Abstract
Objective:
Recently, single nucleotide polymorphisms (SNPs) associated with esophageal adenocarcinoma (EAC) and Barrett's esophagus (BE) were identified; rs10419226 (CRTC1), rs11789015 (BARX1), rs2687201 (FOXP1), rs2178146 (FOXF1), rs3111601 (FOXF1), and rs9936833 (FOXF1). These findings indicate that genetic susceptibility could play a role in the initiation of EAC in BE patients. The aim of this study was to validate the association between these previously identified SNPs and the risk of EAC in an independent and large case–control study.
Design:
Six SNPs found to be associated with EAC and BE were genotyped by a multiplex SNaPshot analysis in 1071 EAC patients diagnosed and treated in the Netherlands. Allele frequencies were compared to a control group derived from the Rotterdam Study, a population-based prospective cohort study (n = 6206). Logistic regression analysis and meta-analysis were performed to calculate odds ratios (OR).
Results:
Rs10419226 (CRTC1) showed a significantly increased EAC risk for the minor allele (OR = 1.17, P = 0.001), and rs11789015 (BARX1) showed a significantly decreased risk for the minor allele (OR = 0.85, P = 0.004) in the logistic regression analysis. The meta-analysis of the original GWAS and the current study revealed an improved level of significance for rs10419226 (CRTC1) (OR = 1.18, P = 6.66 × 10–10) and rs11789015 (BARX1) (OR = 0.83, P = 1.13 × 10–8).
Conclusions:
This independent and large Dutch case–control study confirms the association of rs10419226 (CRTC1) and rs11789015 (BARX1) with the risk of EAC. These findings suggest a contribution of the patient genetic make-up to the development of EAC and might contribute to gain more insight in the etiology of this cancer.
Keywords: Barrett's esophagus, esophageal adenocarcinoma, single nucleotide polymorphisms
INTRODUCTION
Esophageal adenocarcinoma (EAC) is one of the rapidly rising cancers in the Western world.[1,2,3] Despite improvements in multimodality treatment, the prognosis for EAC remains disconcerting.[4] The major risk factor for EAC is the premalignant lesion Barrett's esophagus (BE), in addition to age, male gender, and Caucasian ethnicity.[5] As a consequence of gastro-esophageal reflux disease (GERD), the normal squamous epithelium of the lower esophagus can be replaced by columnar intestinal cells, including goblet cells, representing BE. Per year 0.12–0.5% of the patients diagnosed with BE will develop EAC, following a multimorphological sequence, in which intestinal metaplasia evolves to low-grade dysplasia (LGD), high-grade dysplasia (HGD) and ultimately to invasive adenocarcinoma.[6,7,8]
The prevalence of BE in the general population is estimated at 2%,[9] and among patients with GERD even at 10%.[10] Since the prognosis of advanced EAC is relatively poor,[3] patients with BE are subjected to intensive endoscopic surveillance with biopsy sampling to identify those patients with neoplastic progression at an early stage.[11] However, because the annual risk of developing EAC from BE is relatively low, most BE patients will not progress to cancer and do not benefit from this surveillance.[7,8]
It can be anticipated that genetic susceptibility could play a role in the initiation of EAC in BE patients. From this perspective the identification of single nucleotide polymorphisms (SNPs), which identifies high-risk patients, could make the surveillance of BE patients more cost-effective and could be helpful by diagnosing patients with EAC in an early and curable stage and thereby increase the prognosis remarkably.
Recently, the first Genome-Wide Association Study (GWAS) on EAC and the premalignant lesion BE was published. This study revealed three SNPs associated with EAC: rs10419226 (CRTC1), rs11789015 (BARX1), and rs2687201 (FOXP1).[12] In addition, evidence was found that rs9936833 (FOXF1), previously associated with BE,[13] was also associated with EAC and that the SNPs: rs2178146 and rs3111601 near rs9936833 had even a stronger association with EAC.[12] rs9936833 was first identified in a GWAS on BE performed by Su et al. in 2012, simultaneously rs9257809 (MHC) was found to be associated with BE.[13] The aim of the present study was to validate the association between these six previously identified SNPs and the risk of EAC in an independent and large case–control study.
MATERIALS AND METHODS
Study population
Patients diagnosed with an adenocarcinoma of the distal esophagus or esophagogastric junction (EGJ) and treated at the Department of Surgery, Erasmus MC Cancer institute, University Medical Centre, Rotterdam, between January 1996 and December 2013 (n = 761) were selected for the study. In addition, patients treated in the Academic Medical Centre at the university of Amsterdam, between 1994 and 2004 were included as well (n = 310). All patients underwent an esophagectomy with curative intention.
The control group was derived from the Rotterdam Study, a population-based prospective cohort study. In brief, this is an ongoing large population-based cohort study, which started in January 1990.[14] All inhabitants, who were aged 55 years and older, living in Ommoord, a district in Rotterdam, The Netherlands, were invited to participate. This population-based control group provided reference groups of allele frequencies, which reflect the local general European population. Individuals in the control group diagnosed with EAC were excluded. Patients and controls were of European descent.
DNA isolation
For cases, the tissue samples were obtained from the resection specimens and used according to the Code of Proper Secondary Use of Human Tissue in the Netherlands established by the Dutch Federation of Medical Scientific Societies (http://www.federa.org). Nonmalignant tissue from the resection specimen; tumor negative lymph nodes or tumor negative resection margins, confirmed by an experienced GI-pathologist, were macro dissected from microscopic sections of fresh frozen-or formalin fixed paraffin embedded tissues. DNA was extracted using proteinase K and 5% Chelex 100 resin. For controls, genomic DNA was extracted from whole blood samples using standard methods.[15]
Genotyping
For cases, a multiplex SNaPshot assay was designed; multiplex polymerase chain reaction was used to amplify the regions of the SNPs: rs2178146 (FOXF1), rs10419226 (CRTC1), rs9936833 (FOXF1), rs2687201 (FOXP1), rs11789015 (BARX1), and rs3111601 (FOXF1) (Hg19). The amplified fragments of the normal DNAs were analyzed by SNaPshot using the ABI Prism SNaPshot Multiplex Kit (Life Technologies, Carlsbad, CA, USA). Data analysis was performed using GeneMarker Analysis Software version 2.4.0 (Softgenetics, State College, PA, USA) [Figure 1].[16] Primers and probes sequences are listed in Supplementary Table 1.
Figure 1.
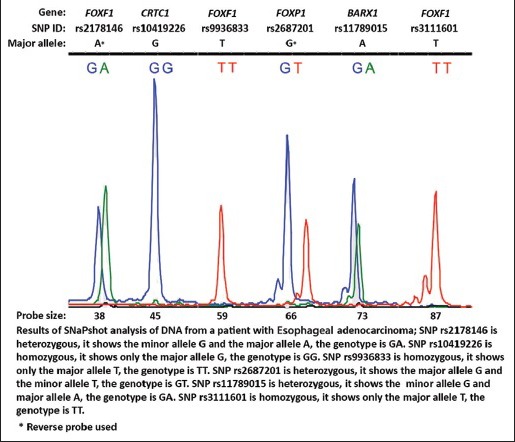
Result of SNaPshot analysis
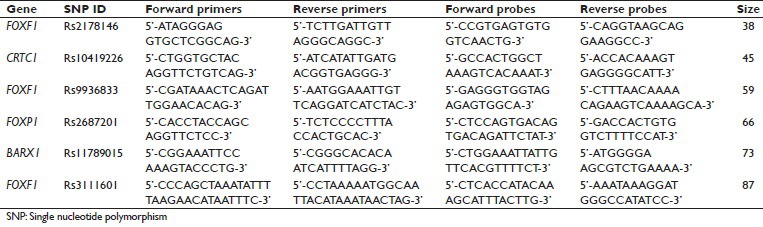
Supplementary Table 1.
Design of primers and probes
For controls, genome-wide SNP genotyping was performed using Infinium II assay on the HumanHap550 and Human 660-quad Genotyping BeadChips (IlluminaInc, San Diego, CA, USA). Approximately 30 million SNPs were imputed using 1000G Phase 1 v3 populations as reference.[17] The imputations were performed using MACH software (http://www.sph.umich.edu/csg/abecasis/MACH/). All variants tested here had an imputation quality of 0.9 or higher, suggesting near perfect imputation. Best-guess genotypes were used for the analyses.
Statistical analysis
Departures from Hardy–Weinberg equilibrium were tested for using the goodness-of-fit χ2 test. Logistic regression analyses were used to calculate odds ratios (OR) with 95% confidence intervals (95% CI). The major allele homozygous was set as a reference and was compared with the minor allele. P values were corrected for multiple testing using Bonferroni correction (P = 0.05/6 = 0.008) before considered significant. Logistic regression was performed with SPSS version 20.0 (SPSS, Chicago, IL, USA) and meta-analysis of the original GWAS,[12] and the current study was performed using R library rmeta.[18] A fixed-effects inverse-variance meta-analysis was performed.
RESULTS
Study population
A total of 1071 cases were initially analyzed, due to technical failure of 972 cases reliable data were obtained that was compared with 6206 controls regarding the six previously mentioned SNPs. Clinical data were available of 550 cases from Rotterdam. The median age of these patients was 63 years (Range: 19–84 years) and 80% was male. Almost half of the patients received some form of neoadjuvant therapy and all patients underwent esophagectomy. Ninety percent was diagnosed with an invasive adenocarcinoma, of which 35% arose clearly from BE. Most tumors were located in the distal esophagus (41.3%) or at the EGJ (40.5%). The majority of the tumors were moderately or poorly differentiated (36.5% and 45.1%, respectively). The most common pathological tumor stage (pT) was pT3 (53.8%) and half of the patients appeared to have positive lymph nodes (pN1-3), whereas four patients had distant metastasis (pM) according to the TNM-classification of the American Joint Committee on Cancer Staging Manual 7th edition. After surgery, 40.4% of the cases developed recurrence of disease (locoregional disease or distant metastasis). The mean overall survival of the 550 cases was 53.8 months (95%CI: 49.5–58.2) and the 5-year overall survival was estimated at 38.2%.
Allelic association analysis
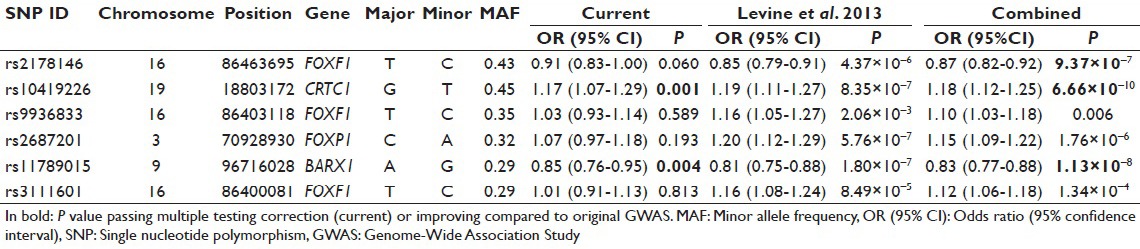
The distribution of genotype frequencies for the investigated SNPs was consistent with Hardy–Weinberg equilibrium (P > 0.05), except for FOXF1 rs3111601 (P = 0.013). The allelic association of the six SNPs with EAC showed significantly increased risk for the minor allele of rs10419226 (CRTC1) (OR = 1.17, P = 0.001) and significantly decreased risk for the minor allele of rs11789015 (BARX1) (OR = 0.85, P = 0.004). None of the other four SNPs were significant in the currently studied population, although direction and effect size were consistent with previous GWAS results. The meta-analysis of the original GWAS and the current study revealed more accurate effect estimate and improved level of significance for rs10419226 (CRTC1) (OR = 1.18, P = 6.66 × 10−10) and rs11789015 (BARX1) (OR = 0.83, P = 1.13 × 10−8), and in addition for rs2178146 (FOXF1) (OR = 0.87, P = 9.37 × 10−7), while there was no significant allelic association with EAC in the currently studied population [Table 1].
Table 1.
Logistic regression and meta-analysis
Genotypic association analysis
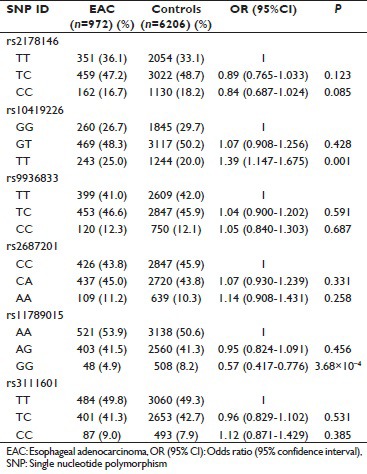
Genotypic association analysis showed a dose effect for the significantly associated SNPs. The GT genotype for rs10419226 (CRTC1) increased the risk of EAC in comparison with the GG genotype (OR = 1.07, P = 0.428), which became significant for the TT genotype (OR = 1.39, P = 0.001). The rs11789015 (BARX1) AG genotype decreased the risk of EAC in comparison with the AA genotype (OR = 0.95, P = 0.456), this decrease in risk became significant for the genotype GG (OR = 0.57, P = 3.68 × 10−4) [Supplementary Table 2].
Supplementary Table 2.
Genotypic association analysis
DISCUSSION
In this study two SNPs, rs10419226 (CRTC1) and rs11789015 (BARX1), were replicated to be associated with EAC. In the performed meta-analysis including the data of the EAC cohort from the original GWAS,[12] the level of significance was improved compared with the original findings of the GWAS, confirming the association of CRTC1 and BARX1 with EAC. This was expected, since the present case–control study revealed a significantly increased risk of EAC for the minor allele T of rs10419226 (CRTC1) and a significantly decreased risk of EAC for the minor allele G of rs11789015 (BARX1). Both SNPs showed a dose-effect in the genotypic analysis; two minor alleles gave a stronger effect than one minor allele.
In addition, rs2178146 (FOXF1) showed an improved level of significance in the meta-analysis while it did not reach significance in the allelic analysis of the present study cohort. This could be explained by a smaller sample size compared to the population used for the original GWAS resulting in a decreased power to detect the association. However, because all cases were retrieved clinically, and controls with EAC were excluded, no attenuation had taken place. Only two of the six previously identified SNPs, appeared to be significantly associated with EAC in the current study, however, a consistency of the direction of effect and effect size was seen for all variants, suggesting that all these variants may play a role in EAC.
Rs10419226 is an intronic variant in the CRTC1 gene, which is encoding for CREB-regulated transcription co-activator that has been found previously to be associated with the oncogenic activity. The down-regulation or loss of LKB1, a tumor suppressor kinase, activates CRTC1 signaling and the transcriptional activity of the downstream targets of CRTC1. In addition, altered LKB1/CRTC1 signaling has been demonstrated to induce a migratory and invasive phenotype in esophageal cancer cell lines.[19,20]
Rs11789015 is located in an intron of BARX1, a homeobox transcription factor. The homolog of BARX1 has been found to be associated with the differentiation of the esophagus and trachea in developing mouse embryos and in addition to be associated with the down-regulation of the Wnt pathway in stomach morphogenesis and differentiation.[21]
Identifying these SNPs associated with EAC suggests that genetic susceptibility might play a role in the initiation of EAC and could be of importance for the surveillance of BE patients. Since the prevalence of BE patients is valued at 2% in the general population,[9] and the annual risk of developing EAC from BE is estimated at 0.12–0.5%,[7,8] most patients with BE will not benefit from endoscopy surveillance. However because of the relatively poor prognosis of EAC,[3] it is of utmost importance to diagnose EAC patients in an early and curable stage of the disease. Therefore, it could be of additional value to incorporate SNPs associated with EAC in the surveillance program of BE, in order to only select the high-risk patients for developing EAC.
CONCLUSIONS
This independent and large Dutch case–control study replicated the association of rs10419226 (CRTC1) and rs11789015 (BARX1) with the risk of EAC. These findings indicate a possible genetic contribution to the development of EAC and might contribute to gain more insight in the etiology of this cancer. In addition, SNPs associated with EAC could be helpful by identifying patients at increased risk for malignant progression during surveillance and/or screening programs aimed to improving the survival of these patients by diagnosing EAC in an early and curable stage.
AUTHOR'S PROFILE
Anna M.J. van Nistelrooij: Departments of Pathology and Surgery, Erasmus MC Cancer Institute, University Medical Centre Rotterdam, The Netherlands.
Hetty A.G.M. van der Korput: Department of Pathology, Erasmus MC Cancer Institute, University Medical Centre Rotterdam, The Netherlands.
Mark I. van Berge Henegouwen: Department of Surgery, Academic Medical Centre, Amsterdam, The Netherlands.
Carel J. van Noesel: Department of Pathology, Academic Medical Centre, Amsterdam, The Netherlands.
Katharina Biermann: Department of Pathology, Erasmus MC Cancer Institute, University Medical Centre Rotterdam, The Netherlands.
Manon C.W. Spaander: Department of Gastroenterology and Hepatology, Erasmus MC Cancer Institute, University Medical Centre Rotterdam, The Netherlands.
Hugo W. Tilanus: Department of Surgery, Erasmus MC Cancer Institute, University Medical Centre Rotterdam, The Netherlands.
J. Jan B. van Lanschot: Department of Surgery, Erasmus MC Cancer Institute, University Medical Centre Rotterdam, The Netherlands.
Albert Hofman: Department of Epidemiology, Erasmus MC Cancer Institute, University Medical Centre Rotterdam, The Netherlands.
André G. Uitterlinden: Departments of Internal Medicine and Epidemiology, Erasmus MC Cancer Institute, University Medical Centre Rotterdam, The Netherlands.
Bas P.L. Wijnhoven: Department of Surgery, Erasmus MC Cancer Institute, University Medical Centre Rotterdam, The Netherlands.
Winand N.M. Dinjens: Department of Pathology, Erasmus MC Cancer Institute, University Medical Centre Rotterdam, The Netherlands.
Ronald van Marion: Department of Pathology, Erasmus MC Cancer Institute, University Medical Centre Rotterdam, The Netherlands.
Linda Broer: Department of Internal Medicine, Erasmus MC Cancer Institute, University Medical Centre Rotterdam, The Netherlands.
ACKNOWLEDGMENTS
The generation and management of GWAS genotype data for the Rotterdam Study are supported by the Netherlands Organization of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) project nr. 050-060-810. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters for their help in creating the GWAS database, and Karol Estrada and Maksim V. Struchalin for their support in creation and analysis of imputed data. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists.
Footnotes
Source and Support: This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) project nr. 050-060-810. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam.
Conflict of Interest: None declared.
References
- 1.Brown LM, Devesa SS, Chow WH. Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J Natl Cancer Inst. 2008;100:1184–7. doi: 10.1093/jnci/djn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosetti C, Levi F, Ferlay J, Garavello W, Lucchini F, Bertuccio P, et al. Trends in oesophageal cancer incidence and mortality in Europe. Int J Cancer. 2008;122:1118–29. doi: 10.1002/ijc.23232. [DOI] [PubMed] [Google Scholar]
- 3.Dikken JL, Lemmens VE, Wouters MW, Wijnhoven BP, Siersema PD, Nieuwenhuijzen GA, et al. Increased incidence and survival for oesophageal cancer but not for gastric cardia cancer in the Netherlands. Eur J Cancer. 2012;48:1624–32. doi: 10.1016/j.ejca.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 4.van Hagen P, Hulshof MC, van Lanschot JJ, Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. 2012;366:2074–84. doi: 10.1056/NEJMoa1112088. [DOI] [PubMed] [Google Scholar]
- 5.Cameron AJ. Epidemiology of columnar-lined esophagus and adenocarcinoma. Gastroenterol Clin North Am. 1997;26:487–94. doi: 10.1016/s0889-8553(05)70308-3. [DOI] [PubMed] [Google Scholar]
- 6.Jankowski JA, Wright NA, Meltzer SJ, Triadafilopoulos G, Geboes K, Casson AG, et al. Molecular evolution of the metaplasia-dysplasia-adenocarcinoma sequence in the esophagus. Am J Pathol. 1999;154:965–73. doi: 10.1016/S0002-9440(10)65346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhat S, Coleman HG, Yousef F, Johnston BT, McManus DT, Gavin AT, et al. Risk of malignant progression in Barrett's esophagus patients: Results from a large population-based study. J Natl Cancer Inst. 2011;103:1049–57. doi: 10.1093/jnci/djr203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hvid-Jensen F, Pedersen L, Drewes AM, Sørensen HT, Funch-Jensen P. Incidence of adenocarcinoma among patients with Barrett's esophagus. N Engl J Med. 2011;365:1375–83. doi: 10.1056/NEJMoa1103042. [DOI] [PubMed] [Google Scholar]
- 9.Ronkainen J, Aro P, Storskrubb T, Johansson SE, Lind T, Bolling-Sternevald E, et al. Prevalence of Barrett's esophagus in the general population: An endoscopic study. Gastroenterology. 2005;129:1825–31. doi: 10.1053/j.gastro.2005.08.053. [DOI] [PubMed] [Google Scholar]
- 10.Rex DK, Cummings OW, Shaw M, Cumings MD, Wong RK, Vasudeva RS, et al. Screening for Barrett's esophagus in colonoscopy patients with and without heartburn. Gastroenterology. 2003;125:1670–7. doi: 10.1053/j.gastro.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 11.Wang KK Sampliner RE; Practice Parameters Committee of the American College of Gastroenterology. Updated guidelines 2008 for the diagnosis, surveillance and therapy of Barrett's esophagus. Am J Gastroenterol. 2008;103:788–97. doi: 10.1111/j.1572-0241.2008.01835.x. [DOI] [PubMed] [Google Scholar]
- 12.Levine DM, Ek WE, Zhang R, Liu X, Onstad L, Sather C, et al. A genome-wide association study identifies new susceptibility loci for esophageal adenocarcinoma and Barrett's esophagus. Nat Genet. 2013;45:1487–93. doi: 10.1038/ng.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su Z, Gay LJ, Strange A, Palles C, Band G, Whiteman DC, et al. Common variants at the MHC locus and at chromosome 16q24.1 predispose to Barrett's esophagus. Nat Genet. 2012;44:1131–6. doi: 10.1038/ng.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofman A, Darwish Murad S, van Duijn CM, Franco OH, Goedegebure A, Ikram MA, et al. The Rotterdam Study: 2014 objectives and design update. Eur J Epidemiol. 2013;28:889–926. doi: 10.1007/s10654-013-9866-z. [DOI] [PubMed] [Google Scholar]
- 15.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allory Y, Beukers W, Sagrera A, Flández M, Marqués M, Márquez M, et al. Telomerase reverse transcriptase promoter mutations in bladder cancer: High frequency across stages, detection in urine, and lack of association with outcome. Eur Urol. 2014;65:360–6. doi: 10.1016/j.eururo.2013.08.052. [DOI] [PubMed] [Google Scholar]
- 17.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria; 2013. Available from: http://www.R-project.org/
- 19.Gu Y, Lin S, Li JL, Nakagawa H, Chen Z, Jin B, et al. Altered LKB1/CREB-regulated transcription co-activator (CRTC) signaling axis promotes esophageal cancer cell migration and invasion. Oncogene. 2012;31:469–79. doi: 10.1038/onc.2011.247. [DOI] [PubMed] [Google Scholar]
- 20.Liu K, Luo Y, Tian H, Yu KZ, He JX, Shen WY. The tumor suppressor LKB1 antagonizes WNT signaling pathway through modulating GSK3ß activity in cell growth of esophageal carcinoma. Tumour Biol. 2014;35:995–1002. doi: 10.1007/s13277-013-1133-0. [DOI] [PubMed] [Google Scholar]
- 21.Woo J, Miletich I, Kim BM, Sharpe PT, Shivdasani RA. Barx1-mediated inhibition of Wnt signaling in the mouse thoracic foregut controls tracheo-esophageal septation and epithelial differentiation. PLoS One. 2011;6:e22493. doi: 10.1371/journal.pone.0022493. [DOI] [PMC free article] [PubMed] [Google Scholar]


