Abstract
Background:
Telomeres are TTAGGG tandem repeats capping chromosomal ends and partially controlled by the telomerase enzyme. The EGFR pathway putatively regulates telomerase function, prompting an investigation of telomere length (TL) and its association with anti-epidermal growth factor receptor (EGFR) therapy in metastatic colorectal cancer (mCRC).
Methods:
Colorectal cancer cell lines were treated with multiple drugs and sensitivity determined. Clinical information was gathered from 75 patients who had received anti-EGFR drugs. Telomere length was measured using a validated qRT–PCR technique.
Results:
In CRC cell lines, TL independently predicted cetuximab sensitivity. Cells with shorter TL had growth inhibition of 18.6±3.41% as compared with 41.39±8.58% in longer TL (P=0.02). These in vitro findings were validated clinically, in a robust multivariate model. Among patients with KRas WT tumours, those with longer TL had a superior median progression-free survival (PFS) of 24.9 weeks than those with shorter TL; median 11.1 weeks, HR 0.31; P=0.048.
Conclusion:
Telomere length could be a potential unique biomarker predictive of clinical benefit (PFS) of mCRC patients treated with anti-EGFR therapy. This is the novel demonstration of a complex hitherto undescribed interaction, placing anti-EGFR therapy, EGFR pathway, and the telomerase complex within a clinical context.
Keywords: epidermal growth factor receptor (EGFR), telomere length, cetuximab, panitumumab, metastatic colorectal cancer, predictive biomarker
Colorectal cancer (CRC) will account for over 50 000 deaths in United States in 2014 (Siegel et al, 2014). The median survival of patients with metastatic CRC (mCRC) has improved to 24–30 months (Goldberg et al, 2007) mainly because of the availability of newer therapeutic option of epidermal growth factor receptor (EGFR)-targeted monoclonal antibodies (mAb) such as cetuximab and panitumumab (Tournigand et al, 2004; Jonker et al, 2007). Incorporation of KRas mutation as a negative predictive biomarker for anti-EGFR therapy in mCRC was a landmark advancement in the pursuit of personalised care for patients with cancer (Lièvre et al, 2006; Di Fiore et al, 2007; De Roock et al, 2008; Karapetis et al, 2008). Among the KRas wild-type (WT) patients, multiple studies evaluating EGFR-based therapy have documented highly variable response rates ranging from 17% to 60% that may be because of the presence of other predictive variables that determine responsiveness to EGFR antibodies. Expanding the repertoire of Ras mutations has further narrowed the spectrum with better identification of patients (Douillard et al, 2013), but there still remains ample room for improvement.
Telomeres are DNA sequences at the ends of linear chromosomes consisting of hundreds to thousands of TTAGGG tandem repeats in the leading strand (Moyzis et al, 1988). The continued proliferation of a cell that has escaped the protective cellular mechanism can lead to extremely short telomeres. Telomerase is an enzyme that is specifically expressed in immortal cells, such as stem cells, cancer cells, and in germ tissues, where it compensates for telomere shortening during DNA replication and stabilises telomere length (TL). It gives cancerous cells the potential for unlimited cell division, and has been implicated in immortalisation and carcinogenesis (Shay and Bacchetti, 1997; Rampazzo et al, 2010).
The EGFR pathway is stimulated upon ligand binding at the receptor level and subsequent canonical transmission of signals to the nucleus through predominantly two parallel pathways: Ras/Braf/MEK/ERK (MAP kinase pathway) and PI3K/AKT/PTEN/mTOR pathway (Jorissen et al, 2003; Quesnelle et al, 2007). The role of EGFR pathway in regulating telomerase complex is increasingly being recognised. Antisense oligonucleotides directed against EGFR have been shown to inhibit telomerase activity up to 54-fold in U87MG glioblastoma cell lines and reduce the TL (Tian et al, 2002). In human uterine cervix epithelial cells, when dissociated into distinctive cellular subsets, EGFR expression and telomerase activity were found to coexist (Kunimura et al, 1998). Furthermore, the HIF1-α and PI3K/AKT pathway are downstream targets of transcriptional regulation of telomerase reverse transcriptase (hTERT) activated through EGFR overexpression (Heeg et al, 2011). Moreover, in a partial hepatectomy mouse model, regenerating hepatocytes showed upregulation of telomerase activity, and preoperative treatment with EGF increased this activity whereas MEK inhibitors repressed it (Inui et al, 2002).
Collectively, based upon these data, we hypothesised that TL may be a biomarker predictive of benefit from anti-EGFR therapy in management of mCRC. We utilised an array of CRC cell lines, determined TL and sensitivity to cetuximab, and identified TL as a biomarker of sensitivity. These data were validated in a robust multivariate model in patients treated with the anti-EGFR monoclonal antibodies, namely cetuximab and panitumumab.
Materials and Methods
Cell lines
Response to cetuximab was determined in 21 human-derived CRC cell lines using the MTT assay (Jhawer et al, 2008). We next treated 18 CRC cell lines with 8 other drugs that may or definitely have a role in the clinical management of mCRC at various concentrations to achieve a 50% growth inhibition (GI50). The drugs were oxaliplatin (DNA-damaging platinum agent, Arango et al, 2004), 5-FU (inhibitor of thymidylate synthase), camptothecin (topoisomerase 1 inhibitor, Mariadason et al, 2003), curcumin (chemoprevention), butyrate (HDAC inhibitor, possible therapeutic), sulindac (chemoprevention), trichostatin (possible therapeutic), and aspirin (chemoprevention, possibly lower recurrence). Genomic DNA was isolated from these cells using the DNeasy blood and tissue kit (no. 69504; Qiagen, Germantown, MD, USA).
Patient selection and ethical practices
We identified 75 consecutive patients with mCRC who received EGFR inhibitors at Montefiore Medical Center from 2004 to 2009 and obtained DNA from 118 archived tumour samples: 66 primary, 52 metastatic, and 29 matched pairs (Sood et al, 2012). Clinical and demographic data were obtained from the MMC Tumour Registry and retrospective chart review was performed using Clinical Information System (Centricity Enterprise, version 6.6.3, Piscataway, NJ, USA) and Electronic Patient Folder (McKesson Information Solutions LLC, Alpharetta, GA, USA). All patient information was de-identified and stored as a Microsoft 2010 Excel file that was encrypted and password protected. An institutional review board approved the study, including a waiver of informed consent.
Telomere length and mutation analysis
Genomic DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tissue blocks using the Qiamp FFPE DNA kit (Qiagen, Valencia, CA, USA) and stored at −80 °C (Sood et al, 2012). The average TL in tumour samples was estimated using a quantitative PCR (qPCR) method (Supplementary Table 1) (Cawthon, 2009). This approach provides relative average TL in genomic DNA by determining the telomere (T) repeat copy number to single-copy (S) β-globin (HGB) gene copy number, or T/S ratio in experimental samples relative to a reference sample. One T/S ratio unit is equivalent to a mean TL of 4270 base pairs (bp) (Atzmon et al, 2010). Samples were also analysed for mutations in the KRas, BRaf, and PI3K oncogenes.
Statistics
Descriptive statistics were used for patient characteristics. Overall survival (OS) was the period from time of diagnosis of metastatic disease to death. Progression-free survival (PFS) was the period from initiation of anti-EGFR therapy to the date of disease progression, death, or censoring. The primary hypothesis being tested was that the TL determines clinical outcome when patients are treated with anti-EGFR inhibitors. Kaplan–Meier method was used to report PFS curves, and log-rank test was used to compare them. A Cox proportional-hazard model was used to assess PFS differences between patients with different TLs. Modelling included the examination of univariate associations between PFS and clinically relevant variables. A priori decision was made to force the variables gender, age at diagnosis, and race into the model. Continuous variables were checked for linearity assumption and, when violated, were categorised using clinically meaningful cut points. One final model was selected that included the variable of interest ‘telomere length dichotomised at the median', gender, age as ≤/> 60 years at diagnosis, and other variables in which P-values were <0.20 in the multivariate analysis. The final model was checked for interactions and confounding. Diagnostics of the final model was assessed by using graphical methods (log–log plots and observed vs expected plots) and the goodness-of-fit test. All analyses were done using STATA 11.2 software (StataCorp LP, College Station, TX, USA).
Results
Telomere length predicts response to cetuximab, a monoclonal antibody to EGFR but not to non-EGFR-targeting drugs in vitro
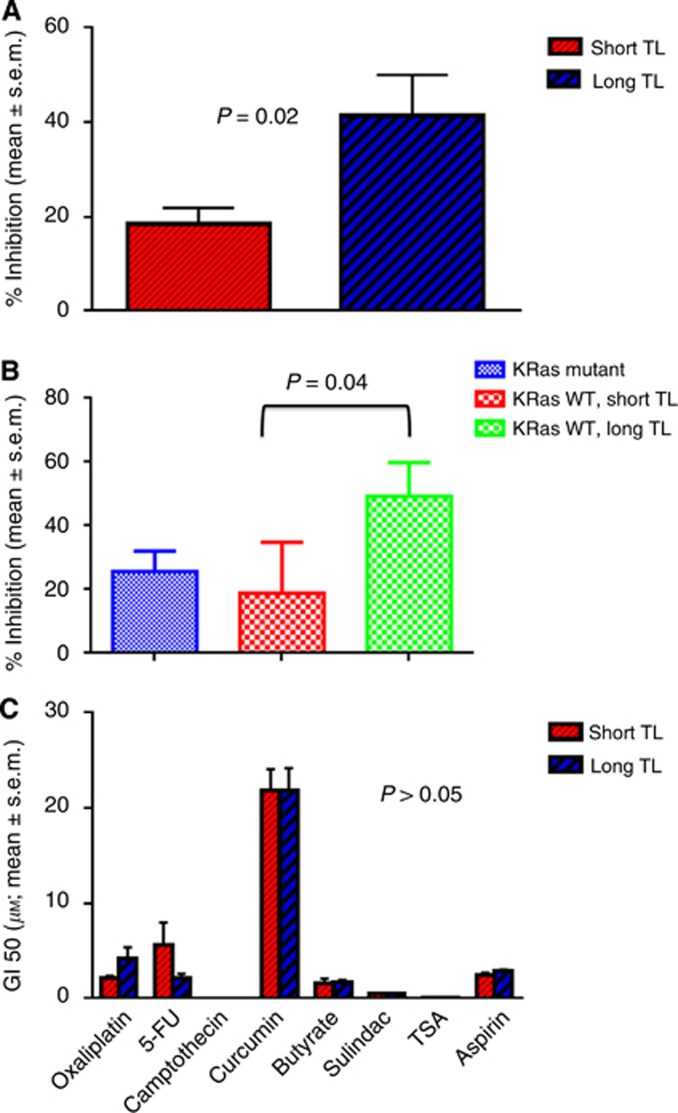
We measured TL in 21 CRC cell lines. When the TL was split at the median, it was independently predictive of cetuximab sensitivity. In cell lines with a shorter TL, cetuximab inhibited cell growth by 18.6%±3.41% (mean±standard error of the mean (s.e.m.)) compared with 41.39±8.58% (mean±s.e.m.) in cell lines with longer TL (P=0.02; Figure 1A). In keeping with the conclusive literature that anti-EGFR therapy does not benefit KRas-mutant tumours, we narrowed the findings in the cell lines that were WT for KRas (Figure 1B). Similar to above, in the cells with a shorter TL, cetuximab inhibited cell growth by 18.96±7.08% compared with 49.2%±10.6% (mean±s.e.m.) in cell lines with longer TL (P=0.04; (Figure 1B). As a comparison, the mean growth inhibition in the KRas-mutant cells was 25.78±6.25% (Figure 1B).
Figure 1.
(A) Sensitivity of CRC cell lines to cetuximab based on TL independent of KRas mutation status. Exponentially growing human-derived CRC cells were treated with cetuximab at 20 μM for 72 h, and cell viability determined. The cell lines with short TL were less sensitive than cell lines with long TL (P=0.02). (B) Sensitivity of CRC cell lines to cetuximab based on TL in KRas WT cell lines. Sensitivity to cetuximab was determined specifically in the KRas WT cell lines. The cell lines with short TL were less sensitive than those with long TL (P=0.04). (C) Sensitivity of CRC cell lines to chemotherapy/preventive agents based on TL. Cell lines were treated with 8 other drugs and GI50 determined. We failed to find any association of the response to these drugs with TL with all P-values of >0.05.
In order to determine the specificity with the EGFR pathway, we analysed TL in 18 cell lines with response data to 8 drugs that are used or have a possible role in the management of CRC (all part of the 21 cell line panel). We failed to find any association of the response to these drugs with TL with all P-values of >0.05 (Figure 1C).
Telomere length in patients' tumour samples is a novel biomarker of clinical outcome following cetuximab/panitumumab treatment, but not to oxaliplatin or irinotecan
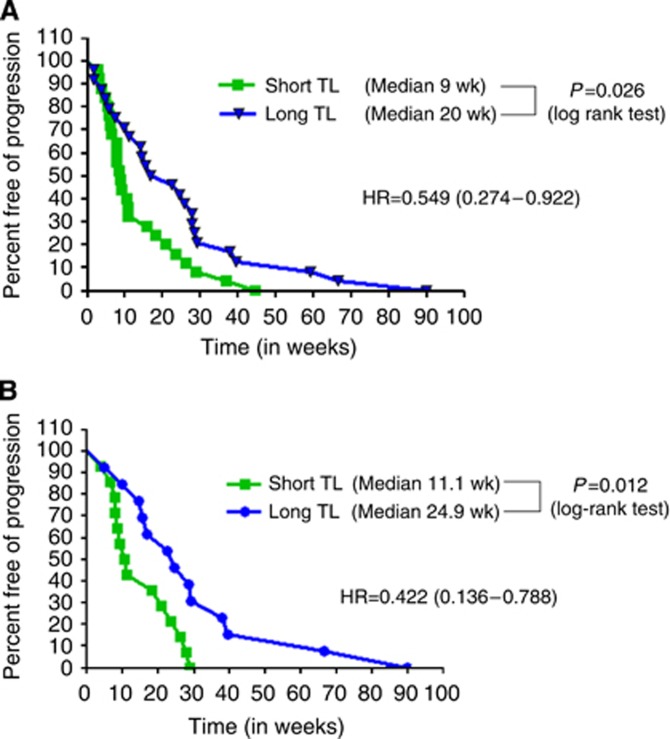
In order to validate our in vitro findings, we next proceeded to validate our findings in vivo in human samples from patients. The analysis of all 75 patients, irrespective of KRas status, demonstrated a statistical significant difference in the PFS to anti-EGFR therapy, with a hazard ratio (HR) of 0.549 (95% CI 0.274–0.922); P=0.026. The median PFS for patients with TL<median was 9 weeks vs 20 weeks for those patients with the TL>median (Figure 2A).
Figure 2.
(A) Progression-free survival with anti-EGFR therapy based on TL independent of KRas status. Data on TL and clinical outcome in 75 patients treated with anti-EGFR therapy, independent of KRas status. The patients with longer TL had a superior PFS than patients with shorter TL (HR 0.549, P=0.026). (B) Progression-free survival with anti-EGFR therapy in KRas WT patients based on TL. The analysis was narrowed down to the patients who were WT for KRas. Again, patients with longer TL had a superior PFS than patients with shorter TL, but with a better HR of 0.31 (P=0.048).
Consistent with the general literature, 32 (42.67%) patients had a KRas mutation (Table 1). We then evaluated the data specifically in the KRas WT population. In univariate analysis, patients with TL>median had a median PFS of 24.9 weeks, whereas those with TL<median had a median PFS of 11.1 weeks with a HR of 0.422 (95% CI 0.136–0.788; P=0.012; log-rank test; Figure 2B). The median PFS of patients with KRas mutation was 7.85 weeks (Supplementary Figure 1).
Table 1. Patient characteristics.
| N=75 | |
|---|---|
| Age: median (range) |
60 (34–93) |
|
Gender % (N) | |
| Male | 29.3 (22) |
| Female |
70.7 (53) |
|
Race % (N) | |
| White | 22.7 (17) |
| Hispanic | 32.0 (24) |
| Black | 41.3 (31) |
| Other |
4.0 (3) |
|
Diagnosis % (N) | |
| Stage IV | 46.7 (35) |
| Recurrence |
53.3 (40) |
|
Site of primary % (N) | |
| Colon | 73.3 (55) |
| Rectum |
26.7 (20) |
|
Sites of disease % (N) | |
| Liver only | 24.0 (18) |
| Other sites |
76.0 (57) |
| Number of metastatic sites: median (range) |
3 (1–6) |
|
Differentiation % (N) | |
| Well | 16 (12) |
| Moderately | 54.7 (41) |
| Poorly | 20.0 (15) |
| Unknown |
9.3 (7) |
| CEA at diagnosis of stage IV, median (range) |
24.5 (0.5–5500) |
|
Number of chemo lines % (N) | |
| 1 | 20.0 (15) |
| 2 | 40.0 (30) |
| 3 | 33.3 (25) |
| 4 | 5.3 (4) |
| 5 |
1.3 (1) |
|
KRas status % (no.) | |
| Mutant | 42.67 (32) |
| WT | 57.33 (43) |
Abbreviations: CEA=carcinoembryonic antigen; WT=wild type.
The table depicts clinical characteristics of the 75 patients who were included in this study.
We then proceeded (Table 2A and B) to evaluate the data in a multivariate model to ensure clinical relevance, as indicated in the Materials and Methods section, and forced age, gender, and race to ensure that all clinically important prognostic variables were factored. The HR for TL in this multivariate model was further improved to 0.31 (95% CI 0.10–0.98, P=0.048). Therefore, patients with TL above the median had a longer PFS with a 69% reduced risk of progression while on EGFR inhibitors when compared with those with TL lower than the median, after adjusting for gender, race, and age at diagnosis (Figure 2B).
Table 2A. Univariate modelling for PFS in KRas WT patients.
| HR (95% CI) | P-value | |
|---|---|---|
| Age: median |
1.32 (0.58–2.99) |
0.5 |
| Gender |
0.84 (0.35–2.05) |
0.71 |
| Race | 1.56 (0.66–3.71) | 0.36 |
| White vs minorities (Black, Hispanic, Other) |
|
|
| Colon vs rectum |
0.89 (0.41–2.00) |
0.79 |
| Stage IV vs recurrence |
1.51 (0.69–3.30) |
0.3 |
| Mets sites (1 vs 2 or more) |
1.66 (0.73–3.78) |
0.22 |
| Diff (well vs mod/poor) |
0.80 (0.23–2.75) |
0.73 |
| Chemo lines (1 vs 2 or more) |
1.54 (0.36–6.65) |
0.57 |
| CEA at Dx St IV (dichot at median 26) |
0.58 (0.26–1.28) |
0.17 |
| Telomere length | 0.42 (0.14–0.79) | 0.012 |
Abbreviations: CEA at Dx St IV (dichot at median 26)=carcinoembryonic antigen at diagnosis of stage IV disease, and dichotomised at median of 26; CI=confidence interval; diff=differentiation; HR=hazard ratio; Mets=metastases; mod=moderate; PFS=progression-free survival; WT=wild type.
The table depicts results of univariate modelling of all important clinical characteristics that could impact the clinical outcome of patients when treated with anti-EGFR therapy. Each row depicts the characteristic, followed by the HR and the associated P-value for each variable.
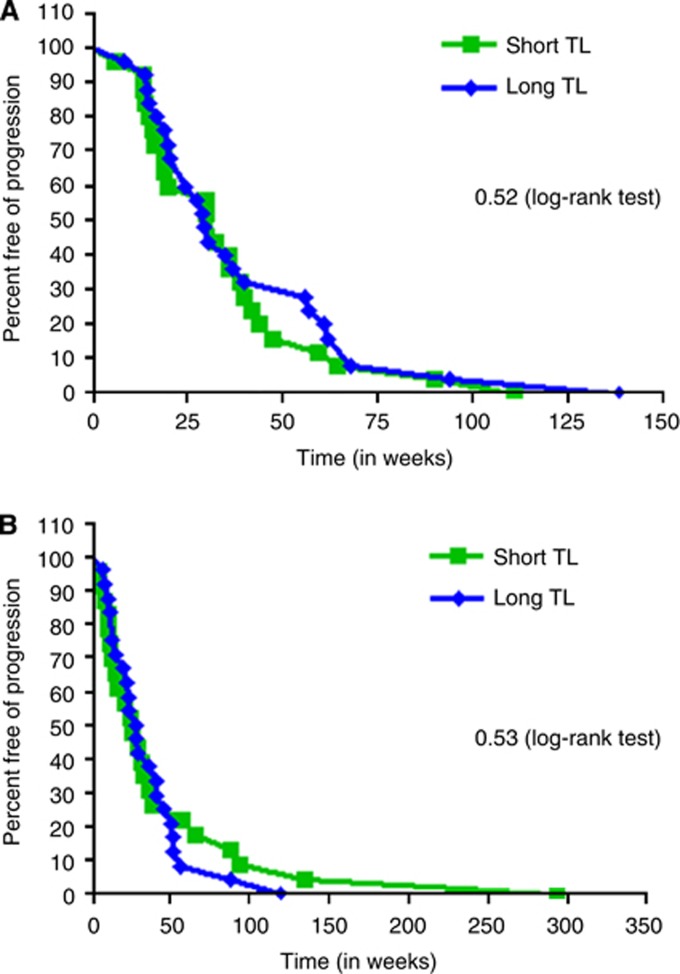
Of the 75 patients, 50 patients had received oxaliplatin in the metastatic setting. The median PFS for patients with shorter TL was 32.57 weeks and for longer TL was 28.85 weeks (HR 1.19, P=0.52; Figure 3A). Next, of the 47 patients who received irinotecan, the median PFS for the patients with short TL was 25.86 weeks and with long TL was 28.79 weeks (HR 0.83, P=0.53; Figure 3B). Therefore, consistent with our in vitro data, TL had no association with clinical outcome when patients were treated with oxaliplatin- or irinotecan-based regimen.
Figure 3.
(A) Progression-free survival with oxaliplatin based on TL. The clinical outcome to oxaliplatin was determined in relation to TL. Similar to preclinical data, there was no association of TL with oxaliplatin. (B) Progression-free survival with irinotecan based on TL. When the data for irinotecan were considered, there was no association between TL and irinotecan.
Association of TL with stage and age
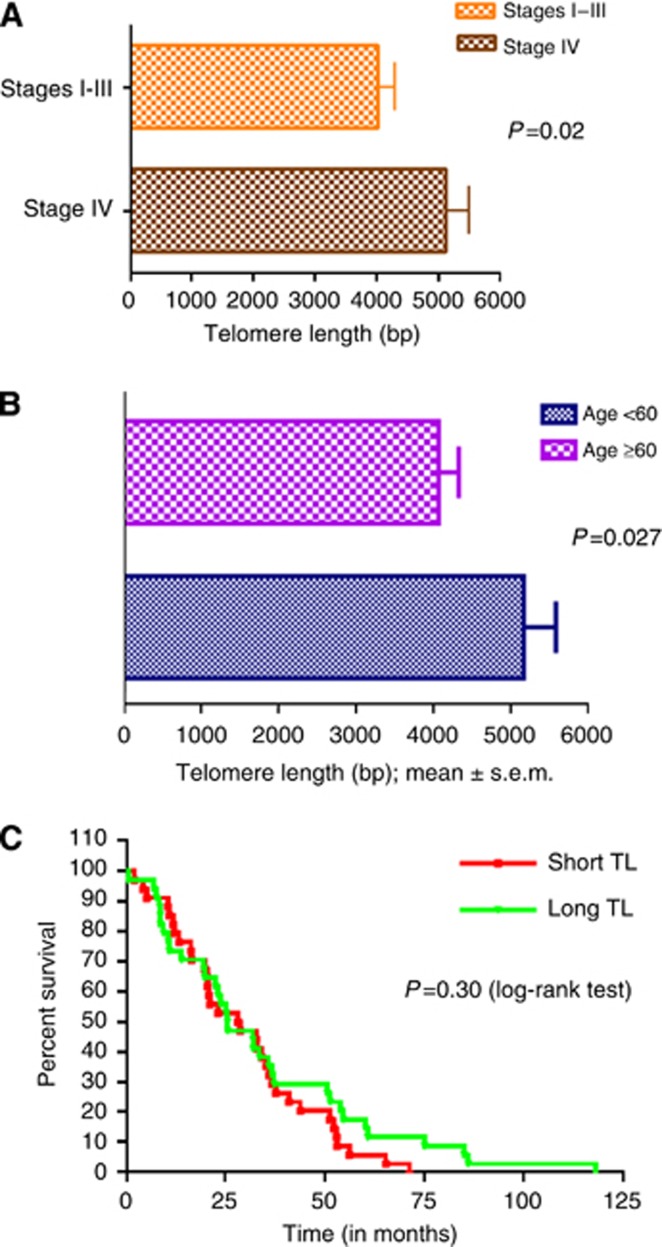
When patients were separated based on stage at presentation, those who had metastatic disease had significantly longer TL (mean TL 5128±389 bp) than those with stages 1–3 (mean TL 4037±264 bp) with P=0.02 (Figure 4A). However, we did not find any difference in OS from time of diagnosis of metastatic disease in both groups. We then studied TL as a function of age at diagnosis in a dichotomous manner based on age 60 years. The mean TL of the younger patients (5165±412 bp) was longer than the older patients (4056±264 bp; P=0.027; Figure 4B).
Figure 4.
(A) Telomere length by stage at diagnosis. Patients with metastatic (stage IV) disease at presentation had longer TL than those who had localised disease or stages 1–3 with P=0.02. (B) Telomere length by age at diagnosis. The data were analysed in a dichotomous manner based on age of 60 years. The mean TL of the younger patients was longer than the older patients (P=0.027). (C) Overall survival of patients based on TL. The OS for patients was determined as the period from time of diagnosis of metastatic disease to death. There was no difference in OS in those patients with TL>median compared with those with TL<median, with a HR of 1.11 (P=0.30).
Telomere length has no association with overall survival or oncogenic mutations
Patients with a TL<median had a median OS of 28.57 months, similar to those with TL>median whose median OS was 25.68 months (P=0.30; Figure 4C). Therefore, in our sample, TL is unlikely to be a prognostic marker in patients with mCRC.
Telomere length was analysed for all the patient samples in relation to mutation status, specifically KRas and PIK3CA. We did not detect any difference in the mean TL in samples with a KRas/PIK3CA mutation or WT status.
Discussion
In this study, we report several novel clinically relevant observations. Most importantly, for the first time, we report that TL has the potential to be a novel and unique predictive biomarker of clinical outcome (PFS) to anti-EGFR therapy in patients with KRas WT metastatic colorectal cancer. The role of KRas as a biomarker of exclusion for therapy with the anti-EGFR monoclonal antibodies is validated and well established in routine clinical practice (Lièvre et al, 2006; Douillard et al, 2013). Expanding the repertoire of Ras mutations to include additional KRas mutations (Douillard et al, 2013), and NRas and BRaf mutations as a screening tool, can further narrow down the spectrum of patients who will benefit from anti-EGFR-based therapy to ∼50%, requiring further improvements in biomarker discovery and validation. Inclusion of TL into this can further focus on the group most likely to benefit. There is extensive literature associating the EGFR pathway with the TL/telomerase complex (Kunimura et al, 1998; Inui et al, 2002; Tian et al, 2002; Heeg et al, 2011). Our finding that longer TL portends for better outcome is therefore not an incidental finding, but indeed has biological plausibility. Further work should clearly focus on the ligand activated EGFR pathway, and the downstream effectors, the MAPK and the PI3K pathway, and study their role in telomerase activity and subsequent effect on TL.
Second, the PFS for both irinotecan- and oxaliplatin-based therapies was essentially overlapping when separated into two cohorts based on TL. This further entrenches the unique interaction between TL and the EGFR pathway. The demonstration of similar observations in the cell line model and in our patient samples is indeed a major strength of our unique finding. Moreover, TL does not appear to be a prognostic biomarker but, rather, simply a predictive one. Third, our data show for the first time that malignant colonic epithelium parallels the age-related decline in TL that is widely observed in nonmalignant cells such as peripheral blood leukocytes as well as colonocytes (O'Sullivan et al, 2006). In contrast, two prior publications that noted the age-related decline in TL in normal mucosa failed to replicate that in cancerous cells (Gertler et al, 2004; Rampazzo et al, 2010). Fourth, we observe that when the patients were categorised as either localised disease (stages 1–3), or stage 4 at diagnosis, there was a statistically significant difference in TL. This finding is in agreement with one prior report (Gertler et al, 2004) and in contrast to one report (Rampazzo et al, 2010). Overall, cancer tissue has lower TL than normal mucosa, and there appears to be a positive correlation between TL and hTERT expression (Rampazzo et al, 2010). Therefore, the enzyme telomerase, which is constitutively expressed in tumour cells, is unable to increase the TL in tumour cells beyond the adjoining normal mucosa.
This study has certain perceived limitations. The use of archived FFPE tissue has limited our study to analysis of TL only (from DNA), thereby preventing us from analysing telomerase activity. However, although higher telomerase activity leads to longer TL, the TL is also controlled by the mechanism of alternate lengthening of telomeres (ALT) (Reddel, 2003), alternate splicing of the hTERT transcripts (Yi et al, 2000), and post-transcriptional changes of hTERT mRNA. Furthermore, in our in vitro analysis, we did not find any correlation between telomerase activity and TL in the CRC cell lines (data not shown), and this is encouraging for further investigations but justifies rationale to choose TL as the appropriate parameter.
Although the number of patients initially may appear to be low, the fact that our findings are based on a valid scientific biologically plausible hypothesis, strong in vitro data, and multivariate validation in a clinical patient cohort should not be overlooked. It is also critical to note that the earliest paper identifying KRas mutation as biomarker of resistance to anti-EGFR therapy had only 30 patients (Lièvre et al, 2006). This initial report prompted further studies leading to multiple publications in larger data sets validating this finding and firmly entrenching KRas mutation as a marker of resistance to anti-EGFR therapy.
In summary, we shed new insights into the complex role of the TL/telomerase system, and the EGFR/MAPK/PI3K pathway within a clinical context. The outcome of patients treated with anti-EGFR therapy seems to be dependent on tumour TL of individual patient. Our data can be used to incite large-scale analysis of TL from large registration studies and cooperative groups with access to patient's DNA and clinical data. This can continue to unravel the mystery of the signalling pathways and the cell survival mechanisms that malignant tissues adapt as they progress.
Table 2B. Multivariate modelling for PFS in KRas WT patients.
| HR (95% CI) | P-value | |
|---|---|---|
| Age: median |
0.68 (0.23–2.0) |
0.49 |
| Gender |
1.35 (0.47–3.83) |
0.57 |
| Race | 1.20 (0.44–3.27) | 0.72 |
| White vs minorities (Black, Hispanic, Other) |
|
|
| Telomere length | 0.31 (0.10–0.98) | 0.048 |
Abbreviations: CI=confidence interval; HR=hazard ratio; PFS=progression-free survival; WT=wild type.
Results of multivariate modelling that included all univariate variables with P<0.20, and with forcing of age, gender, and race. The TL was significant for PFS in this multivariate model with a HR of 0.32 with P=0.048.
Acknowledgments
S Goel is supported by a K-12 award from the National Cancer Institute of the National Institutes of Health 1K12CA132783–01A1 and an Advanced Clinical Research Award (ACRA) in colon cancer by the ASCO (now Conquer) Cancer Foundation.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Arango D, Wilson AJ, Shi Q, Corner GA, Arañes MJ, Nicholas C, Lesser M, Mariadason JM, Augenlicht LH. Molecular mechanisms of action and prediction of response to oxaliplatin in colorectal cancer cells. Br J Cancer. 2004;91 (11:1931–1946. doi: 10.1038/sj.bjc.6602215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzmon G, Cho M, Cawthon RM, Budagov T, Katz M, Yang X, Siegel G, Bergman A, Huffman DM, Schechter CB, Wright WE, Shay JW, Barzilai N, Govindaraju DR, Suh Y. Evolution in health and medicine Sackler colloquium: Genetic variation in human telomerase is associated with telomere length in Ashkenazi centenarians. Proc Natl Acad Sci USA. 2010;107 (Suppl 1:1710–1717. doi: 10.1073/pnas.0906191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37 (3:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roock W, Piessevaux H, De Schutter J, Janssens M, De Hertogh G, Personeni N, Biesmans B, Van Laethem JL, Peeters M, Humblet Y, Van Cutsem E, Tejpar S. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 2008;19 (3:508–515. doi: 10.1093/annonc/mdm496. [DOI] [PubMed] [Google Scholar]
- Di Fiore F, Blanchard F, Charbonnier F, Le Pessot F, Lamy A, Galais MP, Bastit L, Killian A, Sesboüé R, Tuech JJ, Queuniet AM, Paillot B, Sabourin JC, Michot F, Michel P, Frebourg T. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. Br J Cancer. 2007;96 (8:1166–1169. doi: 10.1038/sj.bjc.6603685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocákova I, Ruff P, Błasińska-Morawiec M, Šmakal M, Canon JL, Rother M, Williams R, Rong A, Wiezorek J, Sidhu R, Patterson SD. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369 (11:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- Gertler R, Rosenberg R, Stricker D, Friederichs J, Hoos A, Werner M, Ulm K, Holzmann B, Nekarda H, Siewert JR. Telomere length and human telomerase reverse transcriptase expression as markers for progression and prognosis of colorectal carcinoma. J Clin Oncol. 2004;22 (10:1807–1814. doi: 10.1200/JCO.2004.09.160. [DOI] [PubMed] [Google Scholar]
- Goldberg RM, Rothenberg ML, Van Cutsem E, Benson AB, 3rd, Blanke CD, Diasio RB, Grothey A, Lenz HJ, Meropol NJ, Ramanathan RK, Becerra CH, Wickham R, Armstrong D, Viele C. The continuum of care: a paradigm for the management of metastatic colorectal cancer. Oncologist. 2007;12 (1:38–50. doi: 10.1634/theoncologist.12-1-38. [DOI] [PubMed] [Google Scholar]
- Heeg S, Hirt N, Queisser A, Schmieg H, Thaler M, Kunert H, Quante M, Goessel G, von Werder A, Harder J, Beijersbergen R, Blum HE, Nakagawa H, Opitz OG. EGFR overexpression induces activation of telomerase via PI3K/AKT-mediated phosphorylation and transcriptional regulation through Hif1-alpha in a cellular model of oral-esophageal carcinogenesis. Cancer Sci. 2011;102 (2:351–360. doi: 10.1111/j.1349-7006.2010.01796.x. [DOI] [PubMed] [Google Scholar]
- Inui T, Shinomiya N, Fukasawa M, Kobayashi M, Kuranaga N, Ohkura S, Seki S. Growth-related signaling regulates activation of telomerase in regenerating hepatocytes. Exp Cell Res. 2002;273 (2:147–156. doi: 10.1006/excr.2001.5446. [DOI] [PubMed] [Google Scholar]
- Jhawer M, Goel S, Wilson AJ, Montagna C, Ling YH, Byun DS, Nasser S, Arango D, Shin J, Klampfer L, Augenlicht LH, Perez-Soler R, Mariadason JM. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res. 2008;68 (6:1953–1961. doi: 10.1158/0008-5472.CAN-07-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonker DJ, O'Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, Berry SR, Krahn M, Price T, Simes RJ, Tebbutt NC, van Hazel G, Wierzbicki R, Langer C, Moore MJ. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357 (20:2040–2048. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284 (1:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359 (17:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- Kunimura C, Kikuchi K, Ahmed N, Shimizu A, Yasumoto S. Telomerase activity in a specific cell subset co-expressing integrinbeta1/EGFR but not p75NGFR/bcl2/integrin beta4 in normal human epithelial cells. Oncogene. 1998;17 (2:187–197. doi: 10.1038/sj.onc.1201916. [DOI] [PubMed] [Google Scholar]
- Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66 (8:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- Mariadason JM, Arango D, Shi Q, Wilson AJ, Corner GA, Nicholas C, Aranes MJ, Lesser M, Schwartz EL, Augenlicht LH. Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin. Cancer Res. 2003;63 (24:8791–8812. [PubMed] [Google Scholar]
- Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85 (18:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan J, Risques RA, Mandelson MT, Chen L, Brentnall TA, Bronner MP, Macmillan MP, Feng Z, Siebert JR, Potter JD, Rabinovitch PS. Telomere length in the colon declines with age: a relation to colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2006;15 (3:573–577. doi: 10.1158/1055-9965.EPI-05-0542. [DOI] [PubMed] [Google Scholar]
- Quesnelle KM, Boehm AL, Grandis JR. STAT-mediated EGFR signaling in cancer. J Cell Biochem. 2007;102 (2:311–319. doi: 10.1002/jcb.21475. [DOI] [PubMed] [Google Scholar]
- Rampazzo E, Bertorelle R, Serra L, Terrin L, Candiotto C, Pucciarelli S, Del Bianco P, Nitti D, De Rossi A. Relationship between telomere shortening, genetic instability, and site of tumour origin in colorectal cancers. Br J Cancer. 2010;102 (8:1300–1305. doi: 10.1038/sj.bjc.6605644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddel RR. Alternative lengthening of telomeres, telomerase, and cancer. Cancer Lett. 2003;194 (2:155–162. doi: 10.1016/s0304-3835(02)00702-4. [DOI] [PubMed] [Google Scholar]
- Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33 (5:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64 (1:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- Sood A, McClain D, Maitra R, Basu-Mallick A, Seetharam R, Kaubisch A, Rajdev L, Mariadason JM, Tanaka K, Goel S. PTEN gene expression and mutations in the PIK3CA gene as predictors of clinical benefit to anti-epidermal growth factor receptor antibody therapy in patients with KRAS wild-type metastatic colorectal cancer. Clin Colorectal Cancer. 2012;11 (2:143–150. doi: 10.1016/j.clcc.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian XX, Pang JC, Zheng J, Chen J, To SS, Ng HK. Antisense epidermal growth factor receptor RNA transfection in human glioblastoma cells down-regulates telomerase activity and telomere length. Br J Cancer. 2002;86 (8:1328–1332. doi: 10.1038/sj.bjc.6600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournigand C, André T, Achille E, Lledo G, Flesh M, Mery-Mignard D, Quinaux E, Couteau C, Buyse M, Ganem G, Landi B, Colin P, Louvet C, de Gramont A. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: a randomized GERCOR study. J Clin Oncol. 2004;22 (2:229–237. doi: 10.1200/JCO.2004.05.113. [DOI] [PubMed] [Google Scholar]
- Yi X, White DM, Aisner DL, Baur JA, Wright WE, Shay JW. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia. 2000;2 (5:433–440. doi: 10.1038/sj.neo.7900113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.