Abstract
Brachiopods are a lineage of invertebrates well known for the breadth and depth of their fossil record. Although the quality of this fossil record attracts the attention of paleontologists, geochemists, and paleoclimatologists, modern day brachiopods are also of interest to evolutionary biologists due to their potential to address a variety of questions ranging from developmental biology to biomineralization. The brachiopod shell is a composite material primarily composed of either calcite or calcium phosphate in close association with proteins and polysaccharides which give these composite structures their material properties. The information content of these biomolecules, sequestered within the shell during its construction, has the potential to inform hypotheses focused on describing how brachiopod shell formation evolved. Here, using high throughput proteomic approaches and next generation sequencing, we have surveyed and characterized the first shell-proteome and shell-forming transcriptome of any brachiopod, the South American Magellania venosa (Rhynchonelliformea: Terebratulida). We find that the seven most abundant proteins present in the shell are unique to M. venosa, but that these proteins display biochemical features found in other metazoan biomineralization proteins. We can also detect some M. venosa proteins that display significant sequence similarity to other metazoan biomineralization proteins, suggesting that some elements of the brachiopod shell-forming proteome are deeply evolutionarily conserved. We also employed a variety of preparation methods to isolate shell proteins and find that in comparison to the shells of other spiralian invertebrates (such as mollusks) the shell ultrastructure of M. venosa may explain the effects these preparation strategies have on our results.
Keywords: Brachiopoda, biomineralization, proteome, transcriptome, evolution, Magellania
Introduction
For much of the Phanerozoic, animal life has constructed an amazing diversity of mineralized structures to fulfill a host of biological functions (reviewed in Simkiss and Wilbur 1989). The products of these biomineralization processes, precisely coordinated by macromolecules such as proteins and carbohydrates, range from individual functional elements, for example, magneto-sensors, teeth, armor (Lowenstam and Weiner 1989), to large scale geological structures, for example, coral reefs. Ever since the onset of metazoan biomineralization at the dawn of the Phanerozoic (∼550 Ma; Knoll 2003), their biomineralized structures have most commonly been constructed from one of three minerals: Calcium carbonate (CaCO3), for example, most brachiopod shells, echinoderm spines and tests, molluscan shells, coral skeletons, and sponge spicules; calcium phosphate (CaPO4), for example, some brachiopod shells and vertebrate bones; or silica (SiO2), for example, primarily sponge spicules. Brachiopods were among the first animals to biomineralize in the Cambrian (Knoll 2003), and achieved a high diversity in the Paleozoic (4,200 described genera, all of which constructed biomineralized shells; reviewed in Taylor et al. 2010). Although most brachiopod taxa are now extinct, this historic diversity makes them a significant invertebrate index fossil group, providing a continuous deep-time record of a prevalent biocalcification system (e.g., Rudwick 1970). Morphological (Carlson 1995) as well as recent molecular data (Cohen and Weydmann 2005; Sperling et al. 2011; Cohen 2013) suggest that Brachiopoda is monophyletic either to the inclusion of Phoronida (Cohen and Weydmann 2005) or with robustly supported subphyla Linguliformea and Craniiformea (classically referred to as “Inarticulata”) as a sister group to the Rhynchonelliformea (classically referred to as “Articulata”) (Sperling et al. 2011; see fig. 1).
Fig. 1.—

Simplified phylogeny of the Brachiopoda based on Sperling et al. (2011).
The divergence between these sister groups has been estimated by molecular clock analyses at 547 Ma (Sperling et al. 2011), and importantly, this deep divergence is reflected in the different biomineralizing systems that each group employs; although all brachiopod shells are inorganic/organic composites, within the Rhynchonelliformea and Craniiformea, calcite (CaCO3) dominates the organic content on a w/w basis (∼98%), whereas Linguliformea shells typically possess calcium phosphate in approximately equal ratio to the organic content (∼50%, i.e., “organophosphatic” shells; reviewed in Taylor et al. 2010). Moreover, the “organophosphatic” shells of the Linguliformea display a hierarchically laminated architecture (Williams 1989; Merkel et al. 2007; Schmahl et al. 2008; Merkel et al. 2009), whereas Rhynchonelliformea shells display diverse types of microstructures based on biopolymer reinforced calcite (Goetz et al. 2009, 2011; Schmahl et al. 2012). The dichotomy of shell mineralogies (amongst extinct and extant species) makes brachiopods ideal models for understanding how such diverse biomineralogies evolved. The attraction of this question generated a literature base that described the amino acid compositions of shells from both extinct and extant brachiopods (e.g., Jope 1966a, 1966b). More recent studies by Cusack et al. (1992) reported the N-terminal sequence of a red chromoprotein (ICP-1) from three species of brachiopods, and Gaspard et al. (2008) described the microstructures and general nature of the glycosylated shell-forming proteomes of five rhynchoneliform brachiopods. Geochemical and isotopic techniques have also been applied to brachiopod shells as proxies for climate reconstruction (Curry and Fallick 2002; Brand 2003; Lee et al. 2004; Parkinson et al. 2005; Cusack et al. 2012). Despite the interest from so many disciplines in the organic components of brachiopod shells and how they regulate shell formation, no study has yet conducted a broad scale survey of the proteins that guide brachiopod shell-formation, and no report to our knowledge describes the full-length sequence of any brachiopod biomineralizing protein.
Here, we have surveyed both the shell-forming transcriptome and CaCO3 shell-proteome of Magellania venosa, a rhynchonelliform brachiopod, the first biomineralizing data sets of their kind for any brachiopod. This represents a major step forward in terms of our molecular understanding of how an extant brachiopod constructs a calcified shell. We have compared these data with the relatively extensive shell-forming proteome data sets now available from several mollusks and more distantly related biocalcifying metazoan taxa (corals and sea urchins), providing further insight into how brachiopod shell-formation has evolved. These data will also be invaluable for future comparative studies aimed at revealing the molecular mechanisms that regulate the deposition of calcitic versus phosphatic biominerals.
Materials and Methods
More than 20 specimens of M. venosa were collected by SCUBA diving at the jetty in Huinay (42 °22′29″S, 72 °25′41.58″W) in the Comau Fjord, Southern Chilean fjord region between 2010 and 2013 (6 samples on November 10, 2010; 3 samples on December 11, 2011; 10 samples on January 11, 2013). The shells of whole animals from the first batch were cracked open and preserved in RNAlater overnight at 4 °C and then stored at −20 °C until shipment to Germany. The samples of the second batch were cleaned of soft tissue and the shells air-dried. The samples of the third batch were preserved in absolute ethanol. Scanning electron micrographs were taken from bleached and cracked shells using a JEOL 1430VP Scanning Electron Microscope (SEM) with an accelerator voltage of 15.3 kV at the Zoologische Staatssammlung München.
Organic Matrix Preparation
Shells were cleaned in sodium hypochlorite solution (6–14% active chlorine; Merck, Darmstadt, Germany) for 2 h at room temperature with 5 min ultra sonication and change of hypochlorite solution every 30 min. Cleaned shells were washed with deionized water and dried. One set of three dorsal (D) and three ventral (V) shells was demineralized for matrix extraction. A second set of shells was crushed using mortar and pestle to obtain a homogeneous powder with no visible shell pieces. This powder was treated with approximately 10 volumes of sodium hypochlorite for another 24 h at 4 °C on a roller mixer. Shell particles were collected by centrifugation for 10 min at 3,000 g, washed three times with deionized water, and air-dried. Demineralization of both types of shell preparation was done in 50% acetic acid (20 ml/g of shell) over night at 4–6 °C, and the resulting suspension was dialyzed (Spectra/Por 6 dialysis membrane, molecular weight cut-off 2,000; Spectrum Europe, Breda, The Netherlands) successively against 3 × 1 l of 10% acetic acid and 3 × 1 l of 5% acetic acid at 4–6 °C and lyophilized. Matrices were analyzed by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) using precast 4–12% Novex Bis-Tris gels in MES buffer using reagents and protocols supplied by the manufacturer (Invitrogen, Carlsbad, CA), except that 1% β-mercaptoethanol was used as a reducing agent in the sample buffer. Samples were suspended in sample buffer (200 µg/30 µl), heated to 70 °C for 10 min and centrifuged for 5 min at 15,800 g in a 5415D Eppendorf centrifuge to remove sample buffer-insoluble material. The molecular weight standard was Novex Sharp prestained (Invitrogen).
Peptide Preparation
Reduction, carbamidomethylation, and enzymatic cleavage of matrix proteins were performed using a modification of the FASP (filter-aided sample preparation) method (Wisniewski et al. 2009) as outlined below. Aliquots of 200 µg of matrix were suspended in 300 µl of 0.1 M Tris, pH 8, containing 6 M guanidine hydrochloride, and 0.01 M dithiothreitol (DTT). The mixture was heated to 56 °C for 60 min, cooled to room temperature, and centrifuged at 15,800 g in an Eppendorf bench-top centrifuge 5415D for 15 min. The supernatant was loaded into an Amicon Ultra 0.5 ml 30 K filter device (Millipore; Tullagreen, Ireland). DTT was removed by centrifugation at 15,800 g for 15 min and washing with 2 × 1 vol of the same buffer. Carbamidomethylation was done in the device using 0.1 M Tris buffer, pH 8, containing 6 M guanidine hydrochloride and 0.05 mM iodoacetamide and incubation for 45 min in the dark. Carbamidomethylated proteins were washed with 0.05 M ammonium hydrogen carbonate buffer, pH 8, containing 2 M urea, and centrifuged as before. Trypsin (2 µg, Sequencing grade, modified; Promega, Madison, WI) was added in 40 µl of 0.05 M ammonium hydrogen carbonate buffer containing 2 M urea and the devices were incubated at 37 °C for 16 h. Peptides were collected by centrifugation and the filters were washed twice with 40 µl of 0.05 M ammonium hydrogen carbonate buffer and twice with 1% trifluoroacetic acid in 5% acetonitrile. The acidic peptide solution (pH 1–2) was applied to C18 Stage Tips (Rappsilber et al. 2007) and the eluted peptides were vacuum-dried in an Eppendorf concentrator.
Liquid Chromatography–Mass Spectrometry and Data Analysis
Peptide mixtures were analyzed by on-line nanoflow liquid chromatography (LC) using the EASY-nLC 1000 system (Proxeon Biosystems, Odense, Denmark, now part of Thermo Fisher Scientific) with 20-cm capillary columns of an internal diameter of 75 μm filled with 1.8 μm Reprosil-Pur C18-AQ resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). Peptides were eluted with a linear gradient from 5% to 30% buffer B (80% acetonitrile in 0.1% formic acid) in 90 min, 30–60% B in 5 min, and 60–95% B in 5 min at a flow rate of 250 nl/min and a temperature of 40 °C. The eluate was electro-sprayed into an Orbitrap Q Exactive (Thermo Fisher Scientific, Bremen, Germany) using a Proxeon nanoelectrospray ion source. The instrument was operated in an HCD top 10 mode essentially as described (Michalski et al. 2011). The resolution was 70,000 for full scans and 7,500 for fragments (both specified at m/z 400). Ion target values were 1e6 and 5e4 ms, respectively. Exclusion time was 20 s. Sample runs were monitored using the SprayQC quality monitoring system (Scheltema and Mann 2012). Raw files were processed using the Andromeda search engine-based version 1.4.1.12 of MaxQuant (http://www.maxquant.org/, last accessed May 7, 2015) with enabled second peptide, iBAQ, and match between runs (match time window 0.5 min; alignment time window 20 min) options (Cox and Mann 2008; Cox et al. 2011).
In order to generate an M. venosa peptide database against which the LC–mass spectrometry (MS) data could be searched, we sequenced the mantle transcriptome of M. venosa using the Illumina HiSeq platform. Briefly, total RNA from the mantle tissue of an adult individual was isolated the Qiagen RNeasy Plant Kit according to the manufacturer’s instructions. The resulting RNA was then processed by the sequencing center at the IKMB at the University of Kiel (Germany), where a 230–430 bp paired end insert TrueSeq-RNA library was constructed and sequenced for 101 bases from both ends. More than 370 million reads were returned, quality trimmed, and assembled using the CLC Genomics Workbench 6.5. A total of 27,897 contigs with an N50 of 1,803 bp (average 1,319 bp) were assembled and subsequently translated in all six reading frames.
These data were combined with the sequences of common contaminants, such as human keratins and mammalian cytoskeletal proteins. Carbamidomethylation was set as fixed modification. Variable modifications were oxidation (M), N-acetyl (protein), pyro-Glu/Gln (N-term), and phospho (STY) (Sharma et al. 2014). One batch of replicates was also analyzed tentatively adding hydroxyproline as a variable modification, but no additional collagen peptides containing hydroxyproline were detected. The initial mass tolerance was 7 ppm for full scans and 20 ppm for tandem mass spectrometry. Two missed cleavages were allowed and the minimal length required for a peptide was seven amino acids. Maximal false discovery rate for peptide spectral match, proteins, and site was set to 0.01. The minimal score for peptides was 60 and the minimal delta score for modified peptides was 17. Minimal requirements for final acceptance of identifications were the presence of the protein in at least four biological replicates of 12 (V1–6, D1–6) with at least two sequence-unique peptides in two of the four replicate groups (D1–3, V1–3, D4–6, V4–6). Each of the 12 biological replicates was measured with five technical replicates. Identifications with one sequence-unique peptide were exceptionally accepted if the same protein was identified with more peptides in other biological replicates. Identifications with two sequence-unique peptides were accepted after positive validation using the MaxQuant Expert System software (Neuhauser et al. 2012) considering the assignment of major peaks, occurrence of uninterrupted y- or b-ion series of at least four consecutive amino acids, preferred cleavages N-terminal to proline bonds, the possible presence of a2/b2 ion pairs and immonium ions, and assignment of major fragment peaks. The iBAQ (intensity-based absolute quantification) (Schwanhausser et al. 2011) option of MaxQuant was used to calculate, based on the sum of peak intensities, the approximate share of each protein in the total proteome, including identifications which were not finally accepted. This enabled us to discern between minor and major proteins.
Sequence similarity searches were performed with FASTA (http://www.ebi.ac.uk/Tools/sss/fasta/, last accessed May 7, 2015) (Goujon et al. 2010) against current releases of the Uniprot Knowledgebase. Other bioinformatics tools used were Clustal Omega for sequence alignments (http://www.ebi.ac.uk/Tools/msa/clustalo/, last accessed May 7, 2015) (Sievers et al. 2011), InterPro (http://www.ebi.ac.uk/interpro, last accessed May 7, 2015) (Hunter et al. 2012) for domain predictions, SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/, last accessed May 7, 2015) (Petersen et al. 2011) for signal sequence prediction, and TMHMM (http://www.cbs.dtu.dk/publications/, last accessed May 7, 2015) (Krogh et al. 2001) for prediction of transmembrane helices. Amino acid composition and theoretical pI were determined using the ProtParam tool provided by the Expasy server (http://web.expasy.org/protparam/, last accessed May 7, 2015) (Gasteiger et al. 2005). Intrinsically disordered protein structure was predicted using IUPred (http://iupred.enzim.hu/, last accessed May 7, 2015) (Dosztányi et al. 2005) and PrDOS (http://prdos.hgc.jp/cgi-bin/top.cgi, last accessed May 7, 2015) (Ishida and Kinoshita 2007). Subcellular location predictions were based on sequence similarities to known proteins, domain predictions, signal sequence predictions, and transmembrane segment predictions. Sequence-based comparisons were performed using Circoletto (Darzentas 2010) and Circos (Krzywinski et al. 2009). Tandem repeats were detected using XStream (Newman and Cooper 2007).
Results and Discussion
The Proteome of M. venosa Shells Treated with Hypochlorite for 2 h
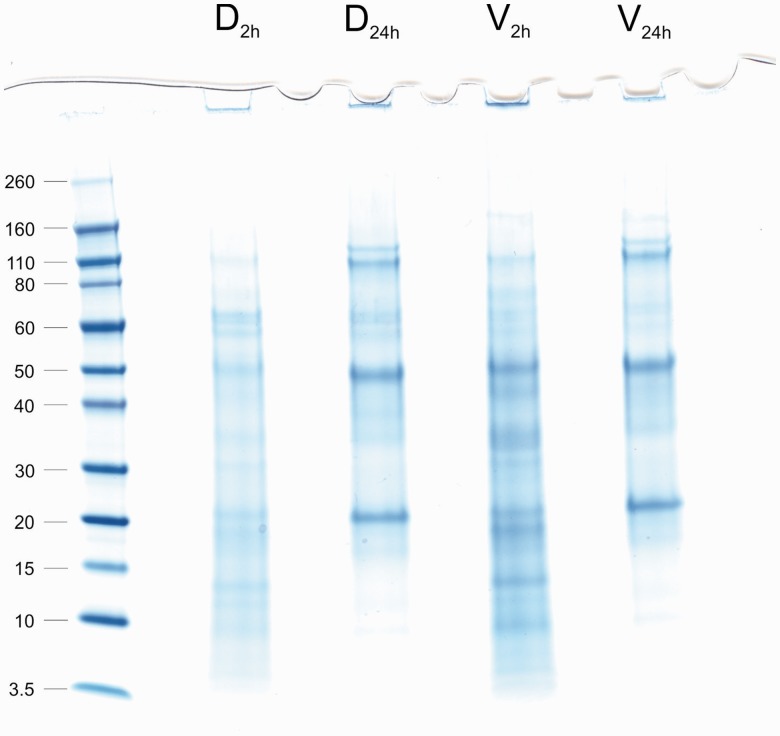
Magellania venosa shells treated with sodium hypochlorite solution for 2 h became soft and crumbly. This was most probably due to some extra-crystalline matrix degradation by the 2-h hypochlorite treatment (see next section). Only that part of the shell around the hinge region remained hard and difficult to crush. Yields of organic matrix from three ventral and three dorsal shells treated with hypochlorite for 2 h were 1.9 ± 0.1 mg of matrix/g dorsal shell (0.19%) and 2.7 ± 0.7 mg/g of ventral shell (0.27%). Comparisons of PAGE patterns of the organic matrices did not indicate any major differences between the proteomes of ventral and dorsal shells (fig. 2, lanes D2h and V2h).
Fig. 2.—
PAGE analysis of M. venosa shell matrices. Lanes D2h (dorsal, 2 h of sodium hypochlorite treatment) and V2h (ventral, 2 h of sodium hypochlorite treatment) show matrix extracted after a 2-h treatment of entire shells with sodium hypochlorite. Lanes D24h and V24h show matrix extracted after an additional treatment of powdered shells for 24 h. The molecular weight of marker proteins is shown in kDa on the left.
Five technical replicates from each of three dorsal and three ventral shells treated with hypochlorite for 2 h were analyzed in two groups (dorsal and ventral). Searching the LC-MS raw-files against the M. venosa sequence database resulted in 496 identifications from the dorsal set of replicates (supplementary file S1, Supplementary Material online) and 591 from ventral set of replicates (supplementary file S2, Supplementary Material online), with an overlap of approximately 70% of each data set. Differences between dorsal and ventral shells concerned minor proteins only, and may be related to the higher protein content of ventral shells (see above) and to experimental variation. Peptide sequences and relevant parameters are shown in supplementary files S3 and S4, Supplementary Material online. After revising the list of identifications according to the rules described above (see Liquid Chromatography–Mass Spectrometry and Data Analysis) and the grouping together of entries suggested by FASTA searches to belong to different regions of the same protein, we obtained 317 proteins/protein groups (supplementary file S5: table S1, Supplementary Material online). Seventeen proteins were identified in ventral samples only and eight were identified in dorsal samples only. Such minor qualitative differences are likely to be due to experimental variation. The higher number of proteins in ventral samples may be related to the higher protein content of the ventral shells (see above).
In contrast to the recently analyzed shell proteomes of the terrestrial snail Cepaea nemoralis (Mann and Jackson 2014) and the marine snail Lottia gigantea (Mann and Edsinger 2014), the proteome of M. venosa contained several intracellular proteins with high abundance (supplementary file S5, Supplementary Material online). The most abundant of the intracellular proteins was actin. Because of the very high sequence conservation among actins from different sources, this cytoskeletal protein shared almost all of its peptides with common mammalian contaminants (one sequence-unique peptide of a total of 16). Therefore, it was difficult to estimate the true abundance of M. venosa actin. The same was true for several other cytoskeletal proteins such as tubulins. However, because such extraordinarily high abundances were not found in our previous studies of molluscan shell proteomes (Mann and Edsinger 2014; Mann and Jackson 2014), our preferred interpretation is that these peptides in fact originated from M. venosa. The formation and growth of calcitic brachiopod shells in rhynchonelliforms have been described by Williams (1989) as the result of the migration of outer mantle epithelial cells “during their secretion of succeeding biomineral components” (Williams 1997). What has been described as the protein boundary of secondary shell fibers, for example, in Notosaria nigricans, may represent organic remnants of this migration process containing, among other components, a considerably large fraction of actin which originally helped the cells to move along their secretional track. Whether this represents extracellular actin as described, for example, in the matrix of mouse smooth muscle cells (Accinni et al. 1983) remains to be investigated. However, the high concentration of actin we detect in M. venosa shell material may be the first molecular evidence that Williams’ hypothesis of shell secretion in the Rhynchonelliformea is indeed correct. Interestingly, other metazoans constructing calcified skeletons have also been found to contain actin in their skeletal matrices even after rigorous washing of powdered skeleton, for example, the stony coral Stylophora pistillata (Drake et al. 2013a, 2013b; Mass et al. 2014), and the sclerites of the calcitic octocoral Sinularia sp. (Rahman et al. 2013), indicating that actin may genuinely have some biomineralization-related function in these organisms.
The Proteome of M. venosa Shell Powder Treated with Hypochlorite for Another 24 h
Despite the apparent accessibility of M. venosa’s organic intercrystalline matrix and the danger of losing it during more aggressive washing treatments prior to matrix extraction, the high abundance of intracellular proteins described above leads us to attempt a more rigorous washing procedure. To this end, shells already treated with sodium hypochlorite for 2 h were reduced to a homogeneous powder using mortar and pestle and treated with hypochlorite solution for another 24 h with constant mixing on a roller mixer at 4 °C. This treatment is equivalent to half of the incubation time previously used for Terebratulina septentrionalis (another brachiopod of the order Terebratulida) shells to destroy the intercrystalline matrix to such an extent that simple shaking was enough to obtain free calcite fibers (Collins et al. 1988). This more aggressive protocol was similar to that used for several other brachiopod shells reported previously (Curry et al. 1991). The yield of organic matrix from this type of preparation was 1.0 ± 0.3 mg/g (0.1%) from both dorsal and ventral shells. Again, PAGE analysis (fig. 2, lanes D24h and V24h) did not reveal any major differences between dorsal and ventral shells. However, there was a distinct difference between the two sodium hypochlorite treatments, likely indicating a loss of extracrystalline matrix and an enrichment of the intracrystalline matrix (fig. 2). Several predominant protein bands disappeared, whereas others, especially three proteins with apparent molecular weights of 110, 50, and 20 kDa were significantly enriched. This three-band pattern was reminiscent to previously published patterns derived from the intracrystalline matrix of the terebratulid brachiopods of the genera Neothyris (Curry et al. 1991), although the molecular weights were not the same (47, 16, and 6.5 kDa), and Calloria (Collins et al. 1991). The latter showed major protein bands between a molecular weight of 30–110 kDa when reacted with antishell matrix antiserum.
Altogether we identified 243 proteins/protein groups from samples processed with this more aggressive sodium hypochlorite treatment (supplementary file S5, Supplementary Material online). This is in comparison to the 317 proteins/protein groups identified using the milder sodium hypochlorite treatment. Additional data about proteins and peptides including identifications not finally accepted are contained in supplementary files S6–S9, Supplementary Material online. Ten and eight minor proteins were found only in dorsal and ventral samples, respectively. The overlap between samples treated for 2 or 24 h with sodium hypochlorite was 205 proteins. Thirty-eight minor proteins appeared in the more rigorously cleaned samples only. This was most probably due to the depletion of some major proteins as compared with the less rigorously cleaned samples. Supplementary file S5 (table S1), Supplementary Material online, shows that the number and abundance of common intracellular proteins decrease dramatically with the more aggressive sodium hypochlorite treatment (e.g., β-tubulin, actin). However, several major proteins that could not be matched to known intracellular proteins were significantly reduced or eliminated with the more aggressive sodium hypochlorite treatment. These proteins may be major components of the extracrystalline matrix (discussed below), in agreement with previous reports indicating that prolonged treatment of brachiopod shells with hypochlorite removes the electron microscopically visible extracrystalline matrix and destroys the cohesion between calcified elements of the shell (Collins et al. 1988).
The Major M. venosa Shell Matrix Proteins
The 66 major shell proteins (arbitrarily defined here as proteins occurring with an average percentage of ≥ 0.1 in either D1–3 and V1–3 [2 h] or D4–6 and V4–6 [24 h] or both) are presented in table 1. The most abundant protein (R20074237) was a novel protein with no sequence similarity to any sequences in SwissProt, GenBank, GenBank nr, or with any recognizable Pfam domains. This protein possessed a signal sequence (suggesting that it is an extracellular protein) and an unusually high proportion of acidic (Asp—12%) amino acids. In addition, this protein shared no sequence similarity with any of the 765 proteins in our targeted comparisons against mollusk, sea urchin, or coral biomineralizing proteomes (see fig. 3). Although the discovery of such novel proteins that are likely to play prominent roles in brachiopod shell formation is exciting, such results are tempered by the fact that an understanding of the mechanisms of biomineralization in many metazoan model systems is in need of high quality in vivo functional assays. Indeed, the six next most abundant proteins present in the shell of M. venosa (F20065642, R30081972, F20050972, R20097001, F30059157, and R10061090), constituting a cumulative total of 53.8% of the dorsal (2 h sodium hypochlorite treated) proteome, shared no similarity with proteins in public databases, nor possessed recognizable Pfam domains. However, these proteins possess unusually high proportions of certain amino acids such as Asp and Gly (a common feature of previously described proteins involved in biomineralization), signal sequences (suggesting that they are secreted from mantle cells and are likely to be directly involved in the biomineralization process), and/or extremely acidic or basic predicted pI values.
Table 1.
Major Proteins of the Magellania venosa Shell Matrix
| Contig ID | Protein ID | BLAST Hits | Interpro Domains; Transmembrane Helices; Disordered Domains; Phosphorylation | % of Total Identified Dorsal Proteome 2/24 h | % of Total Identified Ventral Proteome 2/24 h | pI | SS | Anomalous Amino Acid Contents |
|---|---|---|---|---|---|---|---|---|
| 12373 | R20074237 | Uncharacterized | 2 TMH; DI | 13.8/8.4 | 11.6/8.0 | 4.34 | Y | Asp 12.1% |
| 10941 | F20065642 | Uncharacterized | — | 7.8/13.4 | 7.9/13.6 | 8.88 | N | — |
| 13662 | R30081972 | Uncharacterized | — | 9.6/8.3 | 5.0/8.3 | 9.34 | N | — |
| 8496 | F20050972 | Uncharacterized | — | 7.8/14.4 | 6.4/13.2 | 5.7 | Y | — |
| 16167 | R20097001 | Uncharacterized | TIMP like OB fold | 6.8/9.8 | 6.1/10.4 | 9.96 | N | — |
| 9860 | F30059157 | Uncharacterized | Phosphorylation: S174 (90%) | 4.8/0.6 | 4.4/0.6 | 9.71 | N | Gly 29.3% |
| 10182 | R10061090 | Uncharacterized | DI | 3.2/4.5 | 5.0/4.4 | 4.37 | Y | Asp 15.4% |
| 14565 | R20087389 | Similar to mesenchyme-specific cell surface glycoprotein/calcium and integrin-binding family member 2 | WD40/YVTN repeat like; DI | 4.5/5.3 | 3.1/5.2 | 9.19 | N | — |
| 13892 | R30083352 | Uncharacterized | — | 2.9/3.2 | 3.5/3.0 | 11.38 | Y | — |
| 481 | R20002885 | Uncharacterized | — | 2.8/0.1 | 3.0/0.1 | 9.88 | N | Gly 17.3% |
| 10544 | R20063263 | Uncharacterized | Phosphorylation: S73 (4%) | 2.4/2.7 | 2.9/2.5 | 6.64 | N | Ser 10.4% |
| 21084 a | R10126502 | Uncharacterized | FAD linked oxidase N term | 2.1/<0.01 | 2.9/0.0 | 10.35 | Y | — |
| 6089 b | F10036529 | Similar to complement receptor type 1 | Sushi/SCR/CCP (8×) | 2.4/5.6 | 2.3/5.8 | 6.28 | Y | — |
| 8840 | F20053036 | Uncharacterized | 2 TMH | 2.0/2.2 | 2.6/1.9 | 9.63 | Y | — |
| 25732 | R10154390 | Uncharacterized | — | 1.8/1.4 | 1.2/1.2 | 9.85 | Y | — |
| 17985 c | F20107906 | Similar to peroxidasin | Haem peroxidase, TSP1 repeat | 1.3/0.0 | 1.2/0.0 | 9.12 | N | — |
| 17302 | R20103811 | Uncharacterized | 2 TMH | 1.0/0.1 | 1.1/0.1 | 6.01 | N | Ala 11.8% |
| 16240 d | R10097438 | Uncharacterized | FAD linked oxidase N term, berberine; 3 TMH | 0.5/0.0 | 1.3/0.0 | 9.64 | N | — |
| 8882 | F20053288 | Uncharacterized | 1 TMH | 0.8/0.6 | 1.0/0.7 | 9.99 | Y | Gly 14.5% |
| 10588 | R10063526 | Uncharacterized | DI; phosphorylation: S272 (1%), S277 (9%), doubly phosphorylated peptide (69%) | 0.6/0.0 | 1.2/0.0 | 4.48 | Y | Gly 19.4%, Ser 22.6% |
| 17072 | R30102432 | Uncharacterized | — | 0.7/0.6 | 0.8/0.5 | 9.14 | Y | — |
| 1404 | R10008422 | Uncharacterized | DI | 0.8/1.1 | 0.6/1.3 | 9.49 | N | Ser 10.6% |
| 3349 | F10020089 | Uncharacterized | DI | 0.6/0.0 | 0.8/0.0 | 4.29 | N | Glu 19.5%, Ser 11.9% |
| 25890 | F30155337 | Uncharacterized | DI | 0.6/0.5 | 0.8/0.4 | 12.18 | N | — |
| 12037 | F10072217 | Similar to collagen α5 (VI), nontriple helical region | — | 0.7/0.2 | 0.7/0.2 | 10.01 | Y | Gly 11.5% |
| 1290 | R30007740 | Uncharacterized | VWA | 0.7/0.0 | 0.4/0.0 | 10.14 | N | — |
| 24592 | R20147551 | Uncharacterized | — | 0.4/0.0 | 0.6/0.0 | 9.6 | N | Lys 11.0% |
| 13443 | F30080655 | Uncharacterized | 1 TMH | 0.5/0.2 | 0.5/0.1 | 9.59 | Y | — |
| 3937 | R10023620 | Similar to CRE-TYR-3 | Tyrosinase | 0.3/0.0 | 0.6/0.0 | 9.06 | N | Cys 9.6%, Gly 10.9% |
| 3968 | F10023803 | Similar to mesenchyme-specific cell surface glycoprotein | WD40/YVTN repeat like/cytochrome cd1 (cyt cd1) nitrite reductase like, haem d1; DI | 0.4/1.5 | 0.4/1.7 | 6.47 | N | — |
| 6648 | R30039888 | Similar to lactadherin | Galactose binding like/coagulation factor 5/8 C | 0.3/0.8 | 0.6/0.7 | 9.41 | N | — |
| 12090 | F10072535 | Uncharacterized | DI | 0.4/1.0 | 0.3/1.2 | 8.85 | N | Gly 10.2% |
| 14281 | F10085681 | Similar to hemagglutinin family protein | DI | 0.5/0.4 | 0.3/0.4 | 5.12 | N | Ser 11.3% |
| 23379 | F20140270 | Uncharacterized | — | 0.5/1.9 | 0.3/1.9 | 10.42 | N | Gly 12.8% |
| 10375 | R10062248 | Uncharacterized | 3 TMH; DI | 0.5/0.7 | 0.3/0.8 | 10.91 | Y | Ala 11.2% |
| 22890 | R30137340 | Uncharacterized | ConA-like lectin | 0.1/0.0 | 0.6/0.0 | 9.72 | Y | — |
| 20588 | R20123527 | Uncharacterized | 1 TMH | 0.3/0.3 | 0.4/0.3 | 9.47 | N | — |
| 26985 | F30161907 | Uncharacterized | — | 0.3/0.0 | 0.3/0.0 | 9 | N | Ser 13.7% |
| 14828 e | R20088967 | Similar to complement receptor type 1 | — | 0.3/0.8 | 0.2/0.9 | 6.85 | Y | Gly 10.4% |
| 16143 | F30096855 | Uncharacterized | — | 0.4/0.4 | 0.3/0.4 | 9.02 | Y | — |
| 4638 | R30027828 | Uncharacterized | DOMON, SEA | 0.3/0.3 | 0.1/0.5 | 4.84 | N | — |
| 8473f | R10050836 | Uncharacterized/similar to peroxinectin | Haem peroxidase 3 | 0.1/<0.01 | 0.2/0.0 | 9.25 | N | — |
| 10638 | F10063823 | Uncharacterized | Sushi/SCR/CCP (8×); 1 TMH; DI | 0.2/1.1 | 0.2/1.1 | 9.39 | N | — |
| 14771 | F20088622 | Uncharacterized | 1 TMH; DI | 0.2/0.4 | 0.2/0.4 | 10.07 | N | — |
| 17017 | R20102101 | Similar to noelin-2 | Olfactomedin like | 0.1/0.0 | 0.3/0.0 | 9.14 | N | — |
| 19193 | R30115158 | Uncharacterized | CUB; 1 TMH | 0.3/0.3 | 0.2/0.4 | 8.66 | N | — |
| 19686 | F20118112 | Uncharacterized | — | 0.0/0.0 | 0.3/0.0 | 5.26 | N | — |
| 22374 | F30134241 | Uncharacterized | — | 0.0/0.1 | 0.4/0.1 | 8.91 | N | Cys 8.2%, Gly 18.9% |
| 22496 | F10134971 | Uncharacterized | — | 0.3/0.0 | 0.1/0.0 | 9.3 | N | — |
| 26237 | F30157419 | Uncharacterized | — | 0.2/0.1 | 0.2/0.1 | 7.89 | N | — |
| 1954 | F20011720 | Neutral ceramidase B | Neutral/alkaline nonlysosomal ceramidase | 0.2/0.2 | 0.2/0.2 | 8.26 | Y | — |
| 24553 | R10147316 | Uncharacterized | TSP1 | 0.3/0.1 | 0.2/0.2 | 8 | Y | Cys 10.0%, Gly 11.0% |
| 26581 | F10159481 | Uncharacterized | DI | 0.2/0.1 | 0.2/0.1 | 11.96 | Y | — |
| 1953 | R10011716 | Neutral/alkaline nonlysosomal ceramidase | Neutral/alkaline nonlysosomal ceramidase; 1 TMH | 0.1/0.2 | 0.1/0.2 | 7.77 | N | — |
| 3405 | F30020427 | Similar to cysteine-rich secretory protein LCCL domain-containing 2 | CAP | 0.2/0.0 | 0.1/0.0 | 7.92 | N | — |
| 14004 | R10084022 | Uncharacterized | — | 0.1/0.1 | 0.1/0.1 | 9.27 | N | — |
| 20189 | F20121130 | Uncharacterized | — | 0.1/0.0 | 0.1/0.0 | 9.67 | N | Lys 10.1% |
| 20189 | F30121131 | Uncharacterized | DI | 0.1/0.0 | 0.2/0.0 | 9.69 | N | — |
| 21060 | R20126359 | Similar to MAM domain-containing glycosylphosphatidylinositol anchor protein 1 | ConA like lectin/MAM, peptidase S1 | 0.1/0.2 | 0.1/0.1 | 8.38 | N | — |
| 21061 | R30126366 | Similar to atrial natriuretic peptide-converting enzyme | LDLRA 2, SCRC 2, Peptidase S1/trypsin | 0.1/0.2 | 0.1/0.2 | 8.46 | N | — |
| 14224 | F10085339 | Uncharacterized | EGF; DI | 0.1/0.1 | 0.0/0.1 | 7.04 | Y | — |
| 18776 | R30112656 | Uncharacterized | CUB; 1 TMH | 0.1/0.1 | 0.1/0.1 | 8.02 | Y | — |
| 21109 | F30126651 | Uncharacterized | — | 0.0/0.2 | 0.0/0.2 | 6.69 | N | — |
| 27260 | F20163556 | Similar to SCO-spondin/hemicentin-1 | TSP-1 | 0.0/0.3 | 0.0/0.3 | 8.4 | N | Cys 14.8%, Ser 11.8% |
| 24557 | R30147342 | Uncharacterized | 1 TMH | 0.1/0.8 | <0.010/0.8 | 5.52 | Y | Cys 11.2%, Gly 12.1% |
| 17584 | R20105503 | Uncharacterized | Trypsin inhibitor like Cys-rich | 0.0/0.6 | <0.01/1.1 | 10.03 | Y | Cys 7.6%, Gly 16.0% |
Note.—Major proteins were arbitrarily defined as proteins occurring with an average percentage of ≥0.1 in either D1–3 and V1–3 (2 h) or D4–6 and V4–6 (24 h) or both. Known intracellular proteins or membrane proteins with predominantly intracellular function, some of which showed high percentages in samples treated with hypochlorite for 2 h only, are not included. DI, predicted disordered structure; TMH, predicted transmembrane helices; SS, predicted secretion signal peptide.
aPossibly a fragment of the same protein as peptide R10097438.
bPossibly the same protein as contig 14828 (R20088967), possibly the N-terminal half.
cShares two peptides with R10050836.
dPossibly a fragment of the same protein as peptide R10126502.
ePossibly the same protein as contig 6089 (F10036529)—possibly the C-terminal half.
fShares peptides with F20107906.
Fig. 3.—
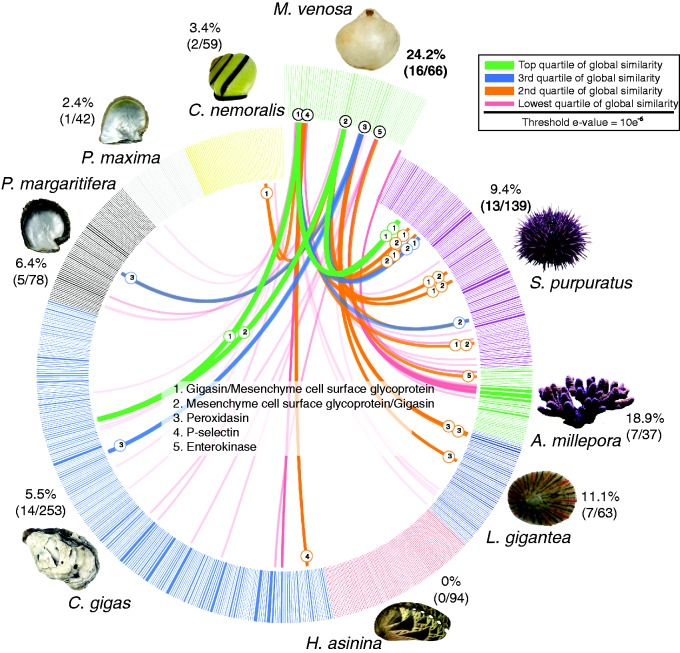
Sequence-based comparisons of M. venosa’s shell-forming proteome against other metazoan data sets. BLASTp based similarity comparison of the 66 most abundant M. venosa shell proteins against 765 shell-forming proteomes derived from eight other biocalcifying metazoans. All 66 major M. venosa shell proteins were searched against a concatenated database of shell-forming proteins derived from P. maxima and P. margaritifera (Marie et al. 2012); H. asinina; (Marie et al. 2010); L. gigantea; (Marie et al. 2013); C. gigas (Zhang et al. 2012), S. purpuratus (Mann et al. 2008), and A. millepora (Ramos-Silva et al. 2014) using BLASTp and an e-value cut-off of 10 e-06. Individual lines spanning the ideogram connect proteins that share significant similarity (e values < 10 e-6). Transparent red lines connect proteins with the lowest quartile of similarity (with a threshold of 10 e-6) and green lines with the highest quartile of similarity. The percentage of each shell proteome that shared similarity with the M. venosa proteome is provided.
One of the moderately abundant proteins (R20087389) shared significant sequence similarity with several other previously identified biomineralization proteins such as “Gigasin,” a protein previously isolated from the shell of the oyster Crassotrea gigas (Zhang et al. 2012), a protein detected in the shell proteome of Cepaea nemoralis (Mann and Jackson 2014), and a mesenchyme specific protein (MSP130) detected in the biomineralizing proteome of the sea urchin Strongylocentrotus purpuratus (Mann et al. 2008) and that was also recently shown to have been horizontally transferred (Ettensohn 2014). In fact, this M. venosa protein and another one (F10023803) were the two proteins to display the highest level of sequence similarity (which could also be interpreted as the highest level of evolutionary conservation) with other metazoan biomineralization proteins in our targeted comparisons (fig. 3). Interestingly, the sea urchin homolog (if this sequence similarity is interpreted as evidence of shared ancestry) of R20087389 has been functionally shown to be critical for the calcification of larval spines (Carson et al. 1985). This suggests that although there has been extensive lineage-specific evolution of M. venosa’s biomineralization strategy, there are some signatures of deep conservation that extend back to a spiralian/deuterostome ancestor. Further functional characterization of these proteins from M. venosa and from a variety of molluscan taxa would be informative from an evolutionary perspective.
Other proteins relatively abundant in the shell of M. venosa that shared similarity with other metazoan biomineralizing proteins were F20107906 and R10050836 which displayed similarity to oyster, Cepaea nemoralis and Lottia gigantea proteins and possess animal heme-dependent peroxidase domains. Peroxidases are well known to be present in biomineralizing tissues in mollusks (Timmermans 1969; Hohagen and Jackson 2013), and are generally thought to function in the cross-linking of proteins present in molluscan periostracum (Kniprath 1977). Another brachiopod shell-forming protein with similarity to proteins in all three oysters surveyed was R10023620. These proteins all possess a tyrosinase domain, an enzyme that catalyzes the production of melanin and is known to be involved in biomineralization in bivalves (Zhang et al. 2006; Clark et al. 2010; Yu et al. 2014).
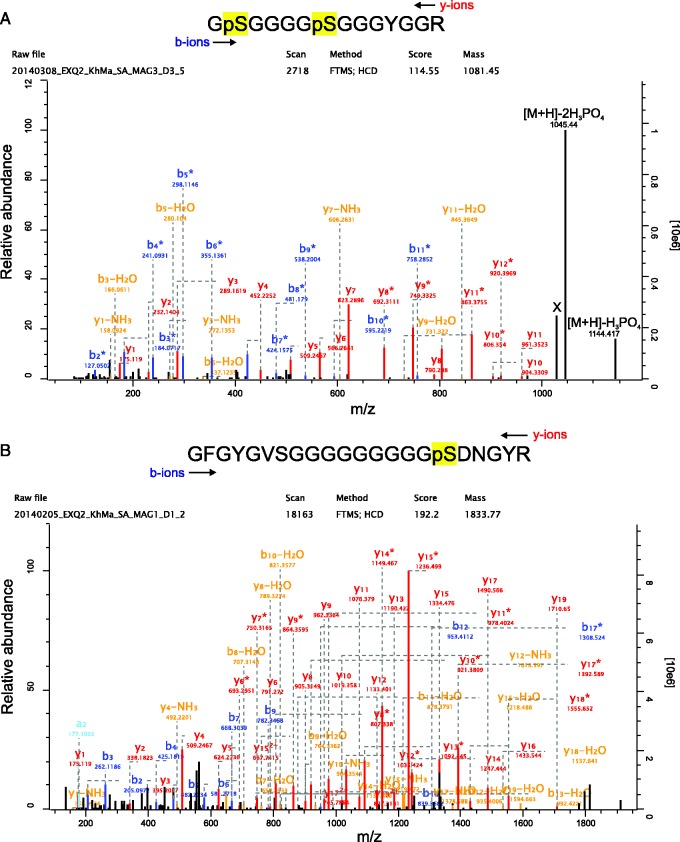
Because many biomineralizing proteins are known to possess repetitive low-complexity domains (Jackson et al. 2010; Marie et al. 2013; Le-Roy et al. 2014), we searched for proteins with tandem repeats in the collection of 66 major M. venosa shell proteins. A total of 15 proteins were identified by XStream to possess tandem repeats and low-complexity domains (supplementary file S10, Supplementary Material online). Of these 15, three are apparently unique to M. venosa (i.e., they possess no sequence similarity to any proteins or nucleotide sequences in GenBank’s nr and nt databases) and are putatively full length. One of these proteins (R10063526) has a striking architecture possessing a signal sequence, extraordinarily high proportions of glycine (19.4%) and serine (22.5%) and phosphorylated residues (table 1). Interestingly, Lustrin-A, one of the first molluscan biocalcification proteins to have its primary structure fully elucidated (Shen et al. 1997), possesses an extensive Gly/Ser-rich domain that has been suggested to function as a molecular shock absorber and to inhibit the propagation of fractures through the biomineral (Smith et al. 1999). The phosphorylation sites of R10063526 (fig. 4A) occur in a single peptide on two closely spaced serines and were identified without prior enrichment of phosphopeptides. Comparison of the number of unmodified copies of this peptide and the phosphorylated forms (among approximately 100 identifications) indicated a high site occupancy and therefore suggested some functional or structural importance of these phospho sites, although this possible function may not be related to interaction with the mineral phase because such phosphoproteins usually show multiple phosphorylation sites. This protein disappeared after 24 h of hypochlorite treatment and may therefore be considered as a potential intercrystalline protein (fig. 5). Another phosphorylated major protein that became less abundant after aggressive hypochlorite treatment was F20107906 (fig. 4B), which contained one single phosphoserine with high site occupancy (fig. 4B). This protein, similar to phosphoprotein R10063526, did not return a significant FASTA hit and its sequence was unusually rich in glycine and alanine (table 1).
Fig. 4.—
Spectrum of the major phospho-peptides of R10063526 and F30059157. (A) The doubly phosphorylated R10063526 peptide containing phosphorylated S272 and S277 of this entry was identified with a posterior error probability (PEP) of 0.0002 and a mass error of 0.7 ppm. The MaxQuant localization probability was 1 for both sites. Expert System advanced annotations (Neuhauser et al. 2012) were omitted to reduce complexity, except for the last two peaks (m/z 1,144.41 and 1,045.44). Peak X was not annotated by the expert system but most probably represents [M+H]-2H3PO4-H2O. Y-ions are shown in red, b-ions are shown in blue, b- or y-ions with a loss of water or ammonia are in orange. Asterisks indicate loss of H3PO4. (B) The singly phosphorylated F30059157 peptide (S174) was identified with a mass error of 0.7 ppm, a PEP of 3.4 e-67, and a phosphosite localization probability of 1. Y-ions are shown in red, b-ions are shown in blue, b- or y-ions with a loss of water or ammonia are in orange. Asterisks indicate loss of H3PO4. Expert System advanced annotations (Neuhauser et al. 2012) are omitted for clarity of presentation.
Fig. 5.—
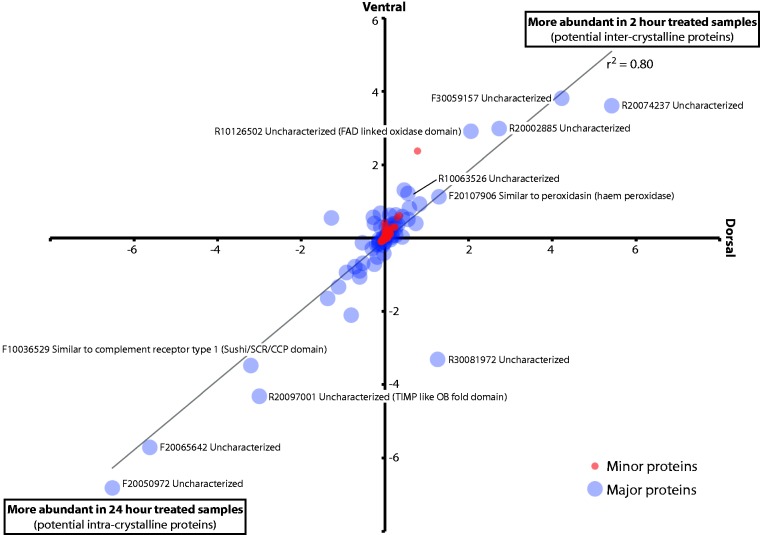
Effect of 2- versus 24-h sodium hypochlorite treatment on the abundance of proteins in dorsal and ventral shell valves. The relative abundances of 333 minor proteins (red circles) and 66 major proteins (blue circles) identified in M. venosa shells were compared following 2- versus 24-h treatment with sodium hypochlorite. iBAQ values for each peptide recorded after 24 h of treatment were subtracted from those recorded after 2 h of treatment. Negative values therefore indicate a higher peptide abundance in 24 h treated samples.
Conspicuous in their absence from the 66 most abundant proteins we detected in shells of M. venosa are any proteins with alpha-carbonic anhydrase (α-CA) domains. As far as we are aware, all full-scale and even most partial proteome surveys of metazoan biominerals have revealed at least one (and usually several) protein with these domains (Jackson et al. 2007; Mann et al. 2008; Marie et al. 2012, 2013; Zhang et al. 2012; Ramos-Silva et al. 2013; Le-Roy et al. 2014; Mann and Edsinger 2014; Mann and Jackson 2014; Voigt et al. 2014). Although we did not detect α-CAs as members of the major proteinaceous component of M. venosa’s shell, we could identify them as minor components (supplementary file S5, Supplementary Material online) and also as sequences present in the mantle transcriptome. We could identify 12 such contigs (140, 2070, 2823, 5097, 5231, 5661, 17483, 17621, 19612, 20103, 20347, and 27050), 6 of which were putatively full length, and 5 of which possessed signal sequences (supplementary file S11, Supplementary Material online). Further analysis of the activity and cellular spatial localization of these transcripts would be informative, especially in a comparative context with other biomineralizing taxa.
As mentioned above, the relative abundances of several major proteins (supplementary file S12, Supplementary Material online) were significantly affected by our extended (24 h cf. 2 h) treatment of M. venosa shells with sodium hypochlorite (fig. 5). Although we have no data that can directly explain this phenomenon, one obvious possibility is that these two “populations” of proteins represent inter- and intracrystalline proteins, respectively. An extended sodium hypochlorite treatment might be expected to degrade more accessible intercrystalline proteins resulting in an increase in the relative abundances of intracrystalline proteins. Also of note is the fact that most proteins are found in equal abundance in dorsal and ventral valves (the line of best fit in fig. 5 has a slope of 0.96 and a correlation coefficient of 0.80), and the majority of the peptides we observe does not appear to be significantly affected by the different sodium hypochlorite treatments (the majority of the data is focused on the origin). The functions of those peptides that are apparently affected by our two different hypochlorite treatments are difficult to infer due to a lack of sequence similarity to previously characterized proteins; however, many of these uncharacterized proteins contain anomalous amino acid contents typical of known biomineralizing proteins.
Relevance of M. venosa’s Shell Ultrastructure to Its Reaction to Sodium Hypochlorite Treatment and Its Shell-Forming Proteome
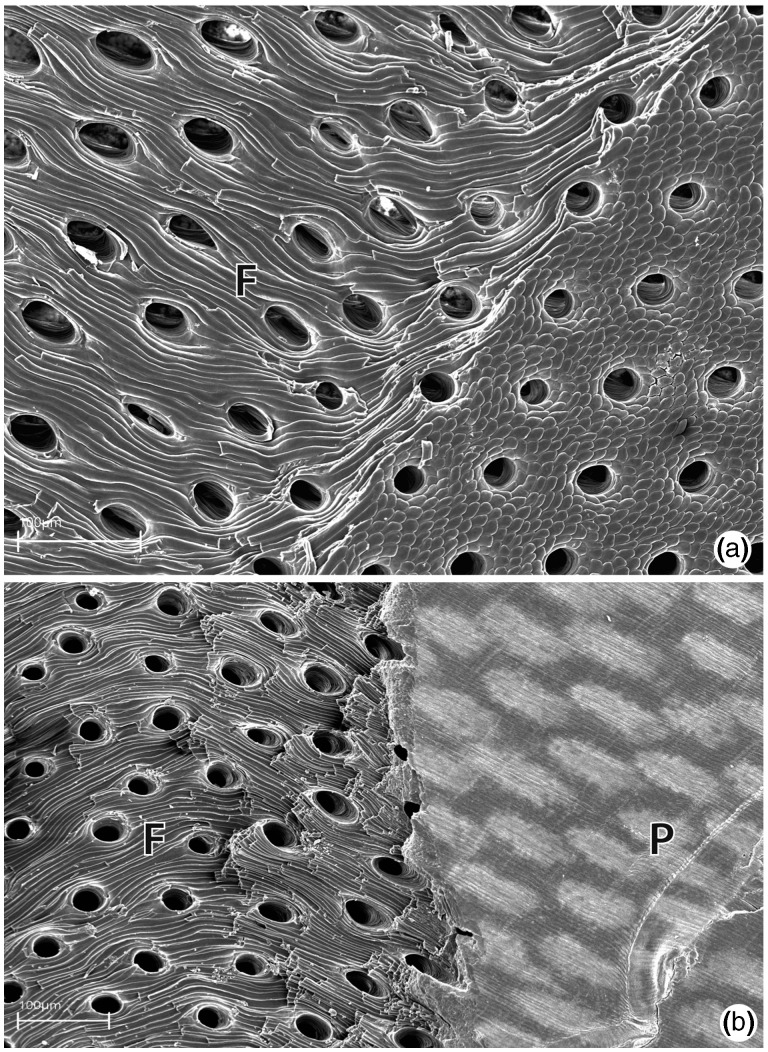
The shell of M. venosa (and of many other brachiopods producing calcitic shells) contains extensive perforations known as punctae which span the entire thickness of the shell (fig. 6) (Pérez-Huerta et al. 2009 and references cited therein). The punctae of M. venosa are typical of terebratulids, with a relatively narrow opening at the side of the shell facing the mantle epithelium and a funnel-shaped end without ramifications adjacent to the periostracum. Although the function of punctae remains a topic of research, a significant result of their presence is the apparent increased exposure of the intercrystalline organic matrix to molecules in the surrounding medium. Punctae are likely relevant to the in vitro preparation methods we employed in this study, and are possibly relevant to microbial and/or predator mediated degradation of calcitic brachiopod shells by proteases (see discussion below). Softening of calcite brachiopod shells as a consequence of hypochlorite treatment has been previously described and attributed to a loss of organic matrix. Collins et al. (1988) demonstrated that prolonged hypochlorite treatment (48 h) is enough to reduce the strength of calcite brachiopod shells to an extent that they can be easily disintegrated by shaking, a phenomenon in our experience not commonly observed with mollusk shells. A similar softening of brachiopod shells was also observed when the shells of dead Terebratulina retusa animals were simply maintained in sea water; after 218 days these shells became extremely fragile and lost more than 90% of their pressure resistance (Collins 1986; Emig 1990). Such studies clearly demonstrate both the important functional role of the organic extracrystalline matrix (those molecules that surround lamellae, fibers, tablets, and all other biomineral substructures vs. occluded intracrystalline molecules) for maintaining shell strength and integrity, and that the accessibility of this matrix to molecules such as microbial proteases may be the cause of extracrystalline matrix degradation in rotting shells (Collins 1986). Conspicuous in their paucity from the M. venosa shell-forming proteomes are proteins possessing protease inhibitor domains (see table 1 and discussion below). Such proteins feature prominently in several molluscan shell-forming proteomes, such as abalone (Marie et al. 2010), limpet (Marie et al. 2013), and the pulmonate Cepaea nemoralis (Mann and Jackson 2014). The relatively abundant presence of these proteins in such molluscan data sets previously led us to suggest that they may function within molluscan shells to prevent degradation of the organic matrix by microbial or shell-boring predatory action (Mann and Jackson 2014). Although such a hypothesis awaits experimental validation, it is interesting to note that brachiopod shells have been documented to be susceptible to such action (Collins 1986; Collins et al. 1988; Emig 1990).
Fig. 6.—
SEM images of punctae in the shell of M. venosa. (a) Punctae in a broken valve of M. venosa (view from inside of shell) with fibrous structure (F) of secondary shell layer clearly visible. (b) Punctae in a broken shell of M. venosa (view from outside of shell) at the interface of the primary (P, right side of image) and fibrous secondary layers (F, left side of image).
Our broad survey of the shell-forming proteome of M. venosa reveals a wealth of novel proteins that are likely to be playing critical roles in determining the mineralogy, ultrastructure and material properties of the mature biomineral, and highlights the need for meaningful in vivo functional assays to be developed for nonmodel organisms such as M. venosa. The data that we present here serve as a platform from which more targeted and in-depth analyses can be performed on M. venosa’s biomineralization strategy. Such efforts would clarify the function of the major elements of M. venosa’s shell-forming proteome, and in turn allow for clearer comparisons to be made between the biomineralizing proteomes of more distant metazoan taxa. Insights gained from such work would guide the development of hypotheses related to how the metazoan ability to biomineralize evolved, an ability of great relevance to the rapid radiation of complex multicellular life in the early Cambrian.
Supplementary Material
Supplementary files S1–S12 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors acknowledge Matthias Mann (MPI of Biochemistry, Martinsried) for generous support, Gaby Sowa (MPI) for preparing the capillary columns, Korbinian Mayr and Igor Paron (both MPI) for keeping the mass spectrometers in excellent condition, Simone Schätzle for assistance with RNA extraction, and Roland Melzer for assistance with the SEM work. They also thank to Günter Försterra, Emma Plotnek, Dan Genter, Ulrich Pörschmann, and Katie McConnell for helping to collect the brachiopods. This work was supported by funding to D.J.J. through the German Excellence Initiative and the Deutsche Forschungsgemeinschaft (DFG) project #JA2108/2-2, as well as funds from the Department of Earth and Environmental Sciences to W.S. and G.W., and from the research department of the Museum für Naturkunde, Berlin to C.L. This is publication nr 121 of Huinay Scientific Field Station. The authors declare no competing interests.
Literature Cited
- Accinni L, Natali PG, Silvestrini M, Martino CD. Actin in the extracellular matrix of smooth muscle cells. An immunoelectron microscopic study. Connect Tiss Res. 1983;11:69–78. doi: 10.3109/03008208309015012. [DOI] [PubMed] [Google Scholar]
- Brand U. Geochemistry of modern brachiopods: applications and implications for oceanography and paleoceanography. Chem Geol. 2003;198:305–334. [Google Scholar]
- Carlson SJ. Phylogenetic relationships among extant brachiopods. Cladistics. 1995;11:131–197. doi: 10.1111/j.1096-0031.1995.tb00084.x. [DOI] [PubMed] [Google Scholar]
- Carson DD, Farachach MC, Earles DS, Decker GL, Lennarz WJ. A monoclonal antibody inhibits calcium accumulation and skeleton formation in cultured embryonic cells of the sea urchin. Cell. 1985;41:639–648. doi: 10.1016/s0092-8674(85)80036-2. [DOI] [PubMed] [Google Scholar]
- Clark MS, et al. Insights into shell deposition in the Antarctic bivalve Laternula elliptica: gene discovery in the mantle transcriptome using 454 pyroseqeuencing. BMC Genomics. 2010;11:362. doi: 10.1186/1471-2164-11-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen BL. Rerooting the rDNA gene tree reveals phoronids to be “brachiopods without shells”; dangers of wide taxon samples in metazoan phylogenetics (Phoronida; Brachiopoda) Zool J Linn Soc. 2013;167:82–92. [Google Scholar]
- Cohen BL, Weydmann A. Molecular evidence that phoronids are a subtaxon of brachiopods (Brachiopoda: Phoronata) and that genetic divergence of metazoan phyla began long before the early Cambrian. Org Div Evol. 2005;5:253–273. [Google Scholar]
- Collins MJ. Post mortality strength loss in shells of the recent articulate brachiopod Terebratulina retusa from the west coast of Scotland. Biostratigr Paléozoïque. 1986;4:209–218. [Google Scholar]
- Collins MJ, et al. Sero-taxonomy of skeletal macromolecules in living Terebratulid brachiopods. Hist Biol. 1988;1:207–224. [Google Scholar]
- Collins MJ, Muyzer G, Curry GB, Sandberg P, Westbroek P. Macromolecules in brachiopod shells: characterization and diagenesis. Lethaia. 1991;24:387–397. [Google Scholar]
- Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Curry GB, et al. Biogeochemistry of brachiopod intracrystalline molecules [and discussion] Philos Trans R Soc Lond B Biol Sci. 1991;333:359–366. [Google Scholar]
- Curry GB, Fallick AE. Use of stable oxygen isotope determinations from brachiopod shells in palaeoenvironmental reconstruction. Palaeogeogr Palaeoclimatol Palaeoecol. 2002;182:133–143. [Google Scholar]
- Cusack M, Curry G, Clegg H, Abbott G. An intracrystalline chromoprotein from red brachiopod shells: implications for the process of biomineralization. Comp Biochem Physiol B. 1992;102:93–95. doi: 10.1016/0305-0491(92)90278-y. [DOI] [PubMed] [Google Scholar]
- Cusack M, Huerta AP, Huerta AP. Brachiopods recording seawater temperature‚ a matter of class or maturation? Chem Geol. 2012;334:139–143. [Google Scholar]
- Darzentas N. Circoletto: visualizing sequence similarity with Circos. Bioinformatics. 2010;26:2620–2621. doi: 10.1093/bioinformatics/btq484. [DOI] [PubMed] [Google Scholar]
- Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- Drake JL, et al. Proteomic analysis of skeletal organic matrix from the stony coral Stylophora pistillata. Proc Natl Acad Sci U S A. 2013a;110:3788–3793. doi: 10.1073/pnas.1301419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JL, et al. Reply to Ramos-Silva et al.: Regarding coral skeletal proteome. Proc Natl Acad Sci U S A. 2013b;110:E2147–E2148. doi: 10.1073/pnas.1304591110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emig CC. Examples of post-mortality alteration in Recent brachiopod shells and (paleo)ecological consequences. Mar Biol. 1990;104:233–238. [Google Scholar]
- Ettensohn CA. Horizontal transfer of the msp130 gene supported the evolution of metazoan biomineralization. Evol Dev. 2014;16:139–148. doi: 10.1111/ede.12074. [DOI] [PubMed] [Google Scholar]
- Gaspard D, et al. Shell matrices of Recent rhynchonelliform brachiopods: microstructures and glycosylation studies. Earth Environ Sci Trans R Soc Edinb. 2008;98:415–424. [Google Scholar]
- Gasteiger E, et al. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. Totowa (NJ): Humana Press; 2005. pp. 571–607. [Google Scholar]
- Goetz AJ, et al. Calcite morphology, texture and hardness in the distinct layers of rhynchonelliform brachiopod shells. Eur J Min. 2009;21:303–315. [Google Scholar]
- Goetz AJ, et al. Interdigitating biocalcite dendrites form a 3-D jigsaw structure in brachiopod shells. Acta Biomater. 2011;7:2237–2243. doi: 10.1016/j.actbio.2011.01.035. [DOI] [PubMed] [Google Scholar]
- Goujon M, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohagen J, Jackson DJ. An ancient process in a modern mollusc: early development of the shell in Lymnaea stagnalis. BMC Dev Biol. 2013;13:27. doi: 10.1186/1471-213X-13-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35:W460–W464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DJ, et al. Parallel evolution of nacre building gene sets in molluscs. Mol Biol Evol. 2010;27:591–608. doi: 10.1093/molbev/msp278. [DOI] [PubMed] [Google Scholar]
- Jackson DJ, Macis L, Reitner J, Degnan BM, Wörheide G. Sponge paleogenomics reveals an ancient role for carbonic anhydrase in skeletogenesis. Science. 2007;316:1893–1895. doi: 10.1126/science.1141560. [DOI] [PubMed] [Google Scholar]
- Jope M. The protein of brachiopod shell—I. Amino acid composition and implied protein taxonomy. Comp Biochem Physiol. 1966a;20:593–600. [Google Scholar]
- Jope M. The protein of brachiopod shell—II. Shell protein from fossil articulates: amino acid composition. Comp Biochem Physiol. 1966b;20:601–605. [Google Scholar]
- Kniprath E. Zur ontogenese des schalenfeldes von Lymnaea stagnalis. Dev Genes Evol. 1977;181:11–30. [Google Scholar]
- Knoll AH. Biomineralization and evolutionary history. Rev Mineralogy Geochem. 2003;54(1):329–356. [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Roy N, Jackson DJ, Marie B, Ramos-Silva P, Marin F. The evolution of metazoan α-carbonic anhydrases and their roles in calcium carbonate biomineralization. Front Zool. 2014;11:75. [Google Scholar]
- Lee X, et al. Ontogenetic trace element distribution in brachiopod shells: an indicator of original seawater chemistry. Chem Geol. 2004;209:49–65. [Google Scholar]
- Lowenstam HA, Weiner S. On biomineralization. New York: Oxford University Press; 1989. [Google Scholar]
- Mann K, Edsinger E. The Lottia gigantea shell matrix proteome: re-analysis including MaxQuant iBAQ quantitation and phosphoproteome analysis. Proteome Sci. 2014;12:28. doi: 10.1186/1477-5956-12-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Jackson DJ. Characterization of the pigmented shell-forming proteome of the common grove snail Cepaea nemoralis. BMC Genomics. 2014;15:249. doi: 10.1186/1471-2164-15-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Poustka AJ, Mann M. The sea urchin (Strongylocentrotus purpuratus) test and spine proteomes. Proteome Sci. 2008;6:22. doi: 10.1186/1477-5956-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, et al. Proteomic analysis of the calcified organic matrix of the tropical abalone Haliotis asinina shell. Prot Sci. 2010;8:54. doi: 10.1186/1477-5956-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, et al. Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. Proc Natl Acad Sci U S A. 2012;109:20986–20991. doi: 10.1073/pnas.1210552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, et al. The shell-forming proteome of Lottia gigantea reveals both deep conservations and lineage specific novelties. FEBS J. 2013;280:214–232. doi: 10.1111/febs.12062. [DOI] [PubMed] [Google Scholar]
- Mass T, Drake JL, Peters EC, Jiang W, Falkowski PG. Immunolocalization of skeletal matrix proteins in tissue and mineral of the coral Stylophora pistillata. Proc Natl Acad Sci U S A. 2014;111:12728–12733. doi: 10.1073/pnas.1408621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel C, et al. Micromechanical properties and structural characterization of modern inarticulated brachiopod shells. J Geophys Res Biogeosci. 2007;112:1–12. [Google Scholar]
- Merkel C, et al. Mechanical properties of modern calcite- (Mergerlia truncata) and phosphate-shelled brachiopods (Discradisca stella and Lingula anatina) determined by nanoindentation. J Struct Biol. 2009;168:396–408. doi: 10.1016/j.jsb.2009.08.014. [DOI] [PubMed] [Google Scholar]
- Michalski A, et al. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics. 2011;10:1–11. doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauser N, Michalski A, Cox J, Mann M. Expert system for computer-assisted annotation of MS/MS spectra. Mol Cell Proteomics. 2012;11:1500–1509. doi: 10.1074/mcp.M112.020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Cooper JB. XSTREAM: a practical algorithm for identification and architecture modeling of tandem repeats in protein sequences. BMC Bioinformatics. 2007;8:382. doi: 10.1186/1471-2105-8-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson D, Curry GB, Cusack M, Fallick AE. Shell structure, patterns and trends of oxygen and carbon stable isotopes in modern brachiopod shells. Chem Geol. 2005;219:193–235. [Google Scholar]
- Pérez-Huerta A, et al. Brachiopod punctae: a complexity in shell biomineralisation. J Struct Biol. 2009;167:62–67. doi: 10.1016/j.jsb.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Rahman MA, Shinjo R, Oomori T, Worheide G. Analysis of the proteinaceous components of the organic matrix of calcitic sclerites from the soft coral Sinularia sp. PLoS One. 2013;8:e58781. doi: 10.1371/journal.pone.0058781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Silva P, et al. The skeletal proteome of the coral Acropora millepora: the evolution of calcification by co-option and domain shuffling. Mol Biol Evol. 2013;30:2099–2112. doi: 10.1093/molbev/mst109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Silva P, et al. The skeleton of the staghorn coral Acropora millepora: molecular and structural characterization. PLoS One. 2014;9:e97454. doi: 10.1371/journal.pone.0097454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Rudwick MJS. Living and fossil brachiopods. London: Hutchinson; 1970. [Google Scholar]
- Scheltema RA, Mann M. SprayQc: a real-time LC-MS/MS quality monitoring system to maximize uptime using off the shelf components. J Proteome Res. 2012;11:3458–3466. doi: 10.1021/pr201219e. [DOI] [PubMed] [Google Scholar]
- Schmahl WW, et al. Hierarchical fibre composite structure and micromechanical properties of phosphatic and calcitic brachiopod shell biomaterials—an overview. Mineral Mag. 2008;72:541–562. [Google Scholar]
- Schmahl WW, et al. Hierarchical structure of marine shell biomaterials: biomechanical functionalization of calcite by brachiopods. Z Kristallogr Cryst Mater. 2012;227:793–804. [Google Scholar]
- Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Sharma K, et al. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- Shen X, Belcher AMB, Hansma PK, Stucky GD, Morse DEM. Molecular cloning and characterization of Lustrin A, a matrix protein from shell and pearl nacre of Haliotis rufescens. J Biol Chem. 1997;272:32472–32481. doi: 10.1074/jbc.272.51.32472. [DOI] [PubMed] [Google Scholar]
- Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkiss K, Wilbur KM. Biomineralization, cell biology and mineral deposition. San Diego (CA): Academic Press; 1989. [Google Scholar]
- Smith BL, et al. Molecular mechanistic origin of the toughness of natural adhesives, fibres and composites. Nature. 1999;399:761–763. [Google Scholar]
- Sperling EA, Pisani D, Peterson KJ. Molecular paleobiological insights into the origin of the Brachiopoda. Evol Dev. 2011;13:290–303. doi: 10.1111/j.1525-142X.2011.00480.x. [DOI] [PubMed] [Google Scholar]
- Taylor PD, Vinn O, Wilson MA. Evolution of biomineralization in lophophorates. Spec Pap Palaeontol. 2010;84:317–333. [Google Scholar]
- Timmermans LPM. Studies on shell formation in molluscs. Neth J Zool. 1969;19:417–523. [Google Scholar]
- Voigt O, Adamski M, Sluzek K, Adamska M. Calcareous sponge genomes reveal complex evolution of alpha-carbonic anhydrases and two key biomineralization enzymes. BMC Evol Biol. 2014;14:230. doi: 10.1186/s12862-014-0230-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A. Biomineralization in the lophophorates. 1989. Skeletal biomineralization: patterns, processes and evolutionary trends. In: Carter JG, editor. Skeletal biomineralization: patterns, processes and evolutionary trends, Vol. 1. New York: Van Nostrand Reinhold. p. 67–82. [Google Scholar]
- Williams A. Shell structure. In: Kaesler RL, editor. Treatise on invertebrate paleontology, Part H: Brachiopoda, revised. Vol. 1. Boulder (CO)/Lawrence (KS): The Geological Society of America/University of Kansas; 1997. pp. 267–320. [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Yu X, et al. Molecular cloning and differential expression in tissues of a tyrosinase gene in the Pacific oyster Crassostrea gigas. Mol Biol Rep. 2014;41:5403–5411. doi: 10.1007/s11033-014-3412-2. [DOI] [PubMed] [Google Scholar]
- Zhang C, Xie L, Huang J, Chen L, Zhang R. A novel putative tyrosinase involved in periostracum formation from the pearl oyster (Pinctada fucata) Biochem Biophys Res Commun. 2006;342:632–639. doi: 10.1016/j.bbrc.2006.01.182. [DOI] [PubMed] [Google Scholar]
- Zhang G, et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012;490:49–54. doi: 10.1038/nature11413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.