stranded DNA, broad-range) assay kit (Life Technologies, Carlsbad, CA). Quality control of extracted gDNA samples prior to library preparation was performed using the TapeStation 2200 system (Agilent, Santa Clara, CA) and gDNA ScreenTape assays. Purified gDNAs were stored at −20°C until further use.
Mutation screening.
Isolates were screened for the presence of a tandem repeat insertion in the promoter region of the cyp51A gene, as well as for the presence of the common mutations (e.g., L98H, G54, and M220), by using a mixed-format real-time PCR as previously described (10).
Library preparation.
Genomic DNA libraries were constructed according to protocols provided by Illumina (TruSeq Nano DNA Sample Preparation Guide) with the TruSeq Nano kit (Illumina, San Diego, CA). Briefly, gDNA was sheared into 350-base-pair fragments using an S220 ultrasonicator (Covaris, Woburn, MA) and AFA fiber Snap-Cap microTubes, followed by end repair and solid-phase reversible immobilization (SPRI) bead-based size selection. The final libraries were quality controlled on the TapeStation 2200 system (Agilent) with D1K ScreenTape assays (Agilent) and quantified with quantitative PCR (qPCR) on an Applied Biosystems 7300 instrument (Life Technologies) using the Kapa library quantification kit (Kapa Biosciences, Boston, MA). DNA libraries were normalized to 10 nM, and randomized indexed samples were pooled into three libraries of 8 samples.
Illumina whole-genome sequencing.
Prepared whole-genome libraries (n = 24) were sequenced on three lanes of the same flow cell using a HiSeq 2500 sequencer (Illumina) at Medical Research Council Clinical Genomics Centre, Imperial College London, Hammersmith, United Kingdom, generating 100-bp paired-end reads in high-output mode. All raw reads and relevant information in this study have been submitted to the European Nucleotide Archive under project accession number PRJEB8623.
Alignment.
Raw Illumina WGS reads were quality checked using FastQC (version 0.10.1; Babraham Institute) and aligned against the AF293 reference genome (59) using the short-read alignment component (aln) of the Burrows-Wheeler Aligner (BWA) alignment tool (65) with a quality threshold of 15. Duplicate reads were marked using Picard (version 1.112).
Identification of SNPs.
Single-nucleotide polymorphism (SNP) detection was conducted using UnifiedGenotyper from the Genome Analysis Toolkit (GATK) package (66, 67). To ensure high confidence in the SNPs called, variants were filtered according to whether they were present in 80% of reads, the mapping quality, and the depth of coverage at each base to provide a list of high-confidence variants, as described in Rhodes et al. (31). SNPs in cyp51A were mapped using VCF-annotator (Broad Institute, Cambridge, MA).
Identification of tandem repeat sequences.
Whole-genome sequence reads for each isolate were used as the input for de novo assembly using SPAdes 3.1.1 (68). The resulting fasta file was mined for the tandem repeat sequences.
Phylogenetic analysis.
Whole-genome SNP data were converted into relaxed interleaved Phylip format. Maximum-likelihood trees were constructed using RAxML (41) and visualized in FigTree version 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/). The rapid bootstrap algorithm was used to bootstrap all phylogenies using 100 replicates.
Analysis of gene diversity.
Three groups, comprising 6 isolates from the United Kingdom, 8 isolates from the Netherlands, and 8 isolates from India, were formed for the purpose of analyzing genetic diversity. The fixation statistic FST was calculated between each population pair using the package BEDASSLE (69) for R (70).
Recombination analysis.
Two statistics commonly used for describing linkage disequilibrium, the index of association (IA) and rBarD, were estimated using Poppr 1.1.2 (71) for the statistical software R (70), using 999 resamplings of data under the null hypothesis of recombination. Only variant sites unique to each population (defined by country) were considered.
The interval program of the LDhat version 2.2 package (42) was also used to estimate population-scale recombination rates using SNP data for all 8 chromosomes of A. fumigatus, partitioned by their country of original isolation. The program was executed for 2 million iterations with sampling every 200 iterations after a 20,000-iteration burn-in period (as recommended in the user manual [http://ldhat.sourceforge.net/manual.pdf]). The output was summarized using LDhat stat.
ACKNOWLEDGMENTS
J.R. and M.A.B. were supported by a Medical Research Council grant (MRC MR/K000373/1) awarded to M.C.F. This study was supported by a grant from Gilead (WMNF P47129) to D.A.-J., The Royal Brompton and Harefield Respiratory Biomedical Research Unit, and the Imperial College London Healthcare Biomedical Research Centre Infectious Diseases Stream.
We thank Elizabeth M. Johnson and Adrien Szekely (National Collection of Pathogenic Fungi and Mycology Reference Laboratory, Public Health England, Bristol, United Kingdom) and Richard C. Barton (Mycology Reference Centre, Leeds Teaching Hospitals National Health Service Trust, Leeds, United Kingdom), who submitted isolates for this study.
J.F.M. has received grants from Astellas, Basilea and Merck. He has been a consultant to Astellas, Basilea, and Merck and has received speaker’s fees from Merck and Gilead. D.A.-J. has received grants from Gilead and Pfizer and is a consultant to Pulmocide.
Footnotes
Citation Abdolrasouli A, Rhodes J, Beale MA, Hagen F, Rogers TR, Chowdhary A, Meis JF, Armstrong-James D, Fisher MC. 2015. Genomic context of azole resistance mutations in Aspergillus fumigatus determined by using whole-genome sequencing. mBio 6(3):e00536-15. doi:10.1128/mBio.00536-15.
REFERENCES
- 1.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong-James D, Meintjes G, Brown GD. 2014. A neglected epidemic: fungal infections in HIV/AIDS. Trends Microbiol 22:120–127. doi: 10.1016/j.tim.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature 484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segal BH. 2009. Aspergillosis. N Engl J Med 360:1870–1884. doi: 10.1056/NEJMra0808853. [DOI] [PubMed] [Google Scholar]
- 5.Walsh TJ, Anaissie EJ, Denning DW, Herbrecht R, Kontoyiannis DP, Marr KA, Morrison VA, Segal BH, Steinbach WJ, Stevens DA, van Burik J-A, Wingard JR, Patterson TF, Infectious Diseases Society of America . 2008. Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clin Infect Dis 46:327–360. doi: 10.1086/525258. [DOI] [PubMed] [Google Scholar]
- 6.Vermeulen E, Lagrou K, Verweij PE. 2013. Azole resistance in Aspergillus fumigatus: a growing public health concern. Curr Opin Infect Dis 26:493–500. doi: 10.1097/QCO.0000000000000005. [DOI] [PubMed] [Google Scholar]
- 7.Chowdhary A, Sharma C, van den Boom M, Yntema JB, Hagen F, Verweij PE, Meis JF. 2014. Multi-azole-resistant Aspergillus fumigatus in the environment in Tanzania. J Antimicrob Chemother 69:2979–2983. doi: 10.1093/jac/dku259. [DOI] [PubMed] [Google Scholar]
- 8.Badali H, Vaezi A, Haghani I, Yazdanparast SA, Hedayati MT, Mousavi B, Ansari S, Hagen F, Meis JF, Chowdhary A. 2013. Environmental study of azole-resistant Aspergillus fumigatus with TR34/L98H mutations in the cyp51A gene in Iran. Mycoses 56:659–663. doi: 10.1111/myc.12089. [DOI] [PubMed] [Google Scholar]
- 9.Chowdhary A, Kathuria S, Xu J, Meis JF. 2013. Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog 9:e1003633. doi: 10.1371/journal.ppat.1003633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pham CD, Reiss E, Hagen F, Meis JF, Lockhart SR. 2014. Passive surveillance for azole-resistant Aspergillus fumigatus, United States, 2011-2013. Emerg Infect Dis 20:1498–1503. doi: 10.3201/eid2009.140142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard SJ, Cerar D, Anderson MJ, Albarrag A, Fisher MC, Pasqualotto AC, Laverdiere M, Arendrup MC, Perlin DS, Denning DW. 2009. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis 15:1068–1076. doi: 10.3201/eid1507.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snelders E, van der Lee HA, Kuijpers J, Rijs AJ, Varga J, Samson RA, Mellado E, Donders AR, Melchers WJ, Verweij PE. 2008. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med 5:e219. doi: 10.1371/journal.pmed.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Linden JW, Snelders E, Kampinga GA, Rijnders BJ, Mattsson E, Debets-Ossenkopp YJ, Kuijper EJ, Van Tiel FH, Melchers WJ, Verweij PE. 2011. Clinical implications of azole resistance in Aspergillus fumigatus 2007-2009. Emerg Infect Dis 17:1846–1854. doi: 10.3201/eid1710.110226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howard SJ, Arendrup MC. 2011. Acquired antifungal drug resistance in Aspergillus fumigatus: epidemiology and detection. Med Mycol 49(Suppl 1):S90–S95. doi: 10.3109/13693786.2010.508469. [DOI] [PubMed] [Google Scholar]
- 15.Verweij PE, Snelders E, Kema GH, Mellado E, Melchers WJ. 2009. Azole resistance in Aspergillus fumigatus: a side-effect of environmental fungicide use? Lancet Infect Dis 9:789–795. doi: 10.1016/S1473-3099(09)70265-8. [DOI] [PubMed] [Google Scholar]
- 16.Snelders E, Camps SM, Karawajczyk A, Schaftenaar G, Kema GH, van der Lee HA, Klaassen CH, Melchers WJ, Verweij PE. 2012. Triazole fungicides can induce cross-resistance to medical triazoles in Aspergillus fumigatus. PLoS One 7:e31801. doi: 10.1371/journal.pone.0031801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alcazar-Fuoli L, Mellado E. 2012. Ergosterol biosynthesis in Aspergillus fumigatus: its relevance as an antifungal target and role in antifungal drug resistance. Front Microbiol 3:439. doi: 10.3389/fmicb.2012.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chowdhary A, Sharma C, Hagen F, Meis JF. 2014. Exploring azole antifungal drug resistance in Aspergillus fumigatus with special reference to resistance mechanisms. Future Microbiol 9:697–711. doi: 10.2217/fmb.14.27. [DOI] [PubMed] [Google Scholar]
- 19.Mellado E, Diaz-Guerra TM, Cuenca-Estrella M, Rodriguez-Tudela JL. 2001. Identification of two different 14-α sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J Clin Microbiol 39:2431–2438. doi: 10.1128/JCM.39.7.2431-2438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nascimento AM, Goldman GH, Park S, Marras SA, Delmas G, Oza U, Lolans K, Dudley MN, Mann PA, Perlin DS. 2003. Multiple resistance mechanisms among Aspergillus fumigatus mutants with high-level resistance to itraconazole. Antimicrob Agents Chemother 47:1719–1726. doi: 10.1128/AAC.47.5.1719-1726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mann PA, Parmegiani RM, Wei SQ, Mendrick CA, Li X, Loebenberg D, Didomenico B, Hare RS, Walker SS, McNicholas PM. 2003. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14α-demethylase. Antimicrob Agents Chemother 47:577–581. doi: 10.1128/AAC.47.2.577-581.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bader O, Weig M, Reichard U, Lugert R, Kuhns M, Christner M, Held J, Peter S, Schumacher U, Buchheidt D, Tintelnot K, Groß U, MykoLabNet-D Partners . 2013. cyp51A-Based mechanisms of Aspergillus fumigatus azole drug resistance present in clinical samples from Germany. Antimicrob Agents Chemother 57:3513–3517. doi: 10.1128/AAC.00167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellado E, Garcia-Effron G, Alcazar-Fuoli L, Cuenca-Estrella M, Rodríguez-Tudela JL. 2004. Substitutions at methionine 220 in the 14α-sterol demethylase (cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. J Clin Microbiol 48:2747–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellado E, Garcia-Effron G, Alcázar-Fuoli L, Melchers WJ, Verweij PE, Cuenca-Estrella M, Rodríguez-Tudela JL. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob Agents Chemother 51:1897–1904. doi: 10.1128/AAC.01092-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van der Linden JW, Camps SM, Kampinga GA, Arends JP, Debets-Ossenkopp YJ, Haas PJ, Rijnders BJ, Kuijper EJ, Van Tiel FH, Varga J, Karawajczyk A, Zoll J, Melchers WJ, Verweij PE. 2013. Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domiciles. Clin Infect Dis 57:513–520. doi: 10.1093/cid/cit320. [DOI] [PubMed] [Google Scholar]
- 26.Vermeulen E, Maertens J, Schoemans H, Lagrou K. 2012. Azole-resistant Aspergillus fumigatus due to TR46/Y121F/T289A mutation emerging in Belgium, July 2012. Euro Surveill 17:1–3. [PubMed] [Google Scholar]
- 27.Astvad KM, Jensen RH, Hassan TM, Mathiasen EG, Thomsen GM, Pedersen UG, Christensen M, Hilberg O, Arendrup MC. 2014. First detection of TR46/Y121F/T289A and TR34/L98H alterations in Aspergillus fumigatus isolates from azole-naive patients in Denmark despite negative findings in the environment. Antimicrob Agents Chemother 58:5096–5101. doi: 10.1128/AAC.02855-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer J, van Koningsbruggen-Rietschel S, Rietschel E, Vehreschild MJ, Wisplinghoff H, Krönke M, Hamprecht A. 2014. Prevalence and molecular characterization of azole resistance in Aspergillus spp. isolates from German cystic fibrosis patients. J Antimicrob Chemother 69:1533–1536. doi: 10.1093/jac/dku009. [DOI] [PubMed] [Google Scholar]
- 29.Chowdhary A, Sharma C, Kathuria S, Hagen F, Meis JF. 2014. Azole-resistant Aspergillus fumigatus with the environmental. TR46/Y121F/T289A mutation in India. J Antimicrob Chemother 69:555–557. [DOI] [PubMed] [Google Scholar]
- 30.Vanhee LM, Symoens F, Jacobsen MD, Nelis HJ, Coenye T. 2009. Comparison of multiple typing methods for Aspergillus fumigatus. Clin Microbiol Infect 15:643–650. doi: 10.1111/j.1469-0691.2009.02844.x. [DOI] [PubMed] [Google Scholar]
- 31.Rhodes J, Beale MA, Fisher MC. 2014. Illuminating choices for library prep: a comparison of library preparation methods for whole genome sequencing of Cryptococcus neoformans using Illumina HiSeq. PLoS One 9:e113501. doi: 10.1371/journal.pone.0113501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peacock S. 2014. Bring microbial sequencing to hospitals. Nature 509:557–559. doi: 10.1038/509557a. [DOI] [PubMed] [Google Scholar]
- 33.Köser CU, Ellington MJ, Peacock SJ. 2014. Whole-genome sequencing to control antimicrobial resistance. Trends Genet 30:401–407. doi: 10.1016/j.tig.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reuter S, Ellington MJ, Cartwright EJ, Köser CU, Török ME, Gouliouris T, Harris SR, Brown NM, Holden MT, Quail M, Parkhill J, Smith GP, Bentley SD, Peacock SJ. 2013. Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Intern Med 173:1397–1404. doi: 10.1001/jamainternmed.2013.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, Henderson DK, Palmore TN, Segre JA, Segre JA. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Köser CU, Bryant JM, Becq J, Török ME, Ellington MJ, Marti-Renom MA, Carmichael AJ, Parkhill J, Smith GP, Peacock SJ. 2013. Whole-genome sequencing for rapid susceptibility testing of M. tuberculosis. N Engl J Med 369:290–292. doi: 10.1056/NEJMc1215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Litvintseva AP, Hurst S, Gade L, Frace MA, Hilsabeck R, Schupp JM, Gillece JD, Roe C, Smith D, Keim P, Lockhart SR, Changayil S, Weil MR, MacCannell DR, Brandt ME, Engelthaler DM. 2014. Whole-genome analysis of Exserohilum rostratum from an outbreak of fungal meningitis and other infections. J Clin Microbiol 52:3216–3222. doi: 10.1128/JCM.00936-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Etienne KA, Gillece J, Hilsabeck R, Schupp JM, Colman R, Lockhart SR, Gade L, Thompson EH, Sutton DA, Neblett-Fanfair R, Park BJ, Turabelidze G, Keim P, Brandt ME, Deak E, Engelthaler DM. 2012. Whole genome sequence typing to investigate the Apophysomyces outbreak following a tornado in Joplin, Missouri, 2011. PLoS One 7:e49989. doi: 10.1371/journal.pone.0049989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engelthaler DM, Chiller T, Schupp JA, Colvin J, Beckstrom-Sternberg SM, Driebe EM, Moses T, Tembe W, Sinari S, Beckstrom-Sternberg JS, Christoforides A, Pearson JV, Carpten J, Keim P, Peterson A, Terashita D, Balajee SA. 2011. Next-generation sequencing of Coccidioides immitis isolated during cluster investigation. Emerg Infect Dis 17:227–232. doi: 10.3201/eid1702.100620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janbon G, Ormerod KL, Paulet D, Byrnes EJ, Yadav V, Chatterjee G, Mullapudi N, Hon C-C, Billmyre RB, Brunel F, Bahn Y-S, Chen W, Chen Y, Chow EW, Coppée J-Y, Floyd-Averette A, Gaillardin C, Gerik KJ, Goldberg J, Gonzalez-Hilarion S, Gujja S, Hamlin JL, Hsueh Y-P, Ianiri G, Jones S, Kodira CD, Kozubowski L, Lam W, Marra M, Mesner LD, Mieczkowski PA, Moyrand F, Nielsen K, Proux C, Rossignol T, Schein JE, Sun S, Wollschlaeger C, Wood IA, Zeng Q, Neuvéglise C, Newlon CS, Perfert JR, Lodge, Idnurm A, Stajich JE, Kronstad JW, Sanyal K, Heitman J, Fraser JA, Cuomo CA, Dietrich FS. 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet 10:e1004261. doi: 10.1371/journal.pgen.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamatakis A. 2006. RAxML-VI HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 42.Mcvean GA, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P. 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304:581–585. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 43.Korte A, Vilhjálmsson BJ, Segura V, Platt A, Long Q, Nordborg M. 2012. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat Genet 44:1066–1071. doi: 10.1038/ng.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lockhart SR, Frade JP, Etienne KA, Pfaller MA, Diekema DJ, Balajee SA. 2011. Azole resistance in Aspergillus fumigatus isolates from the ARTEMIS global surveillance study is primarily due to the TR/L98H mutation in the cyp51A gene. Antimicrob Agents Chemother 55:4465–4468. doi: 10.1128/AAC.00185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selmecki A, Forche A, Berman J. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sionov E, Lee H, Chang YC, Kwon-Chung KJ. 2010. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog 6:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fraczek MG, Bromley M, Buied A, Moore CB, Rajendran R, Rautemaa R, Ramage G, Denning DW, Bowyer P. 2013. The cdr1B efflux transporter is associated with non-cyp51A-mediated itraconazole resistance in Aspergillus fumigatus. J Antimicrob Chemother 68:1486–1496. doi: 10.1093/jac/dkt075. [DOI] [PubMed] [Google Scholar]
- 48.Camps SM, Dutilh BE, Arendrup MC, Rijs AJ, Snelders E, Huynen MA, Verweij PE, Melchers WJ. 2012. Discovery of a HapE mutation that causes azole resistance in Aspergillus fumigatus through whole genome sequencing and sexual crossing. PLoS One 7:e50034. doi: 10.1371/journal.pone.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buied A, Moore CB, Denning DW, Bowyer P. 2013. High-level expression of cyp51B in azole-resistant clinical Aspergillus fumigatus isolates. J Antimicrob Chemother 68:512–514. doi: 10.1093/jac/dks451. [DOI] [PubMed] [Google Scholar]
- 50.Calo S, Shertz-Wall C, Lee SC, Bastidas RJ, Nicolás FE, Granek JA, Mieczkowski P, Torres-Martínez S, Ruiz-Vázquez RM, Cardenas ME, Heitman J. 2014. Antifungal drug resistance evoked via RNAi-dependent epimutations. Nature 513:555–558. doi: 10.1038/nature13575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Müller S, Baldin C, Groth M, Guthke R, Kniemeyer O, Brakhage AA, Valiante V. 2012. Comparison of transcriptome technologies in the pathogenic fungus Aspergillus fumigatus reveals novel insights into the genome and MpkA dependent gene expression. BMC Genomics 13:519. doi: 10.1186/1471-2164-13-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schoustra SE, Debets AJ, Slakhorst M, Hoekstra RF. 2007. Mitotic recombination accelerates adaptation in the fungus Aspergillus nidulans. PLoS Genet 3:e68. doi: 10.1371/journal.pgen.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheppard SK, Didelot X, Meric G, Torralbo A, Jolley KA, Kelly DJ, Bentley SD, Maiden MC, Parkhill J, Falush D. 2013. Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc Natl Acad Sci U S A 110:11923–11927. doi: 10.1073/pnas.1305559110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klaassen CH, Gibbons JG, Fedorova ND, Meis JF, Rokas A. 2012. Evidence for genetic differentiation and variable recombination rates among Dutch populations of the opportunistic human pathogen Aspergillus fumigatus. Mol Ecol 21:57–70. doi: 10.1111/j.1365-294X.2011.05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paoletti M, Rydholm C, Schwier EU, Anderson MJ, Szakacs G, Lutzoni F, Debeaupuis J-P, Latgé J-P, Denning DW, Dyer PS. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr Biol 15:1242–1248. doi: 10.1016/j.cub.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 56.Pringle A, Baker DM, Platt JL, Wares JP, Latgé JP, Taylor JW. 2005. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59:1886–1899. doi: 10.1111/j.0014-3820.2005.tb01059.x. [DOI] [PubMed] [Google Scholar]
- 57.Rydholm C, Szakacs G, Lutzoni F. 2006. Low genetic variation and no detectable population structure in Aspergillus fumigatus compared to closely related Neosartorya species. Eukaryot Cell 5:650–657. doi: 10.1128/EC.5.4.650-657.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henk DA, Shahar-Golan R, Ranjana DK, Zhan N, Fedorova N, Nierman WC, Hsueh PR, Yuen KY, Wertheim H, Day J, Vanittanakom N, Andrianopoulos A, Fisher MC. Clonality despite sex, the evolution of sexual neighborhoods in the pathogenic fungus Penicillium marneffei. PLoS Pathogens 8:e1002851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, García JL, García MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jiménez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Lafton A, Latgé J-P, Li W, Lord A, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 60.Denning DW, Venkateswarlu K, Oakley KL, Anderson MJ, Manning NJ, Stevens DA, Warnock DW, Kelly SL. 1997. Itraconazole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother 41:1364–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chowdhary A, Kathuria S, Xu J, Sharma C, Sundar G, Singh PK, Gaur SN, Hagen F, Klaassen CH, Meis JF. 2012. Clonal expansion and emergence of environmental multiple-triazole-resistant Aspergillus fumigatus strains carrying the TR34/L98H mutations in the cyp51A gene in India. PLoS One 7:e52871. doi: 10.1371/journal.pone.0052871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clinical and Laboratory Standards Institute 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard, 2nd ed CLSI document M38-A2. CLSI, Wayne, PA. [Google Scholar]
- 63.Verweij PE, Howard SJ, Melchers WJ, Denning DW. 2009. Azole-resistance in Aspergillus: proposed nomenclature and breakpoints. Drug Resist Updat 12:141–147. doi: 10.1016/j.drup.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 64.Pfaller JB, Messer SA, Hollis RJ, Diekema DJ, Pfaller MA. 2003. In vitro susceptibility testing of Aspergillus spp.: comparison of Etest and reference microdilution methods for determining voriconazole and itraconazole MICs. J Clin Microbiol 41:1126–1129. doi: 10.1128/JCM.41.3.1126-1129.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van der Auwera GA, Feldgarden M, Kolter R, Mahillon J. 2013. Whole-genome sequence of 94 environmental isolates of Bacillus cereus sensu lato. Genome Announc 1:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bradburd GS, Ralph PL, Coop GM. 2013. Disentangling the effects of geographic and ecological isolation on genetic differentiation. Evolution 67:3258–3273. doi: 10.1111/evo.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R Development Core Team 2008. R:A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 71.Kamvar ZN, Tabima JF, Grünwald NJ. 2014. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. doi: 10.7717/peerj.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Agapow PM, Burt A. 2001. Indices of multilocus disequilibrium. Mol Ecol Notes 1(1–2):101–102. [Google Scholar]