Abstract
Background:
SET and MYND domain-containing protein 2 (SMYD2) is a lysine methyltransferase for histone H3, p53 and Rb and inhibits their transactivation activities. In this study, we tested whether SMYD2 (1q42) acts as a cancer-promoting factor by being overexpressed in gastric cancer.
Methods:
We analysed 7 gastric cancer cell lines and 147 primary tumor samples of gastric cancer, which were curatively resected in our hospital.
Results:
SET and MYND domain-containing protein 2 was detected in these cell lines (five out of seven cell lines; 71.4%) and primary tumor samples (fifty-six out of one hundred and forty-seven cases; 38.1%). Knockdown of SMYD2 using specific small interfering RNA inhibited proliferation, migration and invasion of SMYD2-overexpressing cells in a TP53 mutation-independent manner. Overexpression of SMYD2 protein correlated with larger tumor size, more aggressive lymphatic invasion, deeper tumor invasion and higher rates of lymph node metastasis and recurrence. Patients with SMYD2-overexpressing tumours had a worse overall rate of survival than those with non-expressing tumours (P=0.0073, log-rank test) in an intensity and proportion score-dependent manner. Moreover, multivariate analysis demonstrated that SMYD2 was independently associated with worse outcome (P=0.0021, hazard ratio 4.25 (1.69–10.7)).
Conclusions:
These findings suggest that SMYD2 has a crucial role in tumor cell proliferation by its overexpression and highlight its usefulness as a prognostic factor and potential therapeutic target in gastric cancer.
Keywords: SMYD2, TP53, gastric cancer, prognosis
Gastric cancer is one of the most common causes of death from cancer worldwide (Siegel et al, 2013). Recent advances in diagnostic technologies and peri-operative management have increased early detection of gastric cancer and decreased the morbidity and mortality rates. However, patients with advanced gastric cancer still frequently experience recurrence despite extended radical resections, which results in an extremely poor survival rate (Martin et al, 2002).
Numerous genes have been analysed to understand molecular mechanisms of carcinogenesis and improve clinical outcomes for human gastric cancers; however, only a few genes with frequent alterations have been identified (Ushijima and Sasako, 2004) such as gene amplifications of MET and ERBB2, hypermethylation of p16 (Oue et al, 2002; Ding et al, 2003), mutations of TP53, APC and E-cadherin (Becker et al, 1994; Maesawa et al, 1995; Lee et al, 2002), oncogenic activation of β-catenin and K-ras (Park et al, 1999) and inactivation of the mismatch repair gene hMLH1, which is associated with microsatellite instability (Fang et al, 2003). Including these reports, researchers have attempted to identify biological factors involved in the malignancy of gastric cancer. However, in clinical settings, only a few genes have been used as diagnostic biomarkers and/or therapeutic targets (Bang et al, 2010). We therefore wished to identify novel genes associated with the progression of gastric cancer.
SET and MYND domain-containing protein 2 (SMYD2) (1q42) was recently identified as a lysine methyltransferase for histone H3K36, H3K4 and K370 of p53, K860 and K810 of Rb and K528 of PARP1. It regulates transcription (Brown et al, 2006; Abu-Farha et al, 2008), inhibits tumour suppressor proteins p53 (Huang et al, 2006) and Rb (Saddic et al, 2010; Cho et al, 2012), and enhances the poly (ADP-ribose) activity of the oncogenic protein PARP1 in cancer cells (Piao et al, 2014). Moreover, SMYD2 is a clinically relevant prognostic marker for oesophageal squamous cell carcinoma (ESCC), bladder carcinoma and paediatric acute lymphoblastic leukaemia (Komatsu et al, 2009; Cho et al, 2012; Sakamoto et al, 2014) and it could be used to predict a potentially poor prognosis to plan more aggressive treatment for applicable patients. However, to date, there has been no report on the clinical and prognostic significance of SMYD2 in patients with primary gastric cancer. These findings prompted us to investigate the effects of SMYD2 overexpression and activation in primary gastric cancer.
Consequently, in this study, we demonstrated that SMYD2 was frequently overexpressed in gastric cancer cell lines and primary gastric cancers. Overexpression of SMYD2 was an indicator of poor prognosis independent of other prognostic factors. We also demonstrated that knockdown of SMYD2 suppressed cell proliferation, migration and invasion in gastric cancer cell lines. Our results provide evidence that SMYD2 could be an important molecular marker to determine malignant properties and a target for molecular therapy in patients with gastric cancer.
Materials and Methods
Cell lines and primary tissue samples
A total of seven gastric cancer cell lines (MKN7, MKN74, HGC27, MKN45, KatoIII, NUGC4 and MKN28 cells) and a fibroblast cell line (WI-38) were used in this study. HGC27 and WI-38 cells were cultured in mixture of Dulbecco's Minimum Essential Medium and F12 medium, and the other cell lines were cultured in RPMI-1640 medium. All culture media were purchased from Sigma (St. Louis, MO, USA) and supplemented with 100 ml l−1 foetal bovine serum (Trace Scientific, Melbourne, VIC, Australia). All cell lines were cultured in 5% carbon dioxide at 37 °C in a humidified chamber. Primary tumour samples of gastric cancer were obtained from 147 gastric cancer patients, who underwent curative gastrectomy (R0) at the Division of Digestive Surgery, Department of Surgery, Kyoto Prefectural University of Medicine (Kyoto, Japan) between 2001 and 2003. These samples were embedded in paraffin after 24 h of formalin fixation. Relevant clinical and survival data were available for all patients. Written consent was always obtained in the formal style and after approval by the local ethics committee. None of these patients underwent endoscopic mucosal resection, palliative resection, preoperative chemotherapy or radiotherapy, and none of them had synchronous or metachronous multiple cancers in other organs. Disease stage was defined in accordance with the International Union against Cancer tumour-lymph node-metastases classification (7th edition (Sobin and Wittekind, 2009)). The median follow-up period for surviving patients was 54.5 months (ranging from 0.5 to 84.2 months).
Quantitative real-time RT–PCR
Single-stranded complementary DNA generated from total RNA was amplified with primers specific for each gene, as described below. Abundance of messenger RNA (mRNA) was measured with a quantitative real-time fluorescence detection method (ABI StepOnePlus Sequence Detection System; Applied Biosystems, Foster City, CA, USA) using TaqMan Gene Expression Assays (Hs00220210_m1 for SMYD2; Applied Biosystems) according to the manufacturer's instructions. Results of gene expression were calculated as a ratio between SMYD2 and an internal reference gene (Hs99999903_m1 for β-actin; Applied Biosystems) that provides a normalisation factor for the amount of RNA isolated from a specimen, and the ratio was subsequently normalised by that of the control fibroblast cell line (relative expression). This assay was performed in duplicate for each sample.
Western blotting
Anti-SMYD2 rabbit polyclonal antibody was generated against a 14-amino acid peptide from human SMYD2 (HPYISEIKQEIESH; Operon Biotechnology, Tokyo, Japan) and purified through an affinity column. Anti-β-actin antibody was purchased from Sigma, and anti-p53 (DO-7) and anti-p21 antibodies were from Novocastra Laboratories (Newcastle upon Tyne, UK) and Santa Cruz Biotechnology (Santa Cruz, CA, USA), respectively. Cells were lysed and their proteins were extracted by M-PER Mammalian Protein Extraction Reagent (Thermo Fisher Scientific, Waltham, MA, USA). Twenty micrograms of proteins per lane were loaded for electrophoresis.
Knockdown of SMYD2 by siRNA and analysis of cell proliferation
We knocked down SMYD2 with small interfering RNA (siRNA) targeting SMYD2 (SMARTpool #M-020291-00; Dharmacon, Lafayette, CO, USA) and we used siRNA targeting luciferase as a negative control (Luc, 5′-CGUACGCGGAAUACUUCGA-3′ Sigma, Tokyo, Japan). Gastric cancer cell lines were transfected with each siRNA (10 nmol l−1) with Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Effects of SMYD2 knockdown on protein abundance was confirmed by Western blotting. To measure cell proliferation, the number of viable cells 24, 48 and 72 h after siRNA transfection was assessed by the colorimetric water-soluble tetrazolium salt assay (Cell counting kit-8; Dojindo Laboratories, Kumamoto, Japan). Cell cycle was evaluated 72 h after transfection by fluorescence-activated cell sorting (FACS) as described elsewhere (Komatsu et al, 2009).
Transwell migration and invasion assays
Transwell migration and invasion assays were carried out in 24-well modified Boyden chambers (transwell-chamber, BD Transduction, Franklin Lakes, NJ, USA). For invasion assays, the upper surfaces of the 6.4-mm diameter filters with 8-μm pores that were used for migration assays were precoated with Matrigel (BD Transduction). siRNA transfectants (2 × 104 cells per well) were transferred into the upper chamber. Following 48 h of incubation, migrated or invasive cells on the lower surface of the filters were fixed and stained with the Diff-Quik stain (Sysmex, Kobe, Japan), and stained cell nuclei were counted directly in triplicate. We assessed the ability of the cells to move through extracellular matrices by calculating the number of cells, which is the ratio of the percentage invasion through the matrigel-coated filters relative to migration through the uncoated filters of test cells over that in the control cells as described elsewhere (Kashimoto et al, 2012; Nishimura et al, 2013).
Immunohistochemistry
Primary tumour samples were fixed with 10% formaldehyde in PBS, embedded in paraffin, sectioned into 5-μm-thick slices and subjected to immunohistochemical staining of SMYD2 with the avidin–biotin–peroxidase method as described by Naoi et al (2008). In brief, after deparaffinization, endogenous peroxidases were quenched by incubating the sections for 20 min in 3% H2O2. Antigen retrieval was performed by heating the samples in 10 mmol l−1 citrate buffer (pH 6.0) at 95 °C for 60 min. After treatment with Block Ace (Dainippon Sumitomo Pharmaceutical, Osaka, Japan) for 30 min at room temperature, sections were incubated at 4 °C overnight with the anti-SMYD2 (1 : 200) antibody. The avidin–biotin–peroxidase complex system (Vectastain Elite ABC universal kit; Vector Laboratories Inc., Burlingame, CA, USA) was used for colour development with diaminobenzidine tetrahydrochloride. Tissues were counterstained with Mayer's haematoxylin. To confirm the specificity of the anti-SMYD2 antibody, a formalin-fixed oesophageal cancer cell line overexpressing SMYD2 (KYSE170 cells), in which >50% of cells showed staining of SMYD2 protein, was used as a positive control, whereas KYSE170 cells incubated without the SMYD2 antibody were used as a negative control (Supplementary Figure S1; Komatsu et al, 2009). For scoring SMYD2 abundance, the intensity (intensity score 0=negative, score 1=weak, score 2=moderate, score 3=strong, as described below) and percentage of the SMYD2-expressing cells in the total population (proportion score 0, <10% score 1, 10 to 33% score 2, 34 to 66% score 3, 67 to 100%) were evaluated for each tumour sample using high-power ( × 200) microscopy. In primary tumour samples, SMYD2 protein was not detected in most of the non-tumorous gastric mucosa and stroma. Primary tumours with no detectable SMYD2 (similar to non-tumorous gastric mucosa and stroma) were given an intensity score of 0, whereas those with the greatest SMYD2 abundance were given an intensity score of 3. The remaining tumours were categorised with intensity scores of 1 or 2 according to the intensity of immunohistochemical staining for SMYD2. Finally, expression of SMYD2 was graded based on the sum of the intensity score and the proportion score: high expression (intensity plus proportion scores ⩾4) or low expression (intensity plus proportion scores ⩽3) (Tsuda, 2008).
Statistical analysis
Clinicopathological variables pertaining to the corresponding patients were analysed for statistical significance by the χ2-test or Fisher's exact test. For the analysis of survival, Kaplan–Meier survival curves were constructed for groups based on univariate predictors, and differences between the groups were analysed with the log-rank test. Univariate and multivariate survival analyses were performed using the likelihood ratio test of the stratified Cox proportional hazards model. Differences between subgroups were tested with the non-parametric Mann–Whitney U-test. Differences were assessed with a two-sided test and considered statistically significant at P<0.05.
Results
Overexpression of SMYD2 in gastric cancer cell lines
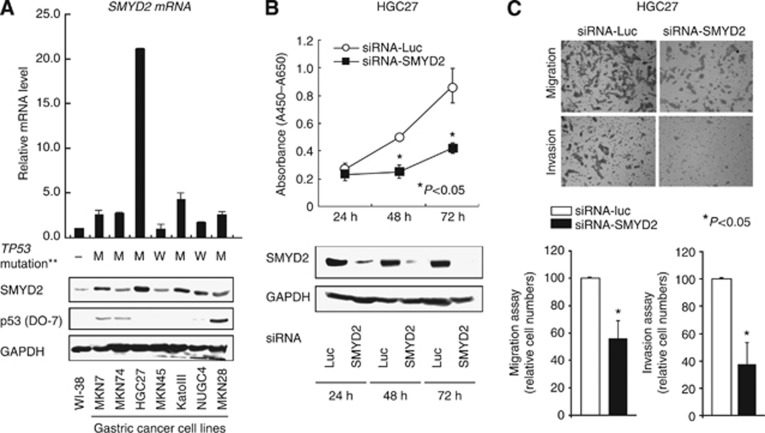
Quantitative RT–PCR and Western blotting analyses were performed to test whether SMYD2 is overexpressed in gastric cancer cell lines compared with the fibroblast cell line WI-38 (Figure 1A). Expression of SMYD2 protein detected by the SMYD2-specific antibody correlated with that of SMYD2 mRNA in gastric cancer cell lines. SMYD2 was overexpressed in almost all gastric cancer cell lines (five out of seven cell lines, 71.4%), suggesting this gene to be a target for activation in gastric cancer cell lines (Figure 1A). Because repression of p53 activity through SMYD2-mediated methylation at K370 was reported previously (Huang et al, 2006), we examined the status of TP53 mutation in the gastric cancer cell lines by Western blotting. These statuses of TP53 mutation in various cell lines are positively associated with their reported status of TP53 mutation in the database (http://p53.free.fr/index.html) (M=mutant TP53; W=wild-type TP53.). Although wild-type p53 is expected to be a more suitable substrate than mutant p53 for SMYD2 (Huang et al, 2006; Komatsu et al, 2009), expression of SMYD2 mRNA and protein was not correlated with the mutation status and expression of p53 (Figure 1A).
Figure 1.
(A) Expression of SMYD2 mRNA (top) and protein (bottom) in seven gastric cancer cell lines compared with that in a fibroblast cell line WI-38. Also, the status of TP53 mutation in each gastric cancer cell line was evaluated by Western blotting. The status of TP53 mutation was positively associated with the reported status of TP53 mutation in the database (http://p53.free.fr/index.html, M=mutant TP53; W=wild-type TP53. )**. (B) HGC27 cells were transfected with siRNA targeting SMYD2 or control luciferase (Luc) for the indicated times. Cell proliferation (top) and SMYD2 protein expression (bottom) were compared between these cells. (C) Migration and invasion of HGC27 cells transfected with siRNA targeting SMYD2. The graphs on the bottom show mean±s.d. (bars) of n=4. The Mann–Whitney U-test was used for statistical analysis. P<0.05 is considered statistically significant.
Suppression of cell proliferation by knockdown of SMYD2
To gain an insight into the potential role of SMYD2 as an oncogene whose overexpression could be associated with gastric carcinogenesis, we first performed cell proliferation assays using siRNA specific for SMYD2 and investigated whether knockdown of SMYD2 would suppress proliferation of gastric cancer cell lines that overexpress SMYD2. We chose the HGC27 cell line for these assays, because it had the highest amount of SMYD2 protein (Figure 1A). Expression of SMYD2 protein in this cell line was efficiently knocked down 24–72 h after the transient introduction of SMYD2-specific siRNA (siRNA-SMYD2) (Figure 1B) than with the control luciferase siRNA (siRNA-Luc). We measured the proliferation of these siRNA-transfected HGC27 cells. The proliferation of the cells transfected with siRNA-SMYD2 was 51.6% lower than that of cells transfected with control siRNA (siRNA-Luc) 72 h after transfection (Figure 1B).
Suppression of cell migration and invasion by knockdown of SMYD2
Next, Transwell migration and invasion assays were performed to examine the ability of HGC27 cells transfected with siRNA-SMYD2 to move through pores under different conditions. Uncoated membrane was used for migration assays, whereas Matrigel-coated membrane was used for invasion assays. In Figure 1C, the number of cells that migrated into the lower chamber was significantly lower for siRNA-SMYD2-transfected cells than for siRNA-Luc-transfected cells under both conditions, suggesting that SMYD2 may increase the ability of gastric cancer cells to migrate.
Correlation between suppression of cell proliferation by knockdown of SMYD2 and TP53 mutation status
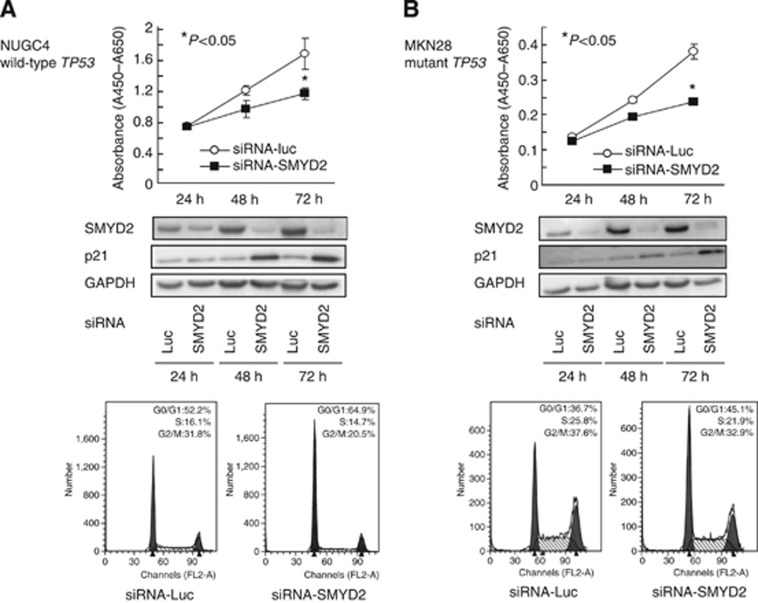
To gain further insight into the potential association of SMYD2 with TP53 mutation status, we performed cell proliferation assays with the NUGC4 cell line, which has wild-type TP53, and the MKN28 cell line, which has mutant TP53. In both NUGC4 and MKN28 cell lines, endogenous SMYD2 protein was efficiently knocked down 24–72 h after the transient introduction of SMYD2-specific siRNA (Figure 2A and B, middle panels). An inhibitory effect of knockdown of SMYD2 on cell proliferation was also observed in both cell lines (Figure 2A and B. top panels), suggesting that this inhibitory effect is not associated with TP53 mutation/expression status. Fluorescence-activated cell sorting analysis demonstrated that transfection of both NUGC4 and MKN28 cell lines with siRNA-SMYD2 resulted in an accumulation of cells in the G0–G1 phase compared with transfection with control siRNA. In addition, p21 protein abundance was also increased at the protein level in siRNA-SMYD2-transfected cells (Figure 2A and B, middle panels), suggesting that the knockdown of SMYD2 directly or indirectly induced the production of p21, which results mainly in G0–G1 arrest.
Figure 2.
Effects of SMYD2 knockdown by siRNA (siRNA-SMYD2) compared with those of control siRNA (siRNA-Luc) in NUGC4 (wild-type TP53, A) and MKN28 (mutant TP53, B) cell lines. Top: effects of knocking down endogenous SMYD2 on cell proliferation at the indicated times. Results are mean±s.d. (bars) for quadruplicate experiments. The Mann–Whitney U-test was used for the statistical analysis: *P<0.05 vs siRNA-Luc-transfected cells. Middle: effects of siRNA-SMYD2 on the abundance of endogenous SMYD2 and p21 proteins 24–72 h after transfection of gastric cancer cell lines. Bottom: representative results of the population in each phase of the cell cycle in gastric cancer cell lines assessed by FACS 72 h after treatment with siRNA.
Immunohistochemical analysis of SMYD2 expression in primary gastric cancer
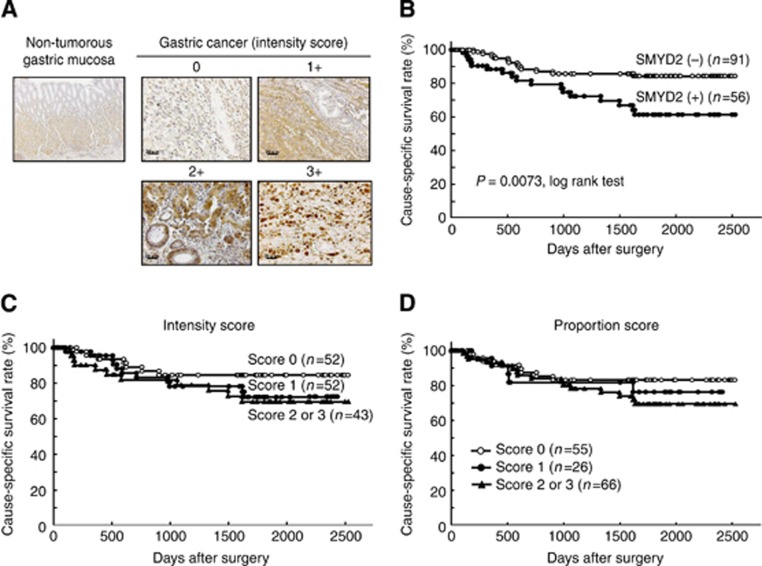
Because SMYD2 protein was overexpressed in almost all gastric cancer cell lines, we hypothesised that SMYD2 would also be highly expressed in primary gastric cancer tissues and would be involved in carcinogenesis and malignant outcomes. We examined the clinicopathological significance of SMYD2 in primary tumour samples based on the immunohistochemical staining pattern of this protein. SMYD2 protein was observed in both the cytoplasm and the nucleus of cancer cells regardless of tumour differentiation (right and left panels, respectively, of Supplementary Figure S2). We classified 147 gastric cancer samples into positive or negative groups according to the intensity and proportion scores of SMYD2 staining as described in the Materials and Methods section. In primary tumours, SMYD2 protein was not detected in most of the non-tumorous gastric mucosal cell population (intensity score 0) (Figure 3A), and it was weakly detected in the gastric fundic gland. Supplementary Table 1 shows the distribution of patients based on the intensity and proportion scores of SMYD2 immunoreactivity of tumour samples. A group of high expression with the combined scores of >4 was considered immunopositive and is shown in grey in this table. Kaplan–Meier survival estimates showed that SMYD2 immunoreactivity in tumour cells was statistically significantly associated with a worse overall survival based on the intensity and proportion scores (Figure 3C and D, respectively). Based on the combined scores, patients in the high expression group had statistically significantly poorer prognoses than did patients in the low expression group (P=0.0073, log-rank test) (Figure 3B).
Figure 3.
(A) Specific immunostaining of SMYD2 protein in a representative primary tumor sample. Based on this result, the intensity scores for SMYD2 staining were determined as follows; 0=negative, 1=weak, 2=moderate, 3=strong). Scale bar: 20 μm. (B–D) Cause-specific survival rates of gastric cancer patients (as determined by Kaplan–Meier plots) depending on the scores of the intensity (C), proportion (D) and in combination (B) based on the immunostaining analysis of their cancers. The log-rank test was used for statistical analysis. P<0.05 is considered statistically significant.
Correlation between SMYD2 protein abundance and clinicopathological characteristics in primary gastric cancer
The association between SMYD2 protein abundance and clinicopathological characteristics is summarised in Table 1. SMYD2 protein abundance was statistically significantly associated with larger tumour size, higher incidence of lymphatic invasion and lymph node metastasis, deeper invasion and higher recurrence rate, and tended to be associated with higher incidence of venous invasion.
Table 1. Association between clinicopathologic characteristics and SMYD2 expression.
| |
SMYD2 immunoreactivity |
|
||
|---|---|---|---|---|
| n | High expression | Low expression | P-valuea | |
| Total |
147 |
56 |
91 |
|
|
Sex | ||||
| Male | 100 | 41 (73%) | 59 (65%) | |
| Female |
47 |
15 (27%) |
32 (35%) |
0.2901 |
|
Age (years) | ||||
| Mean | 63 (range:27–89) | |||
| <60 | 56 | 23 (41%) | 33 (36%) | |
| ⩾60 |
91 |
33 (59%) |
58 (64%) |
0.5036 |
|
Location | ||||
| Upper | 20 | 10 (18%) | 10 (11%) | |
| Middle | 75 | 23 (41%) | 52 (57%) | |
| Lower |
52 |
23 (41%) |
29 (32%) |
0.1505 |
|
Histopathological grading | ||||
| Differentiated | 73 | 32 (57%) | 41 (45%) | |
| Undifferentiated |
74 |
24 (43%) |
50 (55%) |
0.1546 |
|
Tumor size (mm) | ||||
| <40 | 84 | 26 (46%) | 58 (64%) | |
| ⩾40 |
63 |
30 (54%) |
33 (36%) |
0.0394 |
|
Venous invasion | ||||
| 0 | 104 | 35 (63%) | 69 (76%) | |
| 1–3 |
43 |
21 (38%) |
22 (24%) |
0.0846 |
|
Lymphatic invasion | ||||
| 0 | 80 | 20 (36%) | 60 (66%) | |
| 1–3 |
67 |
36 (64%) |
31 (34%) |
0.0003 |
|
TNM classification | ||||
| pT categories | ||||
| pT1 | 83 | 24 (43%) | 59 (65%) | |
| pT2/3 | 34 | 16 (29%) | 18 (20%) | |
| pT4 |
30 |
16 (29%) |
14 (15%) |
0.0290 |
| pN categories | ||||
| N0 | 96 | 34 (61%) | 62 (68%) | |
| N1/2 | 28 | 7 (13%) | 21 (23%) | |
| N3 |
23 |
15 (27%) |
8 (9%) |
0.0204 |
| pStage | ||||
| I | 94 | 31 (55%) | 63 (69%) | |
| II | 16 | 6 (11%) | 10 (11%) | |
| III |
37 |
19 (34%) |
18 (20%) |
0.2117 |
| Rucurrence | ||||
| Absent | 117 | 39 (70%) | 78 (86%) | |
| Present | 30 | 17 (30%) | 13 (14%) | 0.0188 |
Abbreviations: SMYD2=SET and MYND domain-containing protein 2; TNM=tumour-lymph node-metastases.
Statistically significant values are in bold.
P-values are from χ2- or Fisher's exact test and were statistically significant at <0.05.
In the Cox proportional hazard regression model (Table 2), univariate analyses demonstrated that SMYD2 protein abundance, age, location, tumour size, venous and lymphatic invasion, pT category and pN category were statistically significantly associated with cause-specific survival. When data were stratified for multivariate analysis using both the forward and backward stepwise Cox regression procedures, SMYD2 immunoreactivity in tumour cells remained significant, with P<0.05 (hazard ratio, 4.26 (1.69–10.72)) for overall survival in all patients, suggesting that SMYD2 immunoreactivity can be an independent predictor of overall survival.
Table 2. Multivariate analysis using the stepwise Cox regression procedures.
| |
Univariatea |
Multivariateb |
||
|---|---|---|---|---|
| Variables | P-value | HR | 95% CI | P-value |
|
Sex | ||||
| Male vs female |
0.9583 |
2.749 |
1.134–6.665 |
0.0252 |
|
Age | ||||
| ⩾60 vs <60 |
0.0391 |
5.649 |
1.992–16.12 |
0.0011 |
|
Location | ||||
| U vs ML |
0.0262 |
|
— |
|
|
Histological type | ||||
| Undifferentiated vs differentiated |
0.8468 |
|
— |
|
|
Tumor size (mm) | ||||
| ⩾40 vs <40 |
<0.0001 |
|
— |
|
|
Venous invasion | ||||
| Positive vs negative |
<0.0001 |
5.681 |
2.173–14.92 |
0.0004 |
|
Lymphatic invasion | ||||
| Positive vs negative |
<0.0001 |
|
— |
|
|
pT-stage | ||||
| T3-4 vs T1-2 |
<0.0001 |
6.493 |
1.385–27.77 |
0.0177 |
|
pN-stage | ||||
| N2-3 vs N0-1 |
<0.0001 |
17.54 |
4.672–66.66 |
<0.0001 |
|
SMYD2 expression | ||||
| High vs low | 0.0073 | 4.258 | 1.690–10.72 | 0.0021 |
Abbreviations: CI=confidence interval; HR=hazard ratio; ML=middle and lower; SMYD2=SET and MYND domain-containing protein 2; U=upper. Statistically significant values are in bold.
Kaplan–Meier method, and the statistical significance was determined by log-rank test.
Multivariate survival analysis was performed using Cox's proportional hazard model.
Discussion
Lysine methyltransferases for histone or non-histone proteins have emerged as attractive targets for disease treatments (Kelly et al, 2010), and intensive ongoing efforts have been made to clarify their molecular mechanisms and clinical outcomes in cancers (Chang and Hung, 2012; Anglin and Song, 2013; Shankar et al, 2013; Sarris et al, 2014) and to develop novel drugs for this class of enzymes (Wagner and Jung, 2012). The human SMYD family of lysine methyltransferases is composed of five members (SMYD1–5) (Brown et al, 2006), which have various roles in development and cancer. Disruption of SMYD1 leads to perturbed cardiac morphogenesis and embryonic lethality (Gottlieb et al, 2002). SMYD3 is associated with cancer cell proliferation, and it is overexpressed in hepatocellular, colorectal and breast carcinomas (Hamamoto et al, 2004, 2006). SMYD4, as a potential tumour suppressor, has a critical role in breast carcinogenesis at least partly through inhibiting the expression of PDGFRA (Hu et al, 2009). SMYD5, as a negative regulator of inflammatory response genes, is recruited to a subset of TLR4-responsive promoters through its association with NCoR corepressor complexes, where it trimethylates histone H4 K20 (Stender et al, 2012).
SMYD2 methylates both histones (H2B, H3 and H4) and non-histone proteins, including the tumour suppressor proteins, p53 and Rb, and the oncogenic protein, PARP1 (Brown et al, 2006; Huang et al, 2006; Abu-Farha et al, 2008; Wu et al, 2011). Methylation of K370 of p53 impairs its ability to bind to the promoters of target genes (Chuikov et al, 2004; Huang et al, 2006). Methylation of Rb at K860 generates an epitope that is selectively recognised by the transcriptional repressor L3MBTL1, providing a mechanism for recruiting L3MBTL1 to the promoters of specific Rb/E2F target genes, thereby repressing their activities (Saddic et al, 2010). In addition, SMYD2-dependent methylation of Rb at K810 promotes cell cycle progression of cancer cells (Cho et al, 2012). Moreover, methylation of K528 on PARP1 enhances its poly(ADP-ribose) activity in cancer cells (Piao et al, 2014). In agreement with these observations, overexpression of SMYD2 is associated with various human malignancies, such as ESCC (Komatsu et al, 2009), bladder carcinoma (Cho et al, 2012) and paediatric acute lymphoblastic leukaemia (Sakamoto et al, 2014), indicating that SMYD2 acts as a cancer-promoting factor. Moreover, SMYD2 is involved in maintaining the self-renewal activity of MLL-AF9-induced acute myeloid leukaemia (Abu-Farha et al, 2011) and might be related to the response to chemotherapy in breast cancer (Barros Filho et al, 2010). These findings prompted us to determine the clinicopathological and prognostic significance of SMYD2 overexpression activation in primary gastric cancer.
Here, we hypothesised that overexpression/activation of SMYD2 may promote tumour cell proliferation and/or poor survival of gastric cancer patients. To test this hypothesis, we examined the expression status of SMYD2 and clinicopathological and biological significance of its expression in cell lines and primary tumours of gastric cancer. Consequently, we demonstrated that SMYD2 was overexpressed in 38.1% (56/147) of primary gastric cancers as well as in 71.4% (5/7) of gastric cancer cell lines and this overexpression was an indicator of poor prognosis independent of other prognostic factors. The intensity and proportion of SMYD2 abundance detected by immunohistochemical tissue staining was correlated with the prognosis of gastric cancer patients. In addition, knockdown of SMYD2 suppressed cell proliferation, migration and invasion in the gastric cancer cell lines.
Because p53 and its target molecules, which regulate cell cycle and trigger apoptosis after DNA damage, have a key role in a wide range of human cancers, including gastric cancer (Caelles et al, 1994; Fenoglio-Preiser et al, 2003; Bellini et al, 2012), it is possible that overexpression of SMYD2 promotes cell proliferation and/or survival by inhibiting the transactivation activity of p53. Based on this hypothesis, we performed siRNA-mediated knockdown of SMYD2 which had an inhibitory effect on cell proliferation in gastric cancer cells, which overexpress SMYD2, more or less independently of TP53 mutation status. Cell cycle analysis by FACS demonstrated that the inhibition of cell proliferation caused by SMYD2 knockdown occurred mainly because of G0–G1 arrest. This result was supported by our finding that increases in expression of p21 protein were caused by SMYD2 knockdown, although it remains unknown how SMYD2 inhibits the transcription of p21 in a p53-independent manner. These results are consistent with our previous findings of an inhibitory effect on cell proliferation upon SMYD2 knockdown in TP53-mutant ESCC cells and p53-null SaOS2 osteosarcoma cells (Komatsu et al, 2009).
More recently, the first SMYD2 inhibitor AZ505 (AstraZeneca, London, UK) was developed, which is highly selective and shows an activity at submicromolar concentrations in vitro (Ferguson et al, 2011). The IC50 of AZ505 for SMYD2 is 0.12 μM, which is >600-fold greater than the IC50s of AZ505 for other histone methyltransferases, such as SMYD3 (IC50>83.3 μM), DOT1L (IC50>83.3 μM) and EZH2 (IC50>83.3 μM), although functional data on the consequences of chemical inhibition of SMYD2 have not been reported (Ferguson et al, 2011). Moreover, anticancer effects of similar lysine methyltransferase inhibitors have already been proven in animals (Schenk et al, 2012; Willmann et al, 2012). Overexpression of SMYD2 protein was detected in 38.1% of patients with gastric cancer and 76.5% of patients with ESCC (Komatsu et al, 2009). These findings suggest that the SMYD2 inhibitor AZ505 may be a key molecule for the treatment of patients with these cancers. This drug is currently being evaluated in vitro and vivo.
In conclusion, this is the first report to show that SMYD2 has a crucial oncogenic role and is a potential therapeutic target in gastric cancer. We demonstrated the frequent overexpression of SMYD2 protein and its prognostic value in patients with gastric cancer. Although studies of larger cohorts are needed to validate these findings before moving to clinical settings, our results provide evidence that SMYD2 could be a pivotal molecular marker to determine malignant properties of gastric cancer cells and that it could be a target for molecular therapy in patients with this cancer.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Abu-Farha M, Lambert JP, Al-Madhoun AS, Elisma F, Skerjanc IS, Figeys D. The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol Cell Proteomics. 2008;7:560–572. doi: 10.1074/mcp.M700271-MCP200. [DOI] [PubMed] [Google Scholar]
- Abu-Farha M, Lanouette S, Elisma F, Tremblay V, Butson J, Figeys D, Couture JF. Proteomic analyses of the SMYD family interactomes identify HSP90 as a novel target for SMYD2. J Mol Cell Biol. 2011;3:301–308. doi: 10.1093/jmcb/mjr025. [DOI] [PubMed] [Google Scholar]
- Anglin JL, Song Y. A medicinal chemistry perspective for targeting histone H3 lysine-79 methyltransferase DOT1L. J Med Chem. 2013;56:8972–8983. doi: 10.1021/jm4007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, Aprile G, Kulikov E, Hill J, Lehle M, Ruschoff J, Kang YK. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- Barros Filho MC, Katayama ML, Brentani H, Abreu AP, Barbosa EM, Oliveira CT, Góes JC, Brentani MM, Folgueira MA. Gene trio signatures as molecular markers to predict response to doxorubicin cyclophosphamide neoadjuvant chemotherapy in breast cancer patients. Braz J Med Biol Res. 2010;43:1225–1231. doi: 10.1590/s0100-879x2010007500135. [DOI] [PubMed] [Google Scholar]
- Becker KF, Atkinson MJ, Reich U, Becker I, Nekarda H, Siewert JR, Hofler H. E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res. 1994;54:3845–3852. [PubMed] [Google Scholar]
- Bellini MF, Cadamuro AC, Succi M, Proença MA, Silva AE. Alterations of the TP53 gene in gastric and esophageal carcinogenesis. J Biomed Biotechnol. 2012;2012:891961. doi: 10.1155/2012/891961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MA, Sims RJ, 3rd, Gottlieb PD, Tucker PW. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer. 2006;28:26. doi: 10.1186/1476-4598-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caelles C, Helmberg A, Karin M. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370:220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 2012;106:243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HS, Hayami S, Toyokawa G, Maejima K, Yamane Y, Suzuki T, Dohmae N, Kogure M, Kang D, Neal DE, Ponder BA, Yamaue H, Nakamura Y, Hamamoto R. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia. 2012;14:476–486. doi: 10.1593/neo.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- Ding Y, Le XP, Zhang QX, Du P. Methylation and mutation analysis of p16 gene in gastric cancer. World J Gastroenterol. 2003;9:423–426. doi: 10.3748/wjg.v9.i3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang DC, Wang RQ, Yang SM, Yang JM, Liu HF, Peng GY, Xiao TL, Luo YH. Mutation and methylation of hMLH1 in gastric carcinomas with microsatellite instability. World J Gastroenterol. 2003;9 (4:655–659. doi: 10.3748/wjg.v9.i4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoglio-Preiser CM, Wang J, Stemmermann GN, Noffsinger A. TP53 and gastric carcinoma: a review. Hum Mutat. 2003;21:258–270. doi: 10.1002/humu.10180. [DOI] [PubMed] [Google Scholar]
- Ferguson AD, Larsen NA, Howard T, Pollard H, Green I, Grande C, Cheung T, Garcia-Arenas R, Cowen S, Wu J, Godin R, Chen H, Keen N. Structural basis of substrate methylation and inhibition of SMYD2. Structure. 2011;19:1262–1273. doi: 10.1016/j.str.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, Maika SD, Kuziel WA, King HL, Olson EN, Nakagawa O, Srivastava D. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- Hamamoto R, Silva FP, Tsuge M, Nishidate T, Katagiri T, Nakamura Y, Furukawa Y. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–118. doi: 10.1111/j.1349-7006.2006.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Zhu YT, Qi C, Zhu YJ. Identification of Smyd4 as a potential tumor suppressor gene involved in breast cancer development. Cancer Res. 2009;69:4067–4072. doi: 10.1158/0008-5472.CAN-08-4097. [DOI] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- Kashimoto K, Komatsu S, Ichikawa D, Arita T, Konishi H, Nagata H, Takeshita H, Nishimura Y, Hirajima S, Kawaguchi T, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H, Otsuji E. Overexpression of TRIM44 contributes to malignant outcome in gastric carcinoma. Cancer Sci. 2012;103:2021–2026. doi: 10.1111/j.1349-7006.2012.02407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu S, Imoto I, Tsuda H, Kozaki KI, Muramatsu T, Shimada Y, Aiko S, Yoshizumi Y, Ichikawa D, Otsuji E, Inazawa J. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis. 2009;30:1139–1146. doi: 10.1093/carcin/bgp116. [DOI] [PubMed] [Google Scholar]
- Lee JH, Abraham SC, Kim HS, Nam JH, Choi C, Lee MC, Park CS, Juhng SW, Rashid A, Hamilton SR, Wu TT. Inverse relationship between APC gene mutation in gastric adenomas and development of adenocarcinoma. Am J Pathol. 2002;161:611–618. doi: 10.1016/S0002-9440(10)64216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maesawa C, Tamura G, Suzuki Y, Ogasawara S, Sakata K, Kashiwaba M, Satodate R. The sequential accumulation of genetic alterations characteristic of the colorectal adenoma-carcinoma sequence does not occur between gastric adenoma and adenocarcinoma. J Pathol. 1995;176:249–258. doi: 10.1002/path.1711760307. [DOI] [PubMed] [Google Scholar]
- Martin RC, 2nd, Jaques DP, Brennan MF, Karpeh M. Extended local resection for advanced gastric cancer: increased survival versus increased morbidity. Ann Surg. 2002;236:159–165. doi: 10.1097/00000658-200208000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoi Y, Miyoshi Y, Taguchi T, Kim SJ, Arai T, Maruyama N, Tamaki Y, Noguchi S. Connexin26 expression is associated with aggressive phenotype in human papillary and follicular thyroid cancers. Cancer Lett. 2008;262:248–256. doi: 10.1016/j.canlet.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Komatsu S, Ichikawa D, Nagata H, Hirajima S, Takeshita H, Kawaguchi T, Arita T, Konishi H, Kashimoto K, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H, Otsuji E. Overexpression of YWHAZ relates to tumor cell proliferation and malignant outcome of gastric carcinoma. Br J Cancer. 2013;108:1324–1331. doi: 10.1038/bjc.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oue N, Motoshita J, Yokozaki H, Hayashi K, Tahara E, Taniyama K, Matsusaki K, Yasui W. Distinct promoter hypermethylation of p16INK4a, CDH1, and RAR-beta in intestinal, diffuse-adherent, and diffuse-scattered type gastric carcinomas. J Pathol. 2002;198:55–59. doi: 10.1002/path.1170. [DOI] [PubMed] [Google Scholar]
- Park WS, Oh RR, Park JY, Lee SH, Shin MS, Kim YS, Kim SY, Lee HK, Kim PJ, Oh ST, Yoo NJ, Lee JY. Frequent somatic mutations of the beta-catenin gene in intestinal-type gastric cancer. Cancer Res. 1999;59:4257–4260. [PubMed] [Google Scholar]
- Piao L, Kang D, Suzuki T, Masuda A, Dohmae N, Nakamura Y, Hamamoto R. The histone methyltransferase SMYD2 methylates PARP1 and promotes poly(ADP-ribosyl)ation activity in cancer cells. Neoplasia. 2014;16:257–264. doi: 10.1016/j.neo.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddic LA, West LE, Aslanian A, Yates JR, 3rd, Rubin SM, Gozani O, Sage J. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem. 2010;285:37733–37740. doi: 10.1074/jbc.M110.137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto LH, Andrade RV, Felipe MS, Motoyama AB, Pittella Silva F. SMYD2 is highly expressed in pediatric acute lymphoblastic leukemia and constitutes a bad prognostic factor. Leuk Res. 2014;38:496–502. doi: 10.1016/j.leukres.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Sarris M, Nikolaou K, Talianidis I. Context-specific regulation of cancer epigenomes by histone and transcription factor methylation. Oncogene. 2014;33:1207–1217. doi: 10.1038/onc.2013.87. [DOI] [PubMed] [Google Scholar]
- Schenk T, Chen WC, Göllner S, Howell L, Jin L, Hebestreit K, Klein HU, Popescu AC, Burnett A, Mills K, Casero RA, Jr, Marton L, Woster P, Minden MD, Dugas M, Wang JC, Dick JE, Müller-Tidow C, Petrie K, Zelent A. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar SR, Bahirvani AG, Rao VK, Bharathy N, Ow JR, Taneja R. G9a, a multipotent regulator of gene expression. Epigenetics. 2013;8:16–22. doi: 10.4161/epi.23331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Sobin LHG, Gospodarowicz M, Wittekind C.(eds) (2009International Union Against Cancer (UICC) TNM Classification of Malignant Tumors7th ed.Wiley-Blackwell: Oxford, UK [Google Scholar]
- Stender JD, Pascual G, Liu W, Kaikkonen MU, Do K, Spann NJ, Boutros M, Perrimon N, Rosenfeld MG, Glass CK. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell. 2012;48:28–38. doi: 10.1016/j.molcel.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H. Individualization of breast cancer based on histopathological features and molecular alterations. Breast cancer. 2008;15:121–132. doi: 10.1007/s12282-008-0032-5. [DOI] [PubMed] [Google Scholar]
- Ushijima T, Sasako M. Focus on gastric cancer. Cancer cell. 2004;5:121–125. doi: 10.1016/s1535-6108(04)00033-9. [DOI] [PubMed] [Google Scholar]
- Wagner T, Jung M. New lysine methyltransferase drug targets in cancer. Nat Biotechnol. 2012;30:622–623. doi: 10.1038/nbt.2300. [DOI] [PubMed] [Google Scholar]
- Willmann D, Lim S, Wetzel S, Metzger E, Jandausch A, Wilk W, Jung M, Forne I, Imhof A, Janzer A, Kirfel J, Waldmann H, Schüle R, Buettner R. Impairment of prostate cancer cell growth by a selective and reversible lysine-specific demethylase 1 inhibitor. Int J Cancer. 2012;131:2704–2709. doi: 10.1002/ijc.27555. [DOI] [PubMed] [Google Scholar]
- Wu J, Cheung T, Grande C, Ferguson AD, Zhu X, Theriault K, Code E, Birr C, Keen N, Chen H. Biochemical characterization of human SET and MYND domain-containing protein 2 methyltransferase. Biochemistry. 2011;50:6488–6497. doi: 10.1021/bi200725p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.