Abstract
Introduction
The serine/threonine protein kinase ataxia telangiectasia mutated (ATM) is critical in maintaining genomic integrity. Upon DNA double-strand breaks, ATM phosphorylates key downstream proteins including p53 and BRCA1/2, thereby orchestrating complex signaling pathways involved in cell cycle arrest, DNA repair, senescence and apoptosis. Although sporadic mutation of ATM occurs rarely in breast cancer, the status of its protein expression and its clinical significance in breast cancer remain not well established. Our study was designed to investigate the influence of ATM protein in both tumor and cancer-associated stroma on clinical outcome in hormone-positive (HPBC) and hormone-negative (HNBC) early-stage breast cancer (EBC).
Methods
Tissue microarrays (TMAs), containing formalin-fixed, paraffin-embedded resected tumors from two cohorts of patients (HPBC cohort: n = 130; HNBC cohort: n = 168) diagnosed at the Tom Baker Cancer Centre, Calgary, Canada, were analyzed for ATM protein expression using fluorescence immunohistochemistry (IHC) and automated quantitative analysis (AQUA). ATM expression levels were measured within the tumor as a whole (tATM) as indicated by pan-cytokeratin expression, tumor nuclear compartment (nATM) as indicated by both DAPI and pan-cytokeratin-positive results, and cancer-associated stroma (csATM) as indicated by vimentin-positive and pan-cytokeratin-negative results. ATM expression levels within these compartments were correlated with clinical outcome.
Results
While tATM and nATM were significantly lower in tumors compared to normal breast epithelial tissues, csATM was significantly higher than the corresponding normal tissue compartment. In addition, the median expression level of both tATM and nATM were two- to threefold lower (P <0.001) in HNBC than in HPBC. In both HNBC and HPBC cohorts, patients with low tATM, nATM and csATM tumors had significantly poorer survival outcomes than those with a high tATM, nATM and csATM, but this effect was more pronounced in HNBC. A multivariate analysis demonstrates that these biomarkers predict survival independent of tumor size and lymph node status, but only in the HNBC cohort (P <0.001).
Conclusions
Low ATM protein expression in both malignant tumor and stromal compartments likely contributes to the aggressive nature of breast cancer and is an independent prognostic factor associated with worse survival in HNBC patients.
Electronic supplementary material
The online version of this article (doi:10.1186/s13058-015-0575-2) contains supplementary material, which is available to authorized users.
Introduction
Breast cancer continues to be the most frequent cancer type and the second leading cause of cancer death in females in Western Europe and North America. Although we have significantly improved diagnostic tools and therapeutic interventions in breast cancer over the last decade, breast oncologists still face paramount challenges in hormone-negative breast cancer, especially triple-negative breast cancer, the most aggressive breast cancer subtype, for which we do not yet have any molecular-targeted treatments apart from cytotoxic chemotherapy. Several large studies have identified RNA expression signatures that are prognostic for disease recurrence and metastasis early-stage hormone-positive breast cancer (HPBC) [1-3]. No such biomarkers exist currently for the more aggressive hormone-negative breast cancer (HNBC). In addition, mounting evidence indicates that levels of proteins that are immediately relevant to cell growth and metabolism are often not particularly well correlated to mRNA levels [4], suggesting that it may be more informative to directly measure protein levels as a indicator of biological behavior. These biomarkers likely reflect genetic signatures of early cancer formation and evolution of cancer aggressiveness.
Ataxia telangiectasia mutated (ATM) belongs to a family of so-called phosphatidyl inositol-3 kinase (PI3K)-like serine/threonine protein kinases (PIKKs), most of which are involved in the cellular response to various stresses [5]. In response to DNA double-strand breaks, ATM phosphorylates multiple key downstream proteins including p53 and BRCA1/2, the most common sporadic and inheritable mutated genes in breast cancer [6], thereby orchestrating complex signaling pathways involved in cell cycle arrest, DNA repair, senescence and apoptosis [7]. Germline mutation of ATM genes causes ataxia telangiectasia, a rare pleiotropic autosomal recessive disorder characterized by heightened radiation sensitivity and a predisposition to cancers including breast cancer [6,8]. The incidence of sporadic ATM mutations in breast cancer is low (2 to 3%), as compared to p53 and PIK3A mutations whose incidence rates are 30 to 40% [9]. Although we have some evidence supporting ATM loss in malignant breast tumors [10-17], and its prognostic significance in breast cancer was recently uncovered [16,17], the protein level of ATM in breast cancer-associated stromal tissues remains unknown.
In this study, using fluorescent immunohistochemistry (IHC) and automated quantitative analysis (AQUA), we measured ATM protein expression in both malignant tumor and stromal compartments of HPBC and HNBC. We hypothesized that reduced ATM expression correlates with increased tumor aggressiveness and poorer clinical outcome. Remarkably, we also identified a strong association between stromal expression of ATM and clinical outcome, suggesting that the tumor microenvironment contributes significantly to the overall course of disease.
Methods
Case selection and clinical data collection
This study was approved by the University of Calgary Conjoint Faculties Research Ethics Board (Ethic ID #24525), in accordance with the Tri-Council Policy Statement on Research with Human Subjects. Consents directly from patients were not required in this study as per Ethics Board guidelines. Two cohorts of clinical data were collected retrospectively through chart review. One cohort consists of 130 hormone-positive resected early-stage breast cancer (EBC) patients diagnosed at the Tom Baker Cancer Centre (TBCC) from 1989 to 2000. The second cohort consists of 168 hormone-negative resected EBC patients diagnosed at the TBCC from 1999 to 2004. All patients had stage I to III breast cancer with no evidence of metastatic disease at diagnosis and had undergone either lumpectomy or mastectomy. Hormone status including estrogen receptor (ER) and progesterone receptor (PR) were initially assessed by local breast pathologists according to the American Society of Clinical Oncology and the College of American Pathologists guidelines at the time and further evaluated on tissue microarray (TMA) using an independent staining method. Samples with discordant results between initial pathological diagnosis and further evaluation on TMA were excluded.
Tissue microarray generation
All archived formalin-fixed paraffin-embedded (FFPE) resected EBC tumor samples were retrieved from Calgary Laboratory Services and reviewed by a pathologist (AMM). TMAs were constructed as previously described [18], consisting of two cohorts of patients (cohort 1: n = 130 HPBC; cohort 2: n = 168 HNBC) as described in Case Selection above. Normal breast epithelial tissue specimens (n = 8), normal tonsil tissue, an ATM-deficient human lymphoblastoid cell line (L3) and an age-matched ATM-expressing human lymphoblastoid cell line (BT/C3ABR) were also included as controls [19].
Four-color fluorescence immunohistochemistry
TMA sections (4 μm) were deparaffinized in xylene, rinsed in ethanol, and rehydrated as previously described [18,20]. Heat-induced epitope retrieval was performed by heating slides to 121°C for 6 minutes in a Tris/EDTA-based buffer (pH 9.0) (Target Retrieval Solution, DAKO, Mississauga, ON, Canada). IHC was performed using a DAKO Autostainer Link 48. Primary antibodies used were a rabbit monoclonal antibody to ATM (clone Y170, 1:1000, Abcam, Cambridge, MA, USA), a mouse monoclonal antibody to vimentin, which marks the stromal compartment (clone V9, 1:5000, DAKO), and a guinea pig monoclonal pan-cytokeratin antibody identifying the tumor compartment (1:100, Acris, San Diego, CA, USA). Briefly, endogenous peroxidase activity was quenched with a 10-minute incubation of peroxidase-blocking reagent (a component of the DAKO EnVision™ + System) followed by a 15-minute protein block (Signal Stain; Cell Signaling, Danvers, MA, USA) to eliminate nonspecific antibody binding. All incubations were performed at room temperature. Slides were washed with a wash buffer containing Tris-buffered saline and 0.05% Tween-20 (TBST, DAKO) and then incubated with primary antibodies for 30 minutes. After three washes, a goat anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody from the DAKO EnVision™ + System was applied for 30 minutes. Slides were again washed in TBST and treated for 5 minutes with TSA-Plus Cy3 tyramide signal amplification reagent (1:100, PerkinElmer, Woodbridge, ON, Canada). To stop enzyme activity in preparation for vimentin staining, slides were treated with peroxidase block for 30 minutes, followed by four washes of TBST, a 10-minute protein block, and three more washes. To identify stromal cells, slides were incubated for 30 minutes with the vimentin antibody, washed in TBST, and then treated for 30 minutes with a goat anti-mouse HRP-conjugated secondary antibody from the DAKO EnVision™ + System. After three TBST washes, slides were incubated for 5 minutes with TSA-Plus Cy5 tyramide signal amplification reagent (PerkinElmer). To identify tumor cells, slides were further incubated for 30 minutes with the pan-cytokeratin antibody, followed by three washes and a 30-minute incubation of a 1:200 dilution of Alexa-488 conjugated goat anti-guinea pig antibody (Life Technologies, Burlington, ON, Canada). After three washes in TBST, the TMA slides were mounted with ProLong™ Gold anti-fade mounting medium containing diamidino-2-phenylindole (DAPI) (Life Technologies) and stored at 4°C overnight to set before scanning.
Automated image acquisition and analysis
Automated image acquisition was performed using an Aperio Scanscope FL (Aperio Inc., Vista, CA, USA). Briefly, seamless high-resolution slide images were acquired using the Scanscope FL 10-bit monochrome TDI line-image capture camera using filters specific for DAPI to define the nuclear compartment, fluoroscein isothiocyanate (FITC) to define cytokeratin for the tumor cytosolic compartment, Cy3 to define the target marker ATM, and Cy5 to define vimentin-positive nonmalignant stromal cells. Images were then analyzed using the HistoRX AQUAnalysis™ program, version 2.4.4.1 as described [13]. Briefly, a tumor-specific binary mask was generated to distinguish the cancer cells from surrounding stromal tissue by thresholding the pan-cytokeratin images. Thresholding created a binary mask that identified the presence or absence of tumor cells by the presence of a pixel that was ‘on’ or ‘off’, respectively. A similar thresholding technique was applied to generate a nonmalignant stroma-specific mask (from the pan-cytokeratin-negative regions of the vimentin images, a proven superior method for identifying stromal regions [12]. To isolate tumor nuclei, mutually exclusive tumor nuclear and cyoplasmic masks were created by subtracting cytokeratin-positive areas from DAPI-positive areas within the tumor mask. All images were processed using optimized threshold values and all subsequent image manipulations involved only image information from the masked areas. Images were manually validated according to the following: 1) >10% of the tissue area is pan-cytokeratin positive, 2) >50% of the image was usable (that is not compromised due to overlapping or out-of-focus tissue). Unusable areas within each image were manually cropped so that they were excluded from the final analysis.
ATM compartment-specific quantification
ATM expression was calculated as the mean Cy3 pixel intensity within each defined compartment. The tumor-specific, tumor nuclear-specific and stroma-specific AQUA scores represent ATM protein expression in the total malignant area, in nuclei within the malignant area, and in the tumor-associated stromal area, respectively. For each patient sample, the average compartment-specific AQUA score over triplicate cores was used to define the ATM expression tumor score (tATM), tumor nuclear score (nATM), and the cancer-associated stromal score (csATM).
Statistical analysis
Summary and descriptive statistics were used to describe demographic and clinicopathological characteristics of patients in the two cohorts. Student’s t test was used to compare the continuous variable age between low and high ATM groups. Fisher’s exact test was used to compare the categorical variables between low and high ATM groups. Histograms were used to evaluate the distribution of ATM expression (tATM, nATM, and csATM) of the two cohorts. Pairwise group comparisons of ATM expression (tATM, nATM, and csATM) between groups (HPBC, HNBC, human epidermal growth factor receptor 2 (HER2)-positive and triple-negative breast cancer (TNBC)) were performed using parametric (Student’s t test) and nonparametric (Wilcoxon) tests. Cut-points to define low and high ATM expression were modeled using X-tile software [21] in the two cohorts independently. Disease-specific survival (DSS) and disease-free survival (DFS) estimates were calculated using the Kaplan-Meier (KM) survival analysis and log-rank tests were used to compare between low and high ATM groups. Cox proportional hazards regression methods were used to assess the prognostic effect of ATM expression (tATM, nATM, and csATM) for DSS multivariably. A multivariate model from selected clinical known prognostic factors such as tumor size, grade, lymph node (LN) status, lymphovascular invasion (LVI) and age was created. The prognostic ability of ATM expression (tATM, nATM, and csATM) was tested using a backward selection approach to obtain the optimal model. All tests were two-sided, and a P value of 0.05 was considered statistically significant. All statistical analyses were conducted using R statistics software (Version 3.1.2).
Results
Patient characteristics
Our study adheres to the REMARK criteria for the study of biomarkers [22]. The clinical and tumor characteristics of patients in the two cohorts (HPBC and HNBC) are shown in Table 1. Briefly, in the HNBC cohort, mean age at diagnosis was 53 years. The majority of cases (72%) were grade 3, consistent with the aggressive biology of hormone-negative tumors. More than half (58%) the cases were LN negative, and a majority of cases (81%) were of stages I or II, indicative of the early staging of this cohort. Two-thirds of the cases were TNBC, and the remaining third were hormone-negative, HER2-positive cases. Eleven cases in the HNBC cohort were deemed hormone positive based on analysis of the TMA spots, and were excluded from final statistical analysis. Sixty percent of patients had adjuvant radiation treatment (RT). Seventy-five percent of patients had adjuvant chemotherapy; the majority of these patients (83%) had anthracycline-based chemotherapy, while the remainder (17%) received taxane-based chemotherapy. In the HPBC cohort, the mean age at diagnosis was 67 years, older than our HNBC cohort that is consistent with clinical observation. Both cohorts shared similar patterns of tumor size, LVI, stage, LN status and adjuvant RT. Almost all patients (99%) in the HPBC cohort received adjuvant hormone treatment instead of chemotherapy.
Table 1.
Patients’ characteristics of the HPBC and HNBC cohorts, and comparison of clinicopathological features of breast tumors in low and high tATM group
| Baseline characteristics | HNBC cohort | HPBC cohort | ||||||
|---|---|---|---|---|---|---|---|---|
| Full cohort | tATM low | tATM high | tATM low vs high | Full cohort | tATM low | tATM high | tATM low vs high | |
| n = 168 (%) | n = 22 (%) | n = 79 (%) | P value | n = 130 (%) | n = 36 (%) | n = 66 (%) | P value | |
| Age median (min-max) | 53 (30–82) | 58 (33–80) | 51(30–82) | 0.145 | 67 (38–88) | 71(41–86) | 68 (38–88) | 0.408 |
| Tumor size | ||||||||
| T1/T2 (≤5 cm) | 143 (85%) | 20 (20%) | 67 (66%) | 1.00 | 116 (89%) | 27 (26%) | 65 (64%) | 0.002 |
| T3/T4 (>5 cm) | 16 (10%) | 2 (2%) | 10 (10%) | 10 (8%) | 7 (7%) | 1 (1%) | ||
| Missing | 9 (5%) | 2 (2%) | 4 (3%) | 2 (2%) | ||||
| Grade | ||||||||
| 1 | 3 (2%) | 0 (0%) | 0 (0%) | 0.295 | 25 (19%) | 6 (6%) | 12 (12%) | 0.029 |
| 2 | 29 (17%) | 5 (5%) | 9 (9%) | 63 (48%) | 10 (10%) | 38 (37%) | ||
| 3 | 121 (72%) | 17 (17%) | 68 (67%) | 32 (25%) | 15(14.5%) | 15(14.5%) | ||
| Missing | 15 (9%) | 2 (2%) | 10 (8%) | 5 (5%) | 1 (1%) | |||
| LVI | ||||||||
| + | 47 (28%) | 7 (7%) | 27 (27%) | 0.621 | 26 (20%) | 7 (7%) | 13 (13%) | 0.584 |
| - | 97 (58%) | 15(15%) | 43 (42%) | 76 (58%) | 17 (16%) | 43 (42%) | ||
| Missing | 24 (14%) | 9 (9%) | 28 (22%) | 12 (12%) | 10 (10%) | |||
| LN | ||||||||
| 0 | 97 (58%) | 15(15%) | 41 (40%) | 0.230 | 67 (51%) | 14 (14%) | 38 (37%) | 0.03 |
| >0 | 64 (38%) | 7 (7%) | 37 (37%) | 49 (38%) | 20 (19.5%) | 20 (19.5%) | ||
| Missing | 7 (4%) | 1 (1%) | 14 (11%) | 2 (2%) | 8 (8%) | |||
| Stage | ||||||||
| I | 55 (33%) | 4 (4%) | 25 (25%) | 0.551 | 40 (31%) | 6 (6%) | 25 (24%) | 0.002 |
| II | 81 (48%) | 13 (13%) | 35 (34%) | 43 (33%) | 13 (13%) | 22 (21.5%) | ||
| III | 23 (14%) | 4 (4%) | 16 (16%) | 25 (19%) | 15 (14.5%) | 7 (7%) | ||
| DCIS and IV | 9 (5%) | 1 (1%) | 3 (3%) | 3 (2%) | 0 (0%) | 1 (1%) | ||
| Missing | NA | 19 (15%) | 2 (2%) | 11 (11%) | ||||
| ER/PR status | ||||||||
| Positive | 11 (7%) | 0 (0%) | 0 (0%) | 130 (100%) | 36 (35%) | 66 (65%) | ||
| Negative | 48 (92%) | 22 (22%) | 79 (78%) | NA | ||||
| Missing | 2 (1%) | NA | ||||||
| HER2 status | NA | |||||||
| Positive | 62 (36%) | 6 (6%) | 33 (33%) | 0.22 | ||||
| Negative | 104 (62%) | 16 (16%) | 46 (45%) | |||||
| Missing | 2 (1%) | |||||||
| RT | ||||||||
| Yes | 100 (60%) | 11 (11%) | 55 (54%) | 0.127 | 70 (54%) | 20 (20%) | 38 (37%) | 0.831 |
| No | 68 (40%) | 11 (11%) | 24 (24%) | 56 (43%) | 15 (15%) | 25 (24%) | ||
| Unknown | NA | 4 (3%) | 1 (1%) | 3 (3%) | ||||
| Chemotherapy | ||||||||
| Anthracycline-based | 105 (62%) | 11 (11%) | 59 (58%) | 0.04 | ||||
| Anthracycline and taxane-based | 11 (7%) | 3 (3%) | 3 (3%) | |||||
| CMF or 5FU | 7 (4%) | 0 (0%) | 4 (4%) | |||||
| Unknown | 3 (2%) | 1 (1%) | 1 (1%) | 1 (1%) | ||||
| None | 42 (25%) | 7 (7%) | 12 (12%) | 129 (99%) | ||||
One hundred and sixty-eight HNBC and 130 HPBC patients diagnosed with EBC were included in the study. Clinical and histopathological features were summarized and described in numeric numbers and proportions in both cohorts. Sixty-seven cases in the HNBC cohort and 28 cases in the HPBC cohort were excluded for failing to obtain ATM staining due to technical issues. Therefore, a total of 101 cases in the HNBC cohort and 102 cases in the HPBC cohort were dichotomized into high and low ATM groups based on tATM AQUA scores. The cut-point was set at 22% in the HNBC cohort and 38% in the HPBC cohort respectively based on X-tile prediction. Student’s t test was used to compare the continuous variable age between low and high ATM groups. Fisher’s exact test was used to compare the clinicopathological features of breast tumors in high and low ATM groups. HPBC, hormone-positive breast cancer; HNBC, hormone-negative breast cancer; tATM, tumor ATM; LVI, lymphvascular invasion; LN, lymph node status; DCIS, ductal carcinoma in situ; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; RT, radiation treatment; CMT, combined modality treatment; 5FU, 5-fluorouracil; EBC, early-stage breast cancer; ATM, ataxia telangiectasia mutated; AQUA, automated quantitative immunofluorescence analysis.
ATM expression analysis
ATM protein expression levels were measured by automated quantitative fluorescence IHC using AQUAnalysis software [23] (Figure 1). Antibody specificity was validated using the L3 ATM-deficient human lymphoblastoid cell line and the BT/C3ABR age-matched ATM-expressing human lymphoblastoid cell line as controls (Figure 1A, left two panels) [19]. Positive and negative (by omission of the primary antibody) tonsil tissue controls were included in each run (Figure 1A, right two panels). Representative examples of the ATM-staining patterns in normal breast epithelium and invasive ductal carcinomas representing preserved or aberrantly reduced ATM expression are shown in Figure 1B. Normal breast epithelial tissue (pan-cytokeratin-positive) and hormone-positive tumors with high AQUA scores demonstrated a relatively uniform and intense nuclear expression pattern of ATM, consistent with the reported cellular distribution of ATM [7] (Figure 1B, top panel). In contrast, hormone-negative tumors with low AQUA scores exhibited only faint and/or sporadic nuclear staining pattern in malignant cells (Figure 1B, bottom panel).
Figure 1.
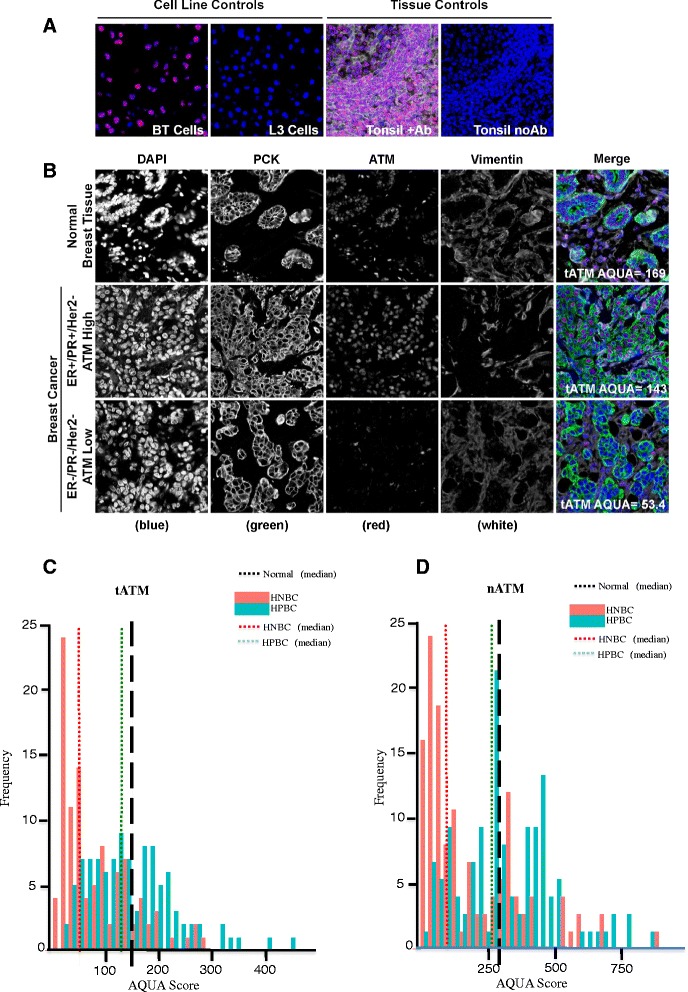
Quantitative fluorescence immunohistochemistry and digital image analysis for ATM in EBC. (A) Control ATM antibody specificity staining in ATM-positive BT cells and ATM-deficient L3 cells, as well as in breast cancer tissue with and without primary anti-ATM antibody. (B) Representative fluorescence images for ATM expression in normal breast epithelial tissue and EBC. DAPI-stained nuclei are depicted in blue, pan-cytokeratin-stained epithelial/malignant cells are depicted in green, vimentin-stained nonmalignant tumor stromal cells are depicted in white, and ATM protein expression is depicted in red. Histogram distributions representing tATM expression (C) and nATM expression (D) in both HPBC cohort (green solid bar) and HNBC cohort (red solid bar). The hashed black line represents median ATM expression (tATM and nATM) in normal breast tissue, the hashed green line represents median ATM expression (tATM and nATM) in the HPBC cohort, and the hashed red line represents median ATM expression (tATM and nATM) in the HNBC cohort. ATM, ataxia telangiectasia mutated; DAPI, diamidino-2-phenylindole; EBC, early-stage breast cancer; HNBC, hormone-negative breast cancer; HPBC, hormone-positive breast cancer; nATM, tumor nuclear ATM; tATM, tumor ATM.
To determine whether ATM expression was altered in EBC compared to normal breast tissue, we compared the median ATM AQUA scores of our two EBC cohorts to the median scores from normal breast tissue (Figures 1C and D). tATM was calculated within each patient tissue core as the average Cy3 pixel intensity within the pan-cytokeratin-positive malignant cell area (Figure 1C). nATM was calculated within each patient tissue core as the average Cy3 pixel intensity within both the pan-cytokeratin and DAPI-positive malignant cell area (Figure 1D). Median tATM and nATM scores in normal breast epithelium were 148 (95% confidence interval (CI): 84 to 19) and 294 (95% CI: 208 to 548) respectively (Figure 1C and 1D, hashed black line). Median tATM scores in the HPBC and HNBC cohorts were 134 (95% CI: 117 to 163) (Figure 1C, hashed green line) and 57 (95% CI: 45 to 85) respectively (Figure 1C, hashed red line). Median nATM scores in the HPBC and HNBC cohorts were 290 (95% CI: 274 to 370) (Figure 1D, hashed green line) and 124 (95% CI: 81 to 187) (Figure 1D, hashed red line) respectively. The median tATM and nATM scores were two- to threefold lower in the HNBC cohort than in the HPBC cohort (P <0.001) (Figure 2A and B). We further classified our HNBC cohort into TNBC and hormone-negative, HER2-positive groups based on HER2 status. We found a persistently significant difference in tATM and nATM scores between the HPBC and the TNBC cohorts, and between the HPBC and the hormone-negative HER2-positive groups (the median tATM scores in TNBC and hormone-negative HER2-positive groups were 52 (95% CI: 37.4 to 69.2) and 85 (95% CI: 44.2 to 117.7) respectively; the median nATM scores in TNBC and hormone-negative HER2-positive groups were 94 (95% CI: 66.9 to 142.2) and 187 (95% CI: 80.7 to 299.0) respectively; Figure 2C and D). It seemed that the TNBC group had the lowest tATM and nATM expression numerically; statistically, the different between the TNBC and the hormone-negative HER2-positive group was not significant, likely due to the small sample sizes of both groups (Figure 2C and D).
Figure 2.
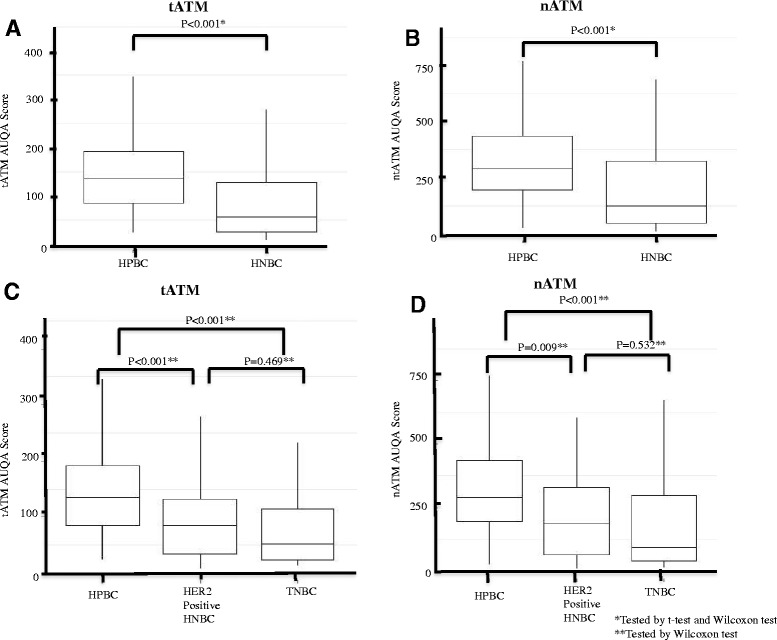
ATM expression in the HPBC and HNBC cohorts measured using malignant cell-specific ATM (tATM) and tumor nuclear ATM (nATM) AQUA scores. (A) Comparison of tATM AQUA scores in the HNBC cohort with the HPBC cohort. (B) Comparison of nATM AQUA scores in the HNBC cohort with the HPBC cohort. (C) Comparison of tATM AQUA scores in the HPBC cohort with the HER2-positive hormone-negative cohort and TNBC cohort. (D) Comparison of nATM AQUA scores in the HPBC cohort with the HER2-positive hormone-negative cohort and TNBC cohort. ATM, ataxia telangiectasia mutated; HER2, human epidermal growth factor receptor 2; HNBC, hormone-negative breast cancer; HPBC, hormone-positive breast cancer; TNBC, triple-negative breast cancer.
We further asked whether there were differences in ATM protein between the cancer-associated stromal compartment adjacent to the malignant tumor compared to the stromal compartment around normal breast epithelium. The csATM score was calculated within each patient tissue core as the average Cy3 pixel intensity of the vimentin-positive but pan-cytokeratin-negative stromal cell area. While median stromal ATM score adjacent to normal breast epithelium was 45 (95% CI: 32 to 72) (Figure 3A, hashed black line), median csATM score in the HPBC cohort was 85 (95% CI: 79 to 95) (Figure 3A, hashed green line) and median csATM was in the HNBC cohort was 62 (95% CI: 51 to 81) (Figure 3A, hashed red line). Interestingly, we found that csATM expression was higher in significant portions of patients in both of our two EBC cohorts, more pronounced in the HPBC than in the HNBC cohort, compared to normal stromal ATM expression surrounding normal breast epithelium (Figure 3B). Our result is consistent with a recent study demonstrating that an abnormally activated ATM-associated pathway had been identified in breast cancer-associated fibroblasts (CAFs) [24], although its biological significance remains elusive. We were not able to identify the biological significance of the increase of csATM compared to normal stroma ATM expression in our study as high csATM expression did not associate with tumor aggressiveness. Rather, low csATM scores appear to be associated with tumor-aggressive features (see below).
Figure 3.
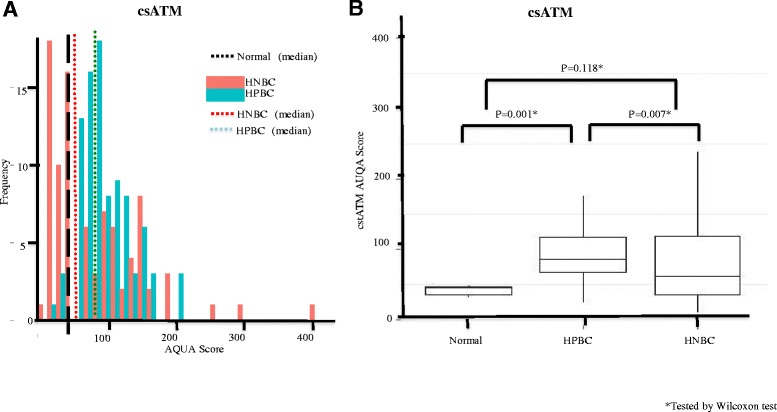
ATM expression in cancer-associated stromal (csATM) in the HPBC and HNBC cohorts compared to its expression in normal stromal adjacent to normal breast epithelium. (A) Histogram distributions representing csATM expression in both the HPBC cohort (green solid bar) and HNBC cohort (red solid bar). The hashed black line represents median csATM expression in normal breast stromal tissue, the hashed green line represents median csATM expression in the HPBC cohort, and the hashed red line represents median csATM expression in the HNBC cohort. (B) Comparison of csATM AQUA scores in normal controls, HPBC cohort and HNBC cohort. AQUA, automated quantitative immunofluorescence analysis; ATM, ataxia telangiectasia mutated; HNBC, hormone-negative breast cancer; HPBC, hormone-positive breast cancer.
Association between ATM protein expression and clinical outcome
Patients were dichotomized within each cohort into those with high and low tATM, nATM and csATM scores, using cut-points identified by X-Tile software [21]. In the HPBC cohort, low tATM and csATM scores correlated with larger tumor size (P = 0.002 and only with a trend P = 0.08 respectively), LN positivity (P = 0.03 and P = 0.004 respectively), higher grade (P = 0.029 and P = 0.033 respectively), and higher stage (P = 0.002 and P = 0.014 respectively), suggesting that low tATM and csATM scores are associated with more aggressive breast tumors in HPBC (Table 1 and Table S1 in Additional file 1). Interestingly, these correlations were not apparent in the HNBC cohort.
Patients were dichotomized into low and high tATM, nATM and csATM scores as described above for further survival analysis using the KM method. As a whole (the HPBC plus the HNBC cohort), patients with low tATM, nATM and csATM scores, compared to patients with high tATM, nATM and csATM scores, had a worse DSS (Figure S2A-C in Additional file 2). Then, patients with HNBC and HPBC were examined separately. In both cohorts, compared to patients with a high tATM, nATM and csATM expression, patients with low tATM, nATM and csATM scores had worse DSS outcome with median follow-up of 7.5 and 15 years in the HNBC and HPBC cohorts respectively (Figures 4A-F). This effect seems greater in the HNBC cohort compared with the HPBC cohort (Figures 4A-F).
Figure 4.
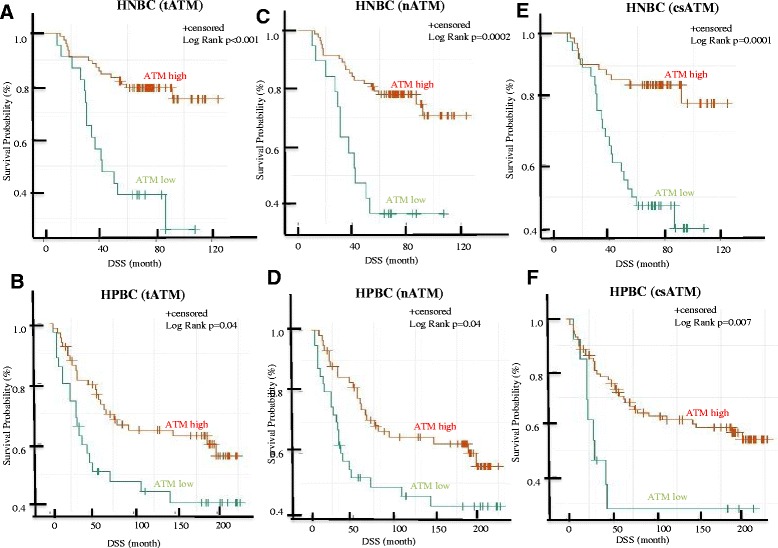
Univariate survival analysis in the HPBC and HNBC cohorts based on tATM, nATM and csATM expression. Kaplan-Meier (KM) analysis of disease-specific survival (DSS) in the HNBC cohort (A, C, E) and in the HPBC cohort (B, D, F) comparing patients with high and low tATM (A, B), nATM (C, D) and csATM (E, F). ATM, ataxia telangiectasia mutated; csATM, cancer-associated stromal ATM; HNBC, hormone-negative breast cancer; HPBC, hormone-positive breast cancer; nATM, tumor nuclear ATM; tATM, tumor ATM.
Cox proportional hazards regression models were used to determine if reduced ATM expression identified by tATM, nATM and csATM scores was an independent prognostic factor for DSS. A multivariate model from selected known clinical prognostic factors such as tumor size, grade, LN status, LVI and age was created. As expected, tumor size and LN status were significantly associated with survival. In the HNBC cohort, the significant association between these biomarkers and DSS persisted in the multivariate model after adjusting for all these known prognostic variables in the optimal model (P <0.001), but not in the HPBC cohort (Table 2A-C and data not shown). Furthermore, within the HNBC cohort, the prognostic value of ATM expression appeared to be similar in the LN-negative and LN-positive subsets (Figure S3A and B in Additional file 3). However, it seemed to have a greater effect in the T1/T2 subset compared to the T3/T4 subset (Figure S3C and D in Additional file 3), although the latter result may be confounded by a very small sample size in the T3/T4 subgroup.
Table 2.
Low ATM expression independently predicts poor disease survival in a multivariate model in the HNBC cohort
| Variables | P value | HR (95%CI) |
|---|---|---|
| A | ||
| tATM (low vs high) | <0.001 | 14.16 (5.17-38.83) |
| Tumor size (T3/4 vs T1/2) | <0.001 | 9.44 (3.26-27.32) |
| LN status (+ vs -) | 0.03 | 4.35 (1.11-17.01) |
| LVI (+ vs -) | 0.08 | 2.51 (0.91-6.91) |
| Grade (3 vs 1/2) | 0.48 | 1.58 (0.44-5.71) |
| Age (>53 vs <53) | 0.51 | 1.34 (0.56-3.21) |
| B | ||
| nATM (low vs high) | <0.001 | 7.43 (2.95-18.76) |
| Tumor size (T3/4 vs T1/2) | <0.001 | 6.39 (2.36-17.30) |
| LN status (+ vs -) | 0.11 | 2.84 (0.80-10.07) |
| LVI (+ vs -) | 0.07 | 2.46 (0.92-6.60) |
| Grade (3 vs 1/2) | 0.55 | 1.47 (0.41-5.25) |
| Age (>53 vs <53) | 0.32 | 1.55 (0.66-3.66) |
| C | ||
| csATM (low vs high) | <0.001 | 4.72 (1.95-11.42) |
| Tumor size (T3/4 vs T1/2) | 0,01 | 3.45 (1.27-9.36) |
| LN status (+ vs -) | 0.24 | 2.07 (0.61-7.00) |
| LVI (+ vs -) | 0.57 | 1.33 (0.50-3.55) |
| Grade (3 vs 1/2) | 0.24 | 2.29 (0.58-9.08) |
| Age (>53 vs <53) | 0.71 | 1.20 (0.46-3.10) |
Independent prognostic value of ATM expression levels (tATM, nATM and csATM) in both malignant tumor and stromal tissues was evaluated in a multivariate model. A multivariate model was created using selected known prognostic factors such as tumor size, lymph node (LN) status, lymphovascular invasion (LVI), grade and age. Cox proportional hazards regression methods were used to calculate P value, hazard ratio (HR) and confidence interval (CI) of each variable in the model. ATM, ataxia telangiectasia mutated; HNBC, hormone-negative breast cancer; tATM, tumor ATM; nATM, tumor nuclear ATM; csATM, cancer-associated stromal ATM.
Discussion
In this study, we show that ATM expression was reduced in HNBC, as compared to either HPBC or normal breast epithelium. In both HPBC and HNBC cohorts, reduced ATM expression was associated with poorer disease-specific outcomes. Interestingly, stromal ATM expression was higher in both cohorts as compared to normal epithelium, though lower expression in this compartment was also associated with poorer clinical outcome. These expression studies were enhanced by fluorescent IHC and AQUAnalysis, which allowed us to describe ATM expression as a continuous variable across a broader range than is possible with brightfield IHC. The dynamic range of protein quantification that is possible in the fluorescent realm allows the analysis, in a research context, of samples with widely varying degrees of protein expression. It should be noted that fluorescent IHC and the associated automated image analysis workflows are not currently used in clinical practice; therefore some data presented in this study may not be directly applicable. It is however possible that they could be translated into clinically relevant assays upon identification of the levels of ATM expression that offer prognostic value. To the best of our knowledge, this is the first study to examine ATM protein expression in malignant tumor and cancer-associated stromal tissues simultaneously.
About 60 to 80% of the EBC patients in both cohorts had ATM scores (tATM or nATM) lower than the mean values for the same compartment in normal breast epithelium. This suggests that loss of ATM expression is an important early event signaling malignant transformation of breast epithelial cells as the majority of EBC has some degree of ATM deficiency. In addition, ATM loss continues when breast cancer evolves and becomes more aggressive, to a certain threshold that significantly impacts on cancer-specific survival. Reduced ATM expression within malignant breast epithelial tissues was predominant in HNBC, especially TNBC, the most aggressive breast cancer type. These results, along with similar outcomes reported by others [10-17], suggest that ATM loss not only occurs early during breast tumorigenesis, but also may promote cancer progression by permitting the accumulation of genetic mutations due to genome instability as a result of ATM loss, thereby selecting cancer cells bearing aggressive phenotypes.
In HPBC, low ATM within malignant breast epithelial cells and stroma is associated with aggressive characteristics of breast cancer including large size, high grade and LN involvement. It is an interesting observation that this association is not seen in HNBC, which is a more aggressive tumor type. It is likely that ATM loss predominates the aggressive nature regardless of other worse features in this aggressive tumor type, which is consistent with our later finding of independent prognostic ability of ATM in only the HNBC cohort. Previous studies [16,17] demonstrated the significant association between low ATM expression and worse clinicopathological features in an undifferentiated breast cancer cohort containing both HPBC and HNBC cases. We believe that our data, consistent with others [10-17], support the notion that ATM loss contributes to the aggressive nature of breast tumors as they evolve.
It is interesting that ATM is higher in cancer-associated stromal tissues compared with stromal tissues around normal breast epithelium. Although this is consistent with another study [24], validation is needed using a larger distinct cohort. To the best of our knowledge, the mechanisms of regulation and functional roles of ATM in malignant stroma are presently unknown. Although it may seem contradictory that the ATM level is increased in cancer-associated stroma compared to normal stroma, albeit the ATM level is decreased in malignant epithelium compared to normal epithelium, we believe that this could be a reflection of the complexity of ATM regulation in diverse cell types. We speculate that the heightened level of DNA damage and oxidative stress [25] that are predominant in malignancy may initially lead to significant induction of ATM in the malignant epithelial tumor as well as in the stroma, Subsequently, a range of other factors may lead to differentially active downregulation of ATM, more so within the epithelial tumor and less so within the stroma. For example, one of such factors may include caspases that are activated particularly in the necrotic cores of aggressive tumors, which were recently shown to proteolytically cleave ATM [26]. Other regulatory factors are discussed later. Furthermore, we speculate that ATM may have context-dependent roles in stroma that are distinct from its well-known conventional function in maintaining genomic stability in response to DNA damage within the epithelial component. For example, it may regulate inflammatory response such as chemokine release (for example interleukin (IL)-6, IL-8) to promote/inhibit tumor growth in a paracrine-mediated fashion [27]. In addition, it may serve as a contributor to extracellular matrix stiffness (in addition to many other cytokines) and regulates tumor invasion and metastasis through bidirectional signaling between tumor cells and stroma microenvironment [28]. Therefore, it is not surprising that low level of ATM in cancer-associated stroma is also associated with a poor survival (Figure 4E and F).
It should be noted that cut-points used to define positive and negative ATM tumors varied widely in previous studies despite all having used conventional IHC as there is no well-defined method for choosing cut-points for biomarker analysis. For example, Bueno’s group [16] defined ATM IHC scores as negative or positive according to the absence or presence of nuclear staining in epithelial cells evaluated by two independent pathologists who had been blinded to the outcome. Abdel-Fatah’s group [17] used a cutoff of <25% cells being classed as low and ≥25% as high for nuclear ATM protein level. Because ATM expression in our study is a continuous variable that is generated automatically by the analysis software, we chose to use the X-tile program [21], which is a validated methodology to define our cut-point in two independent cohorts. The other advantage of using such digital image analysis as compared to conventional IHC is that it can potentially avoid human bias (inter- and intra- observer variability) as the intensity scores are objective representations of protein expression. In addition, this platform allows us to define the specific localization of ATM expression within the tumor. Therefore, we can evaluate tumor nuclear and cytoplasmic expression of ATM concurrently, in a similar fashion as the above tumor and stromal ATM analysis. We examined nuclear versus cytoplasmic ratio of ATM expression (ncATM) in both EBC cohorts as we speculated that aberrant localization of ATM may also contribute to loss of ATM function thereby resulting in tumor aggressiveness and poor prognosis. Although we did observe a significantly decreased ncATM in more aggressive breast tumor, HNBC compared with HPBC (Figure S1 in Additional file 4), the prognostic value of ncATM for clinical outcome (DFS and DSS) was not as significant as tATM, nATM and csATM in both cohorts (data not shown). We speculate that this may be due to our small sample size.
Lastly, our study illustrates the independent prognostic ability of ATM protein expression levels for DSS in both malignant tumor and stromal tissues in HNBC. Bueno et al. found that ATM protein expression associated significantly with both DFS and DSS [16]. We also observed a strong trend that low ATM group (tATM, nATM and csATM) had worse DFS than high ATM (tATM, nATM and csATM) group with a P value of 0.05 to 0.09 based on log-rank tests of KM analysis (data not shown). The lack of statistical significance may be an outcome of relative small sample size as well as the different methodologies of evaluating ATM protein expression and defining cut-points to divide ATM low and high groups. We, and others [16,17], clearly demonstrated that ATM protein has potential use as a novel biomarker in EBC, in addition to the known clinicopathological prognostic factors such as tumor size, grade, LVI and LN status. This could be a much simpler tool than several biomarker tools deployed in HPBC such as Oncotype DX, PAM50 and MammaPrint, which are all RNA-based assays [1-3].
Our study did not seek to investigate the mechanisms underlying the regulation of ATM expression in EBC. ATM gene mutations have been described in the literature, but since the incidence of ATM mutations in breast cancer is low [9], we would not expect this to explain the full range of our results. Other mechanisms such as gene copy number loss [16,29], posttranscriptional microRNA regulation [16,30,31], epigenetic modifications [32,33] and direct caspase cleavage [26] for altering ATM levels have been described. It should also be noted that a simple measure of ATM protein expression by IHC cannot establish the functional capability of ATM within the tumor. Measuring of phosphorylated ATM protein and its targets within the ATM signaling pathways may provide more insight with regard to ATM functions in breast cancer. Our results taken in the context of what is known about the biological role of ATM in protecting the genome suggest that it is very likely that a substantial proportion of breast cancers, which have lost ATM expression at the protein level, are in fact deficient also in ATM function [34].
We attempted to investigate the predictive value of ATM protein for the efficacy of adjuvant RT and chemotherapy in our cohorts. Due to small sample size, we cannot draw any conclusions. We noted that other studies have showed that ATM loss seems to associate with resistance to anthracycline chemotherapy in breast cancer [16,17,29], which seems counterintuitive to many preclinical studies revealing that ATM-deficient cells are sensitive to cytotoxic chemotherapy [7]. More interestingly, we and others have recently shown that ATM loss can predict sensitivity to poly (ADP-ribose) polymerase (PARP) inhibitors, a novel drug that may exploit this deficiency, in breast cancer [35] as well as other cancers [19,36,37]. Prospective clinical trials are warranted to validate its predicative value.
Conclusions
In summary, we report that low ATM protein expression in malignant breast epithelial tissues can be detected in a significant proportion of EBC, and is more pronounced in HNBC compared to HPBC. Our analysis also demonstrates that low ATM protein expression can independently predict a poor DSS in early-stage HNBC. Furthermore, we show that ATM protein expression is increased in cancer-associated stroma compared to normal stroma, although its biological significance remains to be elucidated.
Acknowledgements
We thank Dr Karl Riabowol for his critical review and constructive advice for the manuscript, Dr Don Morris for tremendous institutional support, and the Breast Cancer Society of Canada for financial support.
Abbreviations
- AQUA
automated quantitative immunofluorescence analysis
- ATM
ataxia telangiectasia mutated
- CAFs
cancer-associated fibroblasts
- CI
confidence interval
- csATM
cancer-associated stromal ATM
- DAPI
diamidino-2-phenylindole
- DFS
disease-free survival
- DSS
disease-specific survival
- EBC
early-stage breast cancer
- ER
estrogen receptor
- FFPE
formalin-fixed paraffin-embedded
- FITC
fluoroscein isothiocyanate
- HER2
human epidermal growth factor receptor 2
- HNBC
hormone-negative breast cancer
- HPBC
hormone-positive breast cancer
- HRP
horseradish peroxidase
- IHC
immunohistochemistry
- IL
interleukin
- KM
Kaplan-Meier
- LN
lymph node status
- LVI
lymphvascular invasion
- nATM
tumor nuclear ATM
- ncATM
nuclear versus cytoplasm ATM
- PI3K
phosphatidyl inositol-3 kinase
- PIKKs
PI3K-like serine/threonine protein kinases
- PR
progesterone receptor
- RT
radiation treatment
- tATM
tumor ATM
- TBCC
Tom Baker Cancer Centre
- TBST
Tris-buffered saline with Tween 20
- TMA
tissue microarray
- TNBC
triple-negative breast cancer
Additional files
Comparison of the tumor characteristics in low and high csATM group in the HNBC and HPBC cohorts. One hundred and sixty-eight HNBC and 130 HPBC patients diagnosed with EBC were included in the study. Sixty-seven cases in the HNBC cohort and 28 cases in the HPBC cohort were excluded due to failure to obtain ATM staining due to technical issues. Therefore, a total of 101 cases in the HNBC cohort and 102 cases in the HPBC cohort were dichotomized into the high and low ATM group based on csATM AQUA scores. The cut-point was set at 38% in the HNBC cohort and 13% in the HPBC cohort respectively based on X-tile prediction. Fisher’s exact test was used to compare the tumor characteristics in the high and low csATM groups.
Univariate survival analysis in all breast cancer patients (HNBC + HPBC cohorts) based on tATM, nATM and csATM expression. Kaplan-Meier analysis of disease-specific survival (DSS) in all patients (HNBC + HPBC cohorts) comparing patients with high and low tATM (A), nATM (B) and csATM (C).
Univariate survival analysis in subsets of patients within the HNBC cohort based on tATM expression. Kaplan-Meier analysis of disease-specific survival (DSS) in LN+ subset (A) versus LN- subset (B); T1/T2 subset (C) versus T3/T4 subset (D) comparing HNBC patients with high and low tATM.
ATM expression ratio in malignant epithelial cell-specific nuclear versus cytoplasm (ncATM) in HPBC and HNBC cohorts compared to its expression ratio in normal breast epithelium. (A) Histogram distributions representing ncATM expression in both the HPBC cohort (green solid bar) and the HNBC cohort (red solid bar). The hashed black line represents median ncATM expression in normal breast stromal tissue, the hashed green line represents median ncATM expression in the HPBC cohort, and the hashed red line represents median ncATM expression in the HNBC cohort. (B) Comparison of ncATM AQUA scores in normal controls, HPBC cohort and HNBC cohort.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XF designed the study, interpreted the results, and drafted the manuscript. HL performed the statistical analysis, helped prepare most of the figures and critically revised the manuscript. MD and HW carried out the TMA staining and data analysis in the two cohorts and helped prepare some figures. EK carried out ER/PR/HER2 staining and data analysis in the HPBC cohort. EE participated in optimizing antibodies and techniques for TMA staining, and helped to draft the manuscript. AM generated two TMA cohorts, participated in the study design and critically revised the manuscript. PT participated in generating the HNBC cohort TMA and critically revising the manuscript. AP, SPL-M and GB conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript. In addition, all authors agreed to be accountable for the accuracy and integrity of the results from this study.
Authors’ information
Xiaolan Feng MD PhD FRCPC, Locum Medical Oncologist and Research Scientist.
Haocheng Li PhD, Biostatistician.
Michelle Dean BSc, Technician.
Holly Wilson BSc, Technician.
Elizabeth Kornaga MSc, Senior Technician.
Emeka Enwere PhD, Research Scientist.
Patricia Tang MD FRCPC, Medical Oncologist.
Alexander Paterson MD FRCPC, Medical Oncologist.
Susan P Lees-Miller PhD, Senior Research Scientist.
Anthony M Magliocco MD, Pathologist and Senior Research Scientist.
Gwyn Bebb MD PhD FRCPC, Medical Oncologist and Senior Research Scientist.
Contributor Information
Xiaolan Feng, Email: fxiaolan@ucalgary.ca.
Haocheng Li, Email: Haocheng.Li@ucalgary.ca.
Michelle Dean, Email: Michelle.Dean@albertahealthservices.ca.
Holly E Wilson, Email: Holly.Wilson2@albertahealthservices.ca.
Elizabeth Kornaga, Email: Elizabeth.Kornaga@albertahealthservices.ca.
Emeka K Enwere, Email: Emeka.Enwere@albertahealthservices.ca.
Patricia Tang, Email: Patricia.Tang@albertahealthservices.ca.
Alexander Paterson, Email: Alexander.Paterson@albertahealthservices.ca.
Susan P Lees-Miller, Email: leesmill@ucalgary.ca.
Anthony M Magliocco, Email: anthony.magliocco@moffitt.org.
Gwyn Bebb, Email: Gwyn.Bebb@albertahealthservices.ca.
References
- 1.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 2.Gnant M, Filipits M, Greil R, Stoeger H, Rudas M, Bago-Horvath Z, et al. Predicting distant recurrence in receptor-positive breast cancer patients with limited clinicopathological risk: using the PAM50 Risk of Recurrence score in 1478 postmenopausal patients of the ABCSG-8 trial treated with adjuvant endocrine therapy alone. Ann Oncol. 2014;25:339–45. doi: 10.1093/annonc/mdt494. [DOI] [PubMed] [Google Scholar]
- 3.Mook S, Schmidt MK, Weigelt B, Kreike B, Eekhout I, van de Vijver MJ, et al. The 70-gene prognosis signature predicts early metastasis in breast cancer patients between 55 and 70 years of age. Ann Oncol. 2010;21:717–22. doi: 10.1093/annonc/mdp388. [DOI] [PubMed] [Google Scholar]
- 4.Wu L, Candille SI, Choi Y, Xie D, Jiang L, Li-Pook-Than J, et al. Variation and genetic control of protein abundance in humans. Nature. 2013;499:79–82. doi: 10.1038/nature12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lempiainen H, Halazonetis TD. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 2009;28:3067–73. doi: 10.1038/emboj.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Economopoulou P, Dimitriadis G, Psyrri A. Beyond BRCA: new hereditary breast cancer susceptibility genes. Cancer Treat Rev. 2015;41:1–8. doi: 10.1016/j.ctrv.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. doi: 10.1038/nrm3546. [DOI] [PubMed] [Google Scholar]
- 8.Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38:873–5. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- 9.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waha A, Sturne C, Kessler A, Koch A, Kreyer E, Fimmers R, et al. Expression of the ATM gene is significantly reduced in sporadic breast carcinomas. Int J Cancer. 1998;78:306–9. doi: 10.1002/(SICI)1097-0215(19981029)78:3<306::AID-IJC8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 11.Ding SL, Sheu LF, Yu JC, Yang TL, Chen BF, Leu FJ, et al. Abnormality of the DNA double-strand-break checkpoint/repair genes, ATM, BRCA1 and TP53, in breast cancer is related to tumour grade. Br J Cancer. 2004;90:1995–2001. doi: 10.1038/sj.bjc.6601804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye C, Cai Q, Dai Q, Shu XO, Shin A, Gao YT, et al. Expression patterns of the ATM gene in mammary tissues and their associations with breast cancer survival. Cancer. 2007;109:1729–35. doi: 10.1002/cncr.22592. [DOI] [PubMed] [Google Scholar]
- 13.Tommiska J, Bartkova J, Heinonen M, Hautala L, Kilpivaara O, Eerola H, et al. The DNA damage signalling kinase ATM is aberrantly reduced or lost in BRCA1/BRCA2-deficient and ER/PR/ERBB2-triple-negative breast cancer. Oncogene. 2008;27:2501–6. doi: 10.1038/sj.onc.1210885. [DOI] [PubMed] [Google Scholar]
- 14.Salimi M, Mozdarani H, Majidzadeh K. Expression pattern of ATM and cyclin D1 in ductal carcinoma, normal adjacent and normal breast tissues of Iranian breast cancer patients. Med Oncol. 2012;29:1502–9. doi: 10.1007/s12032-011-0043-5. [DOI] [PubMed] [Google Scholar]
- 15.Bhattacharya N, Mukherjee N, Singh RK, Sinha S, Alam N, Roy A, et al. Frequent alterations of MCPH1 and ATM are associated with primary breast carcinoma: clinical and prognostic implications. Ann Surg Oncol. 2013;20:S424–32. doi: 10.1245/s10434-012-2715-6. [DOI] [PubMed] [Google Scholar]
- 16.Bueno RC, Canevari RA, Villacis RA, Domingues MA, Caldeira JR, Rocha RM, et al. ATM down-regulation is associated with poor prognosis in sporadic breast carcinomas. Ann Oncol. 2014;25:69–75. doi: 10.1093/annonc/mdt421. [DOI] [PubMed] [Google Scholar]
- 17.Abdel-Fatah TM, Arora A, Alsubhi N, Agarwal D, Moseley PM, Perry C, et al. Clinicopathological significance of ATM-Chk2 expression in sporadic breast cancers: a comprehensive analysis in large cohorts. Neoplasia. 2014;16:982–91. doi: 10.1016/j.neo.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otsuka S, Klimowicz AC, Kopciuk K, Petrillo SK, Konno M, Hao D, et al. CXCR4 overexpression is associated with poor outcome in females diagnosed with stage IV non-small cell lung cancer. J Thorac Oncol. 2011;6:1169–78. doi: 10.1097/JTO.0b013e3182199a99. [DOI] [PubMed] [Google Scholar]
- 19.Williamson CT, Kubota E, Hamill JD, Klimowicz A, Ye R, Muzik H, et al. Enhanced cytotoxicity of PARP inhibition in mantle cell lymphoma harbouring mutations in both ATM and p53. EMBO Mol Med. 2012;4:515–27. doi: 10.1002/emmm.201200229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brockton NT, Klimowicz AC, Bose P, Petrillo SK, Konno M, Rudmik L, et al. High stromal carbonic anhydrase IX expression is associated with nodal metastasis and decreased survival in patients with surgically-treated oral cavity squamous cell carcinoma. Oral Oncol. 2012;48:615–22. doi: 10.1016/j.oraloncology.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 21.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–9. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 22.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. REporting recommendations for tumor MARKer prognostic studies (REMARK) Breast Cancer Res Treat. 2006;100:229–35. doi: 10.1007/s10549-006-9242-8. [DOI] [PubMed] [Google Scholar]
- 23.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–7. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 24.Peng Q, Zhao L, Hou Y, Sun Y, Wang L, Luo H, et al. Biological characteristics and genetic heterogeneity between carcinoma-associated fibroblasts and their paired normal fibroblasts in human breast cancer. PLoS One. 2013;8:e60321. doi: 10.1371/journal.pone.0060321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paull TT. Mechanisms of ATM activation. Annual Rev Biochem. 2015;84:12.1–12. doi: 10.1146/annurev-biochem-060614-034335. [DOI] [PubMed] [Google Scholar]
- 26.Hsia SM, Yu CC, Shih YH, Chen MY, Wang TH, Huang YT, et al. Isoliquiritigenin causes DNA damage and inhibits ATM expression leading to G2/M phase arrest and apoptosis in oral squamous cell carcinoma. Head Neck. 2015. [Epub ahead of print]. [DOI] [PubMed]
- 27.Pazolli E, Alspach E, Milczarek A, Prior J, Piwnica-Worms D, Stewart SA. Chromatin remodeling underlies the senescence-associated secretory phenotype of tumor stromal fibroblasts that supports cancer progression. Cancer Res. 2012;72:2251–61. doi: 10.1158/0008-5472.CAN-11-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faurobert E, Bouin AP, Albiges-Rizo C. Microenvironment, tumor cell plasticity, and cancer. Curr Opin Oncol. 2015;27:64–70. doi: 10.1097/CCO.0000000000000154. [DOI] [PubMed] [Google Scholar]
- 29.Knappskog S, Chrisanthar R, Lokkevik E, Anker G, Ostenstad B, Lundgren S, et al. Low expression levels of ATM may substitute for CHEK2 /TP53 mutations predicting resistance towards anthracycline and mitomycin chemotherapy in breast cancer. Breast Cancer Res. 2012;14:R47. doi: 10.1186/bcr3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc Natl Acad Sci U S A. 2010;107:1506–11. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song L, Lin C, Wu Z, Gong H, Zeng Y, Wu J, et al. miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS One. 2011;6:e25454. doi: 10.1371/journal.pone.0025454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai AH, Tong JH, To KF, Chan MW, Man EP, Lo KW, et al. Promoter hypermethylation of tumor-related genes in the progression of colorectal neoplasia. Int J Cancer. 2004;112:846–53. doi: 10.1002/ijc.20485. [DOI] [PubMed] [Google Scholar]
- 33.Rengucci C, De Maio G, Casadei Gardini A, Zucca M, Scarpi E, Zingaretti C, et al. Promoter methylation of tumor suppressor genes in pre-neoplastic lesions; potential marker of disease recurrence. J Exp Clin Cancer Res. 2014;33:65. doi: 10.1186/s13046-014-0065-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–69. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 35.Gilardini Montani MS, Prodosmo A, Stagni V, Merli D, Monteonofrio L, Gatti V, et al. ATM-depletion in breast cancer cells confers sensitivity to PARP inhibition. J Exp Clin Cancer Res. 2013;32:95. doi: 10.1186/1756-9966-32-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bebb DG, Lees-Miller SP. Predicting PARP inhibitor sensitivity and resistance. Cell Cycle. 2012;11:4110. doi: 10.4161/cc.22604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubota E, Williamson CT, Ye R, Elegbede A, Peterson L, Lees-Miller SP, et al. Low ATM protein expression and depletion of p53 correlates with olaparib sensitivity in gastric cancer cell lines. Cell Cycle. 2014;13:2129–37. doi: 10.4161/cc.29212. [DOI] [PMC free article] [PubMed] [Google Scholar]


