Abstract
Two glycoside hydrolase encoding genes (tagh2 and tbgh2) were identified from different Thermus species using functional screening. Based on amino acid similarities, the enzymes were predicted to belong to glycoside hydrolase (GH) family 2. Surprisingly, both enzymes (TaGH2 and TbGH2) showed twofold higher activities for the hydrolysis of nitrophenol-linked β-D-glucopyranoside than of -galactopyranoside. Specific activities of 3,966 U/mg for TaGH2 and 660 U/mg for TbGH2 were observed. In accordance, Km values for both enzymes were significantly lower when β-D-glucopyranoside was used as substrate. Furthermore, TaGH2 was able to hydrolyze cellobiose. TaGH2 and TbGH2 exhibited highest activity at 95 and 90°C at pH 6.5. Both enzymes were extremely thermostable and showed thermal activation up to 250% relative activity at temperatures of 50 and 60°C. Especially, TaGH2 displayed high tolerance toward numerous metal ions (Cu2+, Co2+, Zn2+), which are known as glycoside hydrolase inhibitors. In this study, the first thermoactive GH family 2 enzymes with β-glucosidase activity have been identified and characterized. The hydrolysis of cellobiose is a unique property of TaGH2 when compared to other enzymes of GH family 2. Our work contributes to a broader knowledge of substrate specificities in GH family 2.
Keywords: β-glucosidase, glycoside hydrolase, thermostable, Thermus antranikianii, Thermus brockianus, cellobiose, truncated domains
Introduction
Glycoside hydrolases (GHs) (EC 3.2.1.X) hydrolyze glycosidic bonds between carbohydrates or between carbohydrate and non-carbohydrate components, for instance alcohols or phenols. To date, enzyme classification is based on amino acid similarities rather than substrate specificity (Henrissat, 1991). GHs are assigned to different groups based on conserved domains, and the families are numbered from 1 to 131 that are listed in the Carbohydrate-Active Enzymes Database1 (Lombard et al., 2014). Evolutionary or structurally unrelated β-glucosidases (EC 3.2.1.21) are found in six GH families (1, 3, 5, 9, 30, and 116). The majority of identified β-glucosidases is grouped in GH family 1 (Cairns and Esen, 2010). β-Galactosidase activity can be found in different GH families (1, 2, 3, 16, 20, 35, 42, 43, 50, and 59). In contrast to GH family 1, no β-glucosidase is found in GH family 2. So far, glycoside hydrolases classified as GH2 were described to function as β-galactosidase (EC 3.2.1.23), β-mannosidase (EC 3.2.1.25), β-glucuronidase (EC 3.2.1.31), endo-β-mannosidase (EC 3.2.1.152), or as exo-β-glucosaminidase (EC 3.2.1.165). These enzymes hydrolyze their substrates through a retaining acid/base mechanism with two glutamic acid/glutamate residues involved. One residue acts as acid and base catalyst, whereas the other residue serves as nucleophile (Davies and Henrissat, 1995).
Glycoside hydrolases find a wide range of industrial applications. For instance, β-glucosidases are applied in biorefineries to reduce cellobiose-mediated product inhibition of endoglucanases. Thus, the addition of β-glucosidases leads to increased glucose concentrations during the enzymatic hydrolysis of cellulose, thereby increasing ethanol yields (Viikari et al., 2007). Furthermore, β-glucosidases and β-galactosidases are applied in the food industry to improve the aroma of juices and wine (Bhatia et al., 2002). Many industrial processes run at harsh conditions, e. g., elevated temperatures and extremes of pH. Hence, the demand for novel enzymes which function at high temperatures or acidic or alkaline pH values is growing (Antranikian and Egorova, 2007).
Bacteria belonging to the genus Thermus grow at temperatures between 53 and 86°C, and at pH values between 6.0 and 10.5 (Dworkin et al., 2006). Since the discovery of the type strain of this genus, Thermus aquaticus in 1969, numerous species have been isolated from hot environments (Brock and Freeze, 1969; Oshima and Imahori, 1974). Especially, the DNA polymerase from T. aquaticus (Taq DNA polymerase) became one of the key enzymes in molecular biology (Chien et al., 1976). Furthermore, thermostable DNA-processing enzymes such as ligases, helicases, or endonucleases have been identified (Pantazaki et al., 2002). Although Thermus spp. produce different hydrolytic enzymes like proteases or lipases, only few glycoside hydrolases have been characterized so far (Dion et al., 1999; Fridjonsson et al., 1999; Pantazaki et al., 2002; Kim et al., 2006; Nam et al., 2010; Blank et al., 2014).
In the current report, two genes coding for GHs (TaGH2 and TbGH2) were identified from Thermus antranikianii and Thermus brockianus. Conserved domains of GH2 were detected with incomplete motifs. The recombinant proteins showed highest activity toward 4-NP-β-d-glucopyranoside. The activity of TaGH2 toward cellobiose makes this enzyme unique when compared to the GH family 2.
Materials and Methods
Bacterial strains and plasmids
Thermus antranikianii and Thermus brockianus were used as potential sources for the detection of β-glucosidase-encoding genes. Both strains were obtained from the collection of bacterial strains of the Institute of Technical Microbiology, Hamburg-Harburg. The strains Escherichia coli XL1-Blue MRF’ [Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F’ proAB lacIqZΔM15 Tn10 (TetR)]] and E. coli XLOLR (Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 thi-1 recA1 gyrA96 relA1lac [F’proAB lacIqZΔM15 Tn10 (TetR)] Su-(non-suppressing) λr) were obtained from the “ZAP Express Predigested Vector Kit” (Agilent Technologies, Waldbronn, Germany), and were utilized to construct and screen phagemid libraries using the vector pBK-CMV.
Escherichia coli Nova Blue SingleTM (endA1 hsdR17(r−K12 m+K12) supE44 thi-1 recA1 gyrA96 relA1 lac F’[proA+ B+laclq ZΔM15:Tn10 (TcR)]) (Novagen/Merck, Darmstadt, Germany) and E. coli BL21 StarTM(DE3) [F− ompT hsdSB (rB-mB-) gal dcm rne131 (DE3)] (Invitrogen, Karlsruhe, Germany) were used as hosts for cloned PCR products and expression using the plasmids pJet1.2/blunt (Fermentas, St.Leon-Rot, Germany) and pQE-80L (Qiagen, Hilden, Germany).
Media and culture conditions
The Thermus strains were grown at 70°C and 200 rpm for 16 h in Thermus 162 medium (DSMZ medium 878). E. coli strains were grown in LB medium at 30–37°C and 100–180 rpm for 16 h (Sambrook et al., 2001). LB medium was supplemented with selected antibiotics (ampicillin: 100 μg/mL, kanamycin: 50 μg/mL) when cultivating bacteria harboring appropriate plasmids. For gene expression, isopropyl-β-d-thiogalactopyranoside (IPTG) in concentrations of 0.5–1.0 mM were added.
Isolation of DNA, construction and screening of gene libraries
Genomic DNA from Thermus sp. was isolated according to the “Genomic DNA Handbook” (Qiagen, Hilden, Germany), and further purified by phenol/chloroform/isoamyl alcohol-extraction (25/24/1) and ethanol precipitation. Plasmid DNA was isolated using the “GeneJETTM Plasmid Miniprep Kit” (Fermentas, St. Leon-Rot, Germany).
Isolated genomic DNA from Thermus strains was partially digested with BamHI to generate fragments of 5–10 kb. DNA fragments of the desired size were separated by agarose gel electrophoresis. To prepare an agarose gel of 1%, 1 g agarose was dissolved in 100 mL TAE buffer (40 mM Tris, 1 mM EDTA, 40 mM acetic acid, pH 8.5). The DNA was extracted from the gel by using the “GeneJet Gel Extraction Kit” (Fermentas, St. Leon-Rot, Germany). The ligation into the “ZAP Express Vector,” as well as further steps for gene library construction were carried out according to the manufacturer’s instructions.
To detect β-glucosidase-encoding genes, the gene libraries were plated on LB agar supplemented with 50 μg/mL kanamycin, and after growth over night the colonies were replicated on plates containing additionally 1 mM IPTG. The clones were overlayed with buffer (25 mM sodium acetate, 2.5 mM CaCl2 × 2 H2O, 170 mM NaCl, 1% agarose, pH 6.5) containing 2.5 mM esculin and 0.4 mM ammonium-iron(III)-citrate. After incubation at 70°C for 1–16 h, β-glucosidase positive clones were observed by the formation of a brown halo around the colonies. E. coli XLOLR pBK-CMV clones displaying β-glucosidase activity were conserved as cryostocks containing 25% glycerol.
Sequencing and sequence analysis
Plasmids were isolated (“GeneJETTM Plasmid Miniprep Kit”, Fermentas, St. Leon-Rot, Germany) from selected clones, which conferred β-glucosidase activity. To determine the DNA-sequence of integrated DNA-fragments, the plasmids were sent to Eurofins MWG Operon (Ebersberg, Germany) for sequencing.
Putative open reading frames (ORF) were detected using the program “FramePlot 4.0beta.”2 The DNA- and amino acid-sequences were compared to the database of the “National Centre for Biotechnology Information” (NCBI3) using the “Basic Local Alignment Search Tool” (BLAST) (Altschul et al., 1997). Conserved domains were predicted using “InterProScan” (EMBL-EBI4) and the Conserved Domain Database (Marchler-Bauer et al., 2013). Glycoside hydrolase family 2 signatures were retrieved from http://prosite.expasy.org/PDOC00531. For multiple sequence alignments, ClustalW2 was employed (EMBL-EBI5, 2013). Hypothetical models of protein structures, based on sequence homologies, were prepared with the program “SWISS-MODEL” (Gasteiger et al., 2003).
Cloning of genes
To amplify the β-glucosidase-encoding genes, specific primers were obtained from Eurofins MWG Operon (Ebersberg). For the amplification of the gene tagh2, the primers tagh2_f_PaeI (5′-GCATGCAGGTGGGAAAGAGCTTGGTTTTTG-3′) and tagh2_r_SalIHindIII (5′-AAGCTTGTCGACTCACCAGGCCACCCCCAGGG-3′) were used, while tbgh_f (GGATCCAGGCTAAAAAGCGCCCTTTTC) and tbgh2_r (GTCGACCTACCAAGCCTCTCCAGG) were employed to amplify the gene tbgh2.
The PCRs were carried out in a thermocycler according to the manufacturer’s instructions. A mixture of 20 μL contained 0.4 U “Phusion High-Fidelity DNA-Polymerase” (Fermentas, St. Leon-Rot, Germany), 0.2 mM dATP, dCTP, dGTP, dTTP, 0.5 μM of the forward and reverse primer, 10–300 ng template, reaction buffer, and distilled autoclaved water. The obtained PCR products were cloned into the pJet1.2/blunt vector (Fermentas, St. Leon-Rot, Germany), which was thereafter used to transform competent E. coli Nova Blue SingleTM cells. Positive transformants were identified by colony PCR. Isolated pJet1.2/blunt plasmids were double digested with PaeI/SalI or BamHI/SalI to excise the β-glucosidase-encoding genes. The purified genes were ligated into the PaeI/SalI and BamHI/SalI digested expression vector pQE-80L, which was used to transform competent E. coli BL21 StarTM(DE3) cells. Positive transformants were identified by colony PCR and conserved as cryostocks containing 25% glycerol.
Production and purification of the enzymes
E. coli BL21 StarTM(DE3) harboring the recombinant pQE-80L plasmids were cultivated in 5 mL LB medium containing 100 μg/mL ampicillin at 37°C and 160 rpm for 16 h. The preculture was subcultivated in 500 mL LB medium containing 100 μg/mL ampicillin until the culture reached an optical density of A600 nm = 0.5–0.7. To induce protein production, 0.5 M IPTG was added. The cultivation was continued for 6 h, afterwards the cells were harvested by centrifugation at 13,000 rpm for 20 min at 4°C. The cell pellet was resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8) in the ratio 5 mL buffer/1 g pellet. Cells were disrupted by French press (FrenchR Pressure Cell Press, SLM-Aminco, MD, USA). Crude extracts with protein contents of 1.5 mg/mL (TaGH2) and 1.07 mg/mL (TbGH2) were obtained by centrifugation at 13,000 rpm at 4°C for 20 min.
For heat precipitation, the crude extract was incubated at 70°C for 15 min, and subsequently centrifuged at 13,000 rpm at 4°C for 20 min.
As a second purification step, a Ni2+-nitrilic acid (Ni-NTA) affinity chromatography using a 1.5-mL Ni-NTA superflow column (Qiagen, Hilden, Germany) was conducted. About 2–5 mL of the crude extract was incubated with 1 mL of Ni-NTA-agarose (Qiagen, Hilden, Germany) at 4°C and 200 rpm for 1 h. Two washing steps were performed (50 mM NaH2PO4, 300 mM NaCl, 25 mM imidazole, pH 7.0). To elute the protein, six fractions of 500 μL elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 7.0) were added to the column. Fractions containing the protein were pooled.
In the case of TbGH2, a gel filtration using the “ÄKTATM Fast Protein Liquid Chromatography” (FPLC)-system (GE Healthcare, München, Germany) was carried out. One milliliter of the protein solution was load on a “HiLoad 16/60 Superdex 200 prep grade”-column (GE Healthcare, München, Germany) with a flow rate of 1 mL/min. The protein was eluted with a 1.5-fold column volume of 50 mM sodium phosphate buffer, pH 7.2, which contained 150 mM NaCl. Enzyme fractions were pooled.
The purified proteins were dialyzed against 20 mM citrate buffer (TaGH2) or 20 mM maleate buffer (TbGH2) and stored at 4°C.
The purity of the recombinant β-glucosidases was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, 12%) (Laemmli, 1970).
Protein concentrations were determined using the method described by Bradford (1976).
Determination of enzyme activity
Enzyme activity was determined by the released amounts of 2-nitrophenol (2-Np) and 4-nitrophenol (4-Np) from several nitrophenol-linked substrates (4-Np-β-d-glucopyranoside, 4-Np-α-d-glucopyranoside, 4-Np-β-d-glucuronid, 4-Np-β-d-galactopyranoside, 2-Np-β-d-galactopyranoside, 4-Np-α-d- galactopyranoside, 4-Np-β-d-xylopyranoside, 4-Np-β-d-manno pyranoside, 4-Np-β-d-cellobioside).
The reaction mixture of 1 mL contained 2 mM Np-substrate and 20 mM citrate buffer pH 6.5 (TaGH2), or 20 mM maleate buffer pH 6.5 (TbGH2). After preincubation of the reaction mixture for 5 min at 95°C (TaGH2) or 90°C (TbGH2), 10 μL of diluted enzyme solution were added. The hydrolysis was stopped after 10 min by adding 100 μL of 100 mM Na2CO3. The absorbance was measured at 410 nm. All measurements were determined in triplicates. About 1 U of enzyme activity was defined as the amount of enzyme needed for the release of 1 μmol 4-nitrophenol per minute. Kinetic parameters were determined according to Michaelis and Menten (1913).
Effect of pH and temperature
Relative activities against 4-Np-β-d-glucopyranoside (2 mM) were measured in the range of pH 4.0–10.0 using 20 mM Britton–Robinson buffer (Britton and Robinson, 1931). To determine the pH stability, both enzymes were preincubated in 20 mM Britton–Robinson buffer, pH 3.0–10.0 at 4°C for 24 h. To stabilize the enzymes, the concentration of the solution was adjusted to 0.1 mg/mL with BSA. After preincubation, the relative activity toward 4-Np-β-d-glucopyranoside (2 mM) was determined in 20 mM Britton–Robinson buffer pH 6.5 and 95°C (TaGH2), or in 20 mM maleate buffer at 90°C (TbGH2).
The influence of temperature was determined by measuring relative enzyme activities in the range of 10–115°C with 4-Np-β-d-glucopyranoside (2 mM) in 20 mM citrate buffer (TaGH2) or 20 mM maleate buffer (TbGH2), pH 6.5. To examine the temperature stability, both enzymes were preincubated at 50–90°C for 0–24 h in 20 mM citrate buffer (TaGH2) or 20 mM maleate buffer (TbGH2), pH 6.5. The protein concentration of the solution was adjusted to 0.1 mg/mL with BSA. After preincubation, the relative activity toward 4-Np-β-d-glucopyranoside (2 mM) was determined in 20 mM citrate buffer (TaGH2) or 20 mM maleate buffer (TbGH2), pH 6.5, at 95°C (TaGH2) and 90°C (TbGH2).
Determination of hydrolysis products
Hydrolysis products of 1% (w/v) cellobiose and lactose were examined by HPLC (Agilent Technology 1260 Infinity Quarternary LC system with 1260 ALS sampler, 1260 Quat pump and 1260 Ri detector). The enzymes were incubated with the substrates at 90°C for 1 h in 20 mM citrate buffer (TaGH2) or 20 mM maleate buffer (TbGH2), pH 6.5. After hydrolysis, 20 μL of the centrifuged and filtered solution was applied to a Hi-Plex H column (Agilent Technologies, Waldbronn, Germany). Water was used as solvent with a flowrate of 0.6 mL/min. To identify the hydrolysis products, the retention times (min) were compared to the standards cellobiose (9.433), lactose (9.789), and glucose (11.247).
Nucleotide sequence accession number
DNA sequences of tagh2 and tbgh2 were deposited in GenBank (tagh2: HG969993, tbgh2: HG969994).
Results
Identification of novel GH-encoding genes
Gene libraries were constructed from pure cultures of T. antranikianii and T. brockianus, and screened for genes coding for enzymes active toward esculin. One activity-conferring E. coli clone was detected from each library. The respective inserts of the phagemids (pBK-CMV:Ta and pBK-CMV:Tb) harbored tagh2 and tbgh2. The nucleotide sequences of tagh2 and tbgh2 were 80% identical. GC contents of 68.4 and 66.7% with GC proportions in the third codon position (GC3) of 86.3 and 82.3% were observed.
The deduced amino acid sequences TaGH2 and TbGH2 possessed theoretical isoelectric points (pI) of 6.1 (TaGH2) and 5.99 (TbGH2). Molecular masses of 79.0 kDa (TaGH2) and 78.1 kDa (TbGH2) were computed. The proteins were compared to proteins annotated in databases (GenBank). TaGH2 exhibited highest identity (98%) to a putative β-galactosidase from Thermus scotoductus (YP_004203085, Table 1, Row 3). TbGH2 showed 82% identity to a hypothetical protein from Thermus sp. (YP_005653839, Table 1, Row 4). The annotated proteins were deduced from ORFs identified in the course of whole genome sequencing projects of different Thermus species. Additionally, identities of 86% (TaGH2) and 83% (TbGH2) were obtained by comparison with a putative GH2 β-mannosidase from Thermus thermophilus (ACH89346, Table 1, Row 6). Identities to annotated proteins from Thermus sp. ranged from 83 to 98% (TaGH2) and from 80 to 83% (TbGH2). GH2 enzymes from genera other than Thermus were observed with identities of 38–42% (Caldilinea aerophila, YP_005442964 and Alicyclobacillus pohlia, WP_018133520.1; Table 1, Rows 7 and 8).
Table 1.
Identities of TaGH2 and TbGH2 to other GH2-proteins.
| 1. | 2. | 3. | 4. | 5. | 6. | 7. | 8. | 9. | |
|---|---|---|---|---|---|---|---|---|---|
| 1. T. antranikianii TaGH2 | 100% | ||||||||
| 2. T. brockianus TbGH2 | 80% (100) | 100% | |||||||
| 3. T. scotoductus β-galactosidase (YP_004203085.1) | 98% (100) | 80% (100) | 100% | ||||||
| 4. Thermus sp. hyp. Protein (YP_005653839.1) | 83% (100) | colorno882% (100) | 83% (100) | 100% | |||||
| 5. T. igniterrae β-galactosidase (WP_018110934.1) | 82% (100) | 81% (100) | 81% (100) | 83% (100) | 100% | ||||
| 6. T. thermophilus put. β-mannosidase (ACH89346.1) | 86% (92) | 83% (93) | 85% (99) | 84% (99) | 86% (87) | 100% | |||
| 7. A. pohliae put. Protein (WP_018133520.1) | 38% (97) | 41% (83) | 38% (90) | 38% (95) | 38% (94) | 41% (92) | 100% | ||
| 8. C. aerophila put. GH (YP_005442964.1) | 41% (76) | 42% (82) | 41% (78) | 41% (84) | 42% (83) | 41% (96) | 45% (99) | 100% | |
| 9. E. coli LacZ (1HN1_D) | 27% (35) | 29% (32) | 27% (37) | 27% (32) | 29% (46) | 28% (46) | 23% (61) | 23% (58) | 100% |
Enzymes from Thermus spp. are highlighted in gray. Query-coverage values are indicated in % in brackets.
Numbers at the top are listed in the left column.
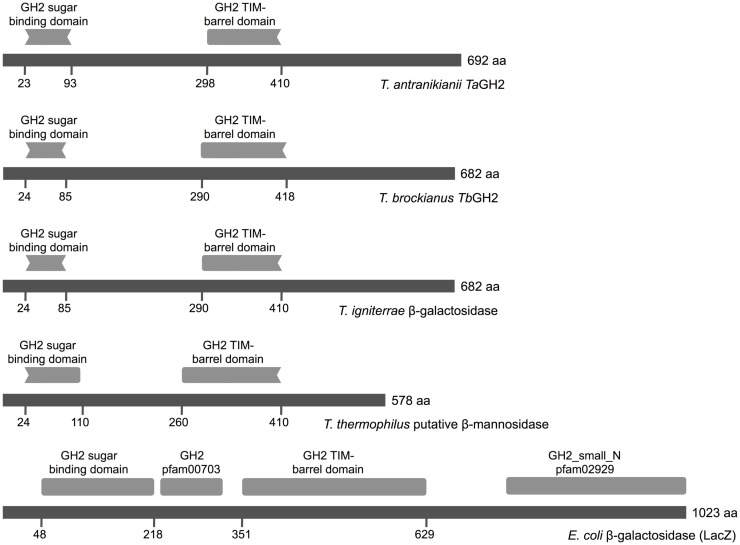
Notably, the Conserved Domain Database indicated two incomplete GH2 domains for both enzymes (Figure 1). A partial GH2 sugar-binding domain (pfam02837) in the protein sequence of TaGH2 (aa 23–93) and TbGH2 (aa 24–85), and an incomplete GH2 TIM-barrel domain (pfam02836) within TaGH2 (aa 298–410) and TbGH2 (aa 290–418) were predicted. Comparable domains were described for E. coli β-galactosidase LacZ in a complete form with low identities to the TIM-barrel domain (≤ 30%) and to the sugar-binding domain (≤ 20%) of TaGH2 and TbGH2 (Figure 1). Within LacZ, the consensus patterns N-X-[LIVMFYWD]-R-[STACN](2)-H-Y-P-X(4)-[LIVMFYWS](2)-x(3)-[DN]-X(2)-G-[LIVMFYW](4) and [DENQLF]-[KRVW]-N-[HRY]-[STAPV]-[SAC]-[LIVMFS]-[LIVMFSA]-[LIVMFS]-W-[GSV]-X(2,3)-N-E, which were described as conserved signatures of GH2, were identified6 (Figure 2). Principally, these conserved signatures were also detected as variations within TaGH2 and TbGH2.
Figure 1.
Conserved domains of GH2 family enzymes. The amino acid sequences of TaGH2 (T. antranikianii), TbGH2 (T. brockianus), β-galactosidase (T. igniterrae, WP_018110934.1), putative β-mannosidase (T. thermophilus, ACH89346), and LacZ (E. coli, 1HN1_D) are depicted in dark gray with homologous predicted sugar-binding domain (pfam02837) and TIM-barrel domain (pfam02836, Conserved Domain Database) in light gray.
Figure 2.
Amino acid signatures of GH2 family enzymes. The amino acid sequences of TaGH2 and TbGH2 are shown in black and the sequence of LacZ (E. coli, 1HN1_D) and the consensus pattern of the two characteristic signatures of GH2 are depicted in gray with possible residue substitutions indicated below (http://prosite.expasy.org/PDOC00531). Corresponding amino acids are shown in bold characters and conformity within all sequences is framed.
Expression of tagh2 and tbgh2 and protein purification
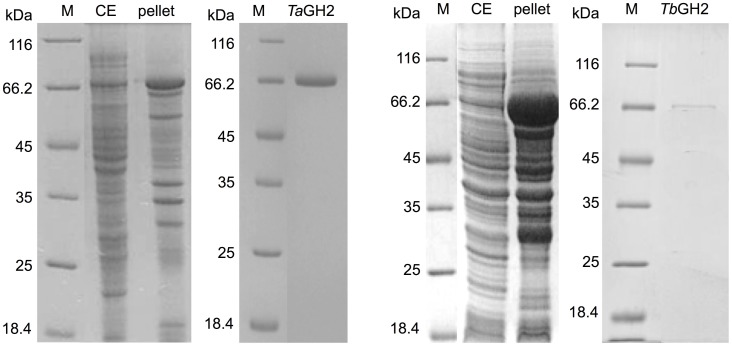
The genes tagh2 and tbgh2 were amplified and expressed in E. coli BL21 StarTM(DE3) using pQE-80L (Figure 3). A high proportion of TbGH2 was produced in insoluble form. TaGH2 and TbGH2 were purified from soluble crude extracts. After heat precipitation and Ni-NTA affinity chromatography, TaGH2 was homogenous with a yield of 17.3%, whereas TbGH2 was subsequently subjected to size exclusion chromatography with a yield of 6.7%. The calculated molecular masses of 79.0 kDa (TaGH2) and 78.1 kDa (TbGH2) were confirmed by SDS gel analysis (Figure 3).
Figure 3.
SDS-PAGE analysis of crude extracts and purified TaGH2 and TbGH2. Soluble (CE) and insoluble (pellet) fractions of crude extracts and purified proteins were separated by SDS-PAGE (200 V, 50 min, M: Marker).
Substrate conversion and kinetics of TaGH2 and TbGH2
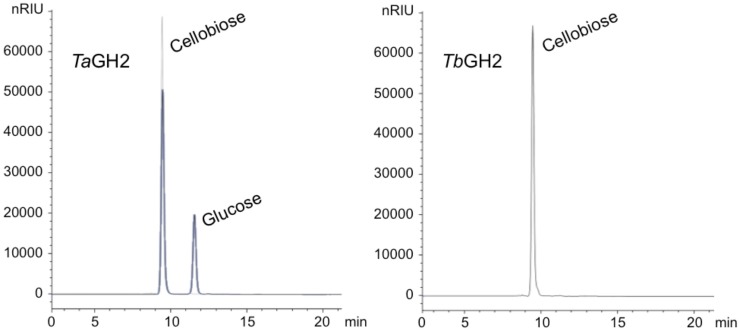
The purified enzymes TaGH2 and TbGH2 were analyzed regarding their substrate specificity. The predicted domains of GH2 and the highest homologies to annotated β-galactosidases suggested both enzymes being β-galactosidases. Nitrophenol-linked substrates were tested and in contrast to expectations, TaGH2 and TbGH2 exhibited highest activities toward 4-Np-β-d-glucopyranoside (TaGH2: 3,966 U/mg and TbGH2: 660 U/mg), and residual activities of 40 and 51% against 4-Np-β-d-galactopyranoside. In accordance, Michaelis constants (Km) were lower when 4-Np-β-d-glucopyranoside was used as substrate (Table 2). The catalytic efficiency was 29,532 mM−1 s−1 for TaGH2 and 7,797 mM−1 s−1 for TbGH2. The maximum reaction rate (V max) of TaGH2 was higher when measured with 4-Np-β-d-glucopyranoside (4,028 ± 21 μmol min−1 mg−1) compared to 4-Np-β-d-galactopyranoside (2,615 ± 3 μmol min−1 mg−1). On the contrary, TbGH2 exhibited a higher maximum reaction rate toward 4-Np-β-d-galactopyranoside (1,333 ± 11 μmol min−1 mg−1) than toward 4-Np-β-d-glucopyranoside (778 ± 22 μmol min−1 mg−1). Other nitrophenol-linked substrates were not converted. Furthermore, non-nitrophenolic substrates, such as cellobiose and lactose (1%, w/v), were tested. Interestingly, TaGH2 converted cellobiose (Figure 4). TbGH2 acted neither on cellobiose nor on lactose.
Table 2.
Kinetic parameters of TaGH2 and TbGH.
| 4-Np-β- glucopyranoside |
4-Np-β- galactopyranoside |
|||
|---|---|---|---|---|
| TaGH2 | TbGH2 | TaGH2 | TbGH2 | |
| Km [mM] | 0.73 ± 0.03 | 0.50 ± 0.02 | 2.20 ± 0.02 | 3.56 ± 0.07 |
| V max [μmol min−1 mg−1] | 4,028 ± 21 | 778 ± 22 | 2,615 ± 3 | 1,333 ± 11 |
| kcat [s−1] | 21,559 | 3,899 | 13,996 | 6,683 |
| kcat/Km [mM−1 s−1] | 29,532 | 7,797 | 6,361 | 1,877 |
Activity of TaGH2 and TbGH2 was measured at 90°C in 20 mM citrate or maleate buffer (pH 6.5) with 0–10 mM 4-Np-β-D-glucopyranoside and 0–25 mM 4-Np-β-D-galactopyranoside for 10 min.
Figure 4.
HPLC analyses of cellobiose degradation. The degradation of cellobiose (1% w/v) by TaGH2 and by TbGH2 is depicted. The hydrolysis was carried out at 90°C for 1 h in 20 mM citrate or maleate buffer (pH 6.5). Subsequently, the samples were subjected to HPLC analyses using an Hi-Plex H column and water with a flow rate of 0.6 mL/min. Standards are shown in gray. nRIU, nano Refractive Index Units.
Influence of temperature and pH
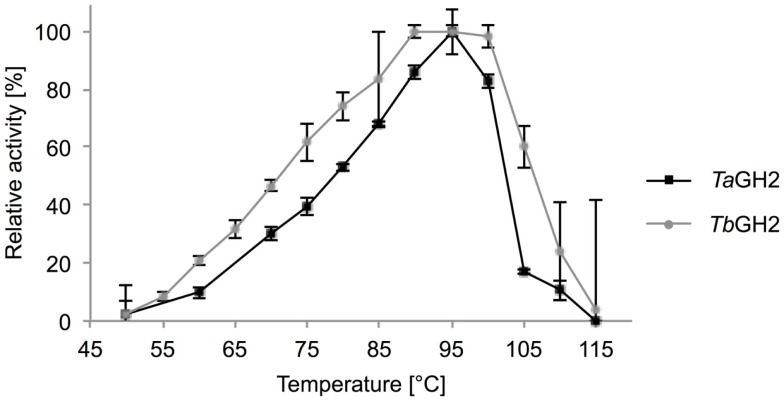
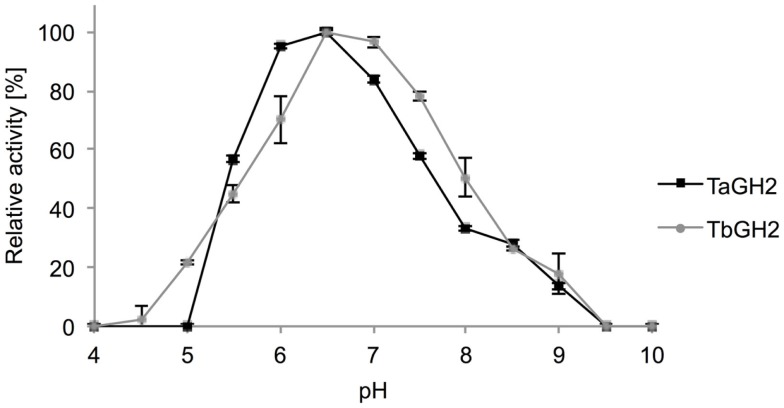
Highest activities toward 4-Np-β-d-glucopyranoside were observed at 95°C (TaGH2) and 90°C (TbGH2) at pH 6.5 (Figures 5 and 6). Residual activities of 83 ± 2% (TaGH2) and 98 ± 4% (TbGH2) were detected at 100°C. TaGH2 showed no loss of activity when incubated at 50–90°C for 3 h (Table 3). By contrast, TbGH2 was less stable and showed 17 ± 6% activity at 80°C after 3 h of incubation. At 70°C, TaGH2 exhibited a half-life time of approximately 22 h and TbGH2 of 12 h. In accordance, the predicted Tm was higher for TaGH2 (>65°C) when compared to TbGH2 (55–65°C) (Ku et al., 2009). Thermal activation was observed for both enzymes at 50 and 60°C. The enzyme TaGH2 was stable when preincubated (4°C, 24 h) at pH values between 3.0 and 10.0 (81 ± 5-100 ± 2%). TbGH2 was stable at pH 3.5–7.5 (71 ± 8-114 ± 13%) with residual activities of 63 ± 7 and 52 ± 11% at higher pH values (pH 8.5, 9.5).
Figure 5.
Effect of temperature on activity of TaGH2 and TbGH2. Activity of TaGH2 and TbGH2 was measured at different temperatures (50–115°C) for 10 min in 20 mM citrate or maleate buffer (pH 6.5) with 2 mM 4-Np-β-D-glucopyranoside. Temperatures above 95°C were measured in heated oil.
Figure 6.
Effect of pH on activity of TaGH2 and TbGH2. Activity of TaGH2 and TbGH2 was measured for 10 min at 95 or 90°C in 20 mM Britton–Robinson buffer (pH 4–10) with 2 mM 4-Np-β-D-glucopyranoside.
Table 3.
Effect of temperature on stability of TaGH2 and TbGH2.
| 3 h |
24 h |
|||
|---|---|---|---|---|
| TaGH2 | TbGH2 | TaGH2 | TbGH2 | |
| 50°C | 127 ± 5 | 129 ± 9 | 151 ± 4 | 246 ± 12 |
| 60°C | 123 ± 2 | 199 ± 7 | 108 ± 8 | 202 ± 10 |
| 70°C | 123 ± 4 | 94 ± 3 | 47 ± 3 | 0 |
| 80°C | 128 ± 2 | 17 ± 6 | 15 ± 3 | 0 |
| 90°C | 127 ± 3 | 0 | 0 | 0 |
Activity of TaGH2 and TbGH2 was measured at 90°C in 20 mM citrate or maleate buffer (pH 6.5) with 2 mM 4-Np-β-D-glucopyranoside for 10 min after preincubation for 3 or 24 h, respectively, at different temperatures (50–90°C). Activities determined at 0 h were set to 100%.
Influence of additives
The influence of different metal ions on the activity of TaGH2 and TbGH2 was tested (Table 4). K+, Mg2+, and Na+ ions (1–10 mM) had no negative influence on the enzymatic performance of both enzymes. Furthermore, Ca2+, Co2+, Mn2+, Ni2+, Rb+, and Sr2+ (1–10 mM) had no significant influence on the hydrolytic action of TaGH2 (88 ± 4-104 ± 4%). The activity of TbGH2 was reduced in presence of 10 mM CaCl2 (56 ± 20%), MnCl2 (64 ± 12%), RbCl (81 ± 5%), and SrCl2 (67 ± 15%), or completely inhibited in presence of 5–10 mM CoCl2 and NiCl2. TbGH2 was inactivated by 1 mM Ag+, Cu2+, and Zn2+. TaGH2 was exclusively inhibited by Ag+. Fe2+ and Fe3+ (1 mM) had no influence on the enzymatic performance of both hydrolases (95 ± 2-120 ± 2%). TbGH2 showed higher activity when 1 mM AlCl3 (149 ±1%), CrCl3 (139 ± 4%), KCl (146 ± 4%), MgCl2 (136 ± 4%), NaCl (138 ± 11%), or RbCl (143 ± 10%) was added.
Table 4.
Effects of metal ions on the activity of TaGH2 and TbGH2.
| Reagent | Relative activity of TaGH2 (%) |
Relative activity of TbGH2 (%) |
||||
|---|---|---|---|---|---|---|
| 1 mM | 5 mM | 10 mM | 1 mM | 5 mM | 10 mM | |
| None | 100 ± 3 | 100 ± 5 | 100 ± 2 | 100 ± 4 | 100 ± 4 | 100 ± 5 |
| AgNO3 | 0 | 0 | 0 | 0 | 0 | 0 |
| AlCl3 | 97 ± 4 | 53 ± 1 | 17 ± 5 | 149 ± 1 | 98 ± 13 | 0 |
| CaCl2 | 100 ± 3 | 100 ± 4 | 103 ± 4 | 93 ± 6 | 72 ± 5 | 56 ± 20 |
| CoCl2 | 99 ± 3 | 101 ± 2 | 97 ± 2 | 52 ± 7 | 0 | 0 |
| CrCl3 | 88 ± 2 | 43 ± 3 | 13 ± 5 | 139 ± 4 | 53 ± 11 | 0 |
| CuCl2 | 98 ± 4 | 64 ± 5 | 20 ± 4 | 0 | 0 | 0 |
| FeCl2 | 97 ± 1 | 42 ± 3 | 0 | 117 ± 3 | 31 ± 14 | 0 |
| FeCl3 | 95 ± 2 | 53 ± 4 | 12 ± 3 | 120 ± 2 | 37 ± 18 | 0 |
| KCl | 99 ± 3 | 98 ± 3 | 102 ± 4 | 146 ± 4 | 144 ± 7 | 83 ± 5 |
| MgCl2 | 99 ± 1 | 103 ± 2 | 103 ± 2 | 136 ± 4 | 117 ± 2 | 113 ± 7 |
| MnCl2 | 96 ± 3 | 98 ± 1 | 88 ± 4 | 94 ± 15 | 86 ± 3 | 64 ± 12 |
| NaCl | 97 ± 2 | 99 ± 4 | 100 ± 4 | 138 ± 11 | 129 ± 6 | 104 ± 10 |
| NiCl2 | 99 ± 3 | 104 ± 4 | 90 ± 3 | 61 ± 13 | 0 | 0 |
| RbCl | 96 ± 2 | 102 ± 1 | 98 ± 1 | 143 ± 10 | 143 ± 2 | 81 ± 5 |
| SrCl2 | 101 ± 3 | 104 ± 4 | 104 ± 2 | 127 ± 4 | 106 ± 9 | 67 ± 15 |
| ZnCl2 | 99 ± 2 | 97 ± 2 | 64 ± 3 | 0 | 0 | 0 |
Activity of TaGH2 and TbGH2 was measured at 90°C in 20 mM citrate or maleate buffer (pH 6.5) with 2 mM 4-Np-β-D-glucopyranoside for 10 min in presence or absence of metal ions.
The presence of 5 mM additives, such as β-mercaptoethanol, DTT, EDTA, urea, iodoacetic acid, sodium azide, and Tween 80 did not decrease the activity of both enzymes. Moreover, Triton X-100 and Tween 20 did not influence the performance of TbGH2, whereas TaGH2 exhibited 49 ± 2 and 76 ± 1% residual activity. Complete loss of activity for both glycoside hydrolases was detected when SDS or CTAB (5 mM) were added.
Discussion
Screening of gene libraries from T. antranikianii and T. brockianus on esculin resulted in the identification of two novel glycoside hydrolases (TaGH2 and TbGH2). Identities of amino acid sequences higher than 80% were observed to proteins from other Thermus species. The glycoside hydrolase family 2 domains (sugar binding domain and TIM-barrel domain) appeared to be incomplete. The domains are either truncated but functional or they are not recognized as domains due to large differences to other representatives of GH2. Comparison with the GH2 LacZ from E. coli (1HN1_D) showed low similarities within the corresponding domains. Additionally, conserved amino acid signatures typical for members of GH2 were partially identified (Figure 2). Similar hypothetical proteins with typical conserved domains from different genera were detected with identities of 39% (TaGH2) and 42% (TbGH2) or below. Thus, a differentiation of proteins produced from Thermus spp. would be suggested. Low sequence similarities within one protein superfamily could be the result of adaptation to different environmental conditions. Different enzymatic activities may have developed from a common ancestor (Galperin and Koonin, 2012). Structural diversification that preserved the active site residues may result in catalytically active enzymes with altered substrate specificity. Evolutionary pressure promotes functional and effective enzymes, which may result in reduction of conserved domains with remaining functionality (Juers et al., 1999). However, substrate specificity can be neglected for classification of glycoside hydrolases, since the structural homologies especially the motif forming the catalytic center is highly conserved (Henrissat and Davies, 1997). Homologies of up to 98 and 82%, respectively, were observed by comparison with annotated β-galactosidases or β-mannosidases from Thermus species. No similar characterized enzyme to TaGH2 or TbGH2 was detected in the NCBI database; thus, classification of similar proteins was achieved by domain prediction rather than by functional analyses. Highest catalytic efficiencies for TaGH2 and TbGH2 were observed toward 4-Np-β-d-glucopyranoside with residual activities toward 4-Np-β-d-galactopyranoside. Cellobiose conversion distinguishes TaGH2 from TbGH2. The hydrolysis characteristics may vary between artificial and natural substrates. This may be due to differences in size, charge, and binding properties of the artificial compound. Higher activity toward artificial substrates was also described as common phenomenon for α-galactosidases (Wang et al., 2014). Hydrolysis of cellobiose or lactose by TbGH2 could not be detected, although high β-glucosidase activity was observed when the artificial substrate was used. β-Glucosidases are reported to often exhibit a broad substrate spectrum. A β-glucosidase from Thermotoga neapolitana hydrolyzed among others 4-Np-β-d-glucopyranoside, cellobiose, and lactose (Park et al., 2005). Likewise, a GH1-β-glucosidase derived from a hot-spring metagenome exhibited activity toward 4-Np-β-d-glucopyranoside, 4-Np-β-d-galactopyranoside, cellobiose, and lactose (Schröder et al., 2014). However, the newly discovered GH2 β-glucosidases TaGH2 and TbGH2 showed a narrow substrate range.
Protein production in the mesophilic host E. coli resulted in notable amount of inclusion bodies due to differences in genomic GC contents of the genus Thermus, as previously reported (Ishida and Oshima, 1994; Fridjonsson et al., 1999). The β-glucosidase TaGH2 and the β-glucosidase/galactosidase TbGH2 showed high activities at elevated temperatures. Other β-glycosidases from Thermus thermophilus also show activity at 88–90°C and pH 5.4–7.0 (Dion et al., 1999; Nam et al., 2010). A thermal activation was detected at 50 and 60°C, especially for TbGH2 with 246 and 202% activity after 24 h, respectively. It was reported in the literature that enzymes from thermophilic organisms produced in mesophilic hosts may require thermal activation as demonstrated for a β-glycosidase from T. thermophilus at 70°C (Gerard et al., 2002). Specific activities were considered high with 3,966 and 660 U/mg, when compared to the majority of previously reported β-glucosidases and β-galactosidases with <1–100 U/mg7. Similar to GH family 1 β-glycosidases from T. thermophilus, significantly lower Km values were observed with 4-Np-β-glucopyranoside when compared to -galactopyranoside (Dion et al., 1999; Nam et al., 2010). Highest substrate affinities for -glucopyranoside were also reported for other β-glucosidases from an uncultured soil bacterium and Cellulomonas flavigena with Km-values of 0.16 and 7.1 mM, respectively (Barrera-Islas et al., 2007; Kim et al., 2007). Hence, the enzymes described here exhibit Km-values in the range of other described GH1 β-glucosidases.
Ag+, Cu2+, and Fe3+ are most frequently described as glycoside hydrolase inhibitor (Cairns and Esen, 2010). By contrast, Fe3+ (1 mM) had no influence on the activity of both enzymes. Zinc ions inhibited TbGH2 completely but had, interestingly, no effect on activity of TaGH2. The surfactant CTAB (5 mM) inhibited both enzymes. The negatively charged catalytic amino acids in the active center might be covered by the positively charged additive. Moreover, negatively charged surface residues might be influenced resulting in structural destabilization of the enzyme. Although three cysteins are present in the amino acid sequence of both enzymes, reducing agents such as DTT and β-mercaptoethanol did not show a negative effect on the activity. Either no disulfide bond is formed or it is not affected by the prevalent conditions. It is a frequently described phenomenon that reducing agents can also have stabilizing effects on enzymes (Park et al., 2005). The influence of additives on β-glucosidases and β-galactosidases does not follow a certain pattern, and appears to be specific for each individual enzyme.
TaGH2 and TbGH2 were classified as members of glycoside hydrolase family 2 based on amino acid sequence similarities. Although GH family 2 comprises enzymes with different substrate specificities, β-glucosidase activity has not been reported in this family, yet (Henrissat, 1991). This finding proves the necessity of function-based screening to identify genes coding for proteins with unusual domain structures or unexpected activities. The substrate specificity was reported to be less relevant than structural homologies for classification of members of glycoside hydrolase families (Henrissat and Davies, 1997).
In conclusion, our study demonstrates that enzymes structurally related to GH family 2 can exhibit more diverse substrate specificities than previously predicted. Therefore, we recommend to incorporate β-glucosidases into GH family 2 and consequently to evaluate activity on glucopyranosides including cellobiose for characterization of enzymes from this family.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the German Federal Ministry of Education and Research (BMBF) in the funding cluster “Biorefinery2021,” and by Clariant Produkte (Deutschland) GmbH. This publication was supported by the German Research Foundation (DFG) and the Hamburg University of Technology (TUHH) in the funding program “Open Access Publishing”. Thanks are also due to Bernard Henrissat for correspondence.
Footnotes
References
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antranikian G., Egorova K. (2007). “Extremophiles, a unique resource of biocatalysts for industrial biotechnlology,” in Physiology and Biochemistry of Extremophiles, eds Gerday C., Glansdorff N. (Washington, DC: ASM Press; ), 361–406. 10.1128/9781555815813.ch27 [DOI] [Google Scholar]
- Barrera-Islas G. A., Ramos-Valdivia A. C., Salgado L. M., Ponce-Noyola T. (2007). Characterization of a beta-glucosidase produced by a high-specific growth-rate mutant of Cellulomonas flavigena. Curr. Microbiol. 54, 266–270. 10.1007/s00284-006-0105-7 [DOI] [PubMed] [Google Scholar]
- Bhatia Y., Mishra S., Bisaria V. S. (2002). Microbial beta-glucosidases: cloning, properties, and applications. Crit. Rev. Biotechnol. 22, 375–407. 10.1080/07388550290789568 [DOI] [PubMed] [Google Scholar]
- Blank S., Schröder C., Schirrmacher G., Reisinger C., Antranikian G. (2014). Biochemical characterization of a recombinant xylanase from Thermus brockianus, suitable for biofuel production. JSM Biotechnol. Biomed. Eng. 2, 1027. [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Britton H. T. S., Robinson R. A. (1931). Universal buffer solutions and the dissociation constant of veronal. J. Chem. Soc. 1456–1473. 10.1039/jr9310001456 [DOI] [Google Scholar]
- Brock T. D., Freeze H. (1969). Thermus aquaticus gen. n. and sp. n., a nonsporulating extreme thermophile. J. Bacteriol. 98, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J. R., Esen A. (2010). β-Glucosidases. Cell. Mol. Life Sci. 67, 3389–3405. 10.1007/s00018-010-0399-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien A., Edgar D. B., Trela J. M. (1976). Deoxyribonucleic acid polymerase from the extreme thermophile Thermus aquaticus. J. Bacteriol. 127, 1550–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G., Henrissat B. (1995). Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859. 10.1016/S0969-2126(01)00220-9 [DOI] [PubMed] [Google Scholar]
- Dion M., Fourage L., Hallet J. N., Colas B. (1999). Cloning and expression of a beta-glycosidase gene from Thermus thermophilus. Sequence and biochemical characterization of the encoded enzyme. Glycoconj. J. 16, 27–37. 10.1023/A:1006997602727 [DOI] [PubMed] [Google Scholar]
- Dworkin M., Falkow S., Rosenberg E., Schleifer K. H., Stackebrandt E. (2006). The Prokaryotes. New York: Springer. [Google Scholar]
- Fridjonsson O., Watzlawick H., Gehweiler A., Rohrhirsch T., Mattes R. (1999). Cloning of the gene encoding a novel thermostable alpha-galactosidase from Thermus brockianus ITI360. Appl. Environ. Microbiol. 65, 3955–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Koonin E. V. (2012). Divergence and convergence in enzyme evolution. J. Biol. Chem. 287, 21–28. 10.1074/jbc.R111.241976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. 10.1093/nar/gkg563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard F., Angelini S., Wu L. F. (2002). Export of Thermus thermophilus cytoplasmic beta-glycosidase via the E. coli Tat pathway. J. Mol. Microbiol. Biotechnol. 4, 533–538. [PubMed] [Google Scholar]
- Henrissat B. (1991). A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280(Pt 2), 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B., Davies G. (1997). Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7, 637–644. 10.1016/S0959-440X(97)80072-3 [DOI] [PubMed] [Google Scholar]
- Ishida M., Oshima T. (1994). Overexpression of genes of an extreme thermophile Thermus thermophilus, in Escherichia coli cells. J. Bacteriol. 176, 2767–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juers D. H., Huber R. E., Matthews B. W. (1999). Structural comparisons of TIM barrel proteins suggest functional and evolutionary relationships between beta-galactosidase and other glycohydrolases. Protein Sci. 8, 122–136. 10.1110/ps.8.1.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Park B. H., Jung B. W., Kim M., Hong S., Lee D. (2006). Identification and molecular modeling of a family 5 endocellulase from Thermus caldophilus GK24, a cellulolytic strain of Thermus thermophilus. Int. J. Mol. Sci. 7, 571–589. 10.3390/i7120571 [DOI] [Google Scholar]
- Kim S. J., Lee C. M., Kim M. Y., Yeo Y. S., Yoon S. H., Kang H. C., et al. (2007). Screening and characterization of an enzyme with beta-glucosidase activity from environmental DNA. J. Microbiol. Biotechnol. 17, 905–912. [PubMed] [Google Scholar]
- Ku T., Lu P., Chan C., Wang T., Lai S., Lyu P., et al. (2009). Predicting melting temperature directly from protein sequences. Comput. Biol. Chem. 33, 445–450. 10.1016/j.compbiolchem.2009.10.002 [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Zheng C., Chitsaz F., Derbyshire M. K., Geer L. Y., Geer R. C., et al. (2013). CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41, D348–D352. 10.1093/nar/gks1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis L., Menten M. L. (1913). Die Kinetik der Invertinwirkung. Biochem. Z. 49, 333–369. [Google Scholar]
- Nam E. S., Kim M. S., Lee H. B., Ahn J. K. (2010). β-Glycosidase of Thermus thermophilus KNOUC202: gene and biochemical properties of the enzyme expressed in Escherichia coli. Appl. Biochem. Microbiol. 46, 515–524. 10.1134/S0003683810050091 [DOI] [PubMed] [Google Scholar]
- Oshima T. I., Imahori K. (1974). Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int. J. Syst. Bacteriol. 24, 102–112. 10.1099/00207713-24-1-102 [DOI] [Google Scholar]
- Pantazaki A., Pritsa D., Kyriakidis A. (2002). Biotechnologically relevant enzymes from Thermus thermophilus. Appl. Microbiol. Biotechnol. 58, 1–12. 10.1007/s00253-001-0843-1 [DOI] [PubMed] [Google Scholar]
- Park T. H., Choi K. W., Park C. S., Lee S. B., Kang H. Y., Shon K. J., et al. (2005). Substrate specificity and transglycosylation catalyzed by a thermostable beta-glucosidase from marine hyperthermophile Thermotoga neapolitana. Appl. Microbiol. Biotechnol. 69, 411–422. 10.1007/s00253-005-0055-1 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E., Maniatis T. (2001). Molecular Cloning, A Laboratory Manual, 3rd Edn New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schröder C., Elleuche S., Blank S., Antranikian G. (2014). Characterization of a heat-active archaeal beta-glucosidase from a hydrothermal spring metagenome. Enzyme Microb. Technol. 57, 48–54. 10.1016/j.enzmictec.2014.01.010 [DOI] [PubMed] [Google Scholar]
- Viikari L., Alapuranen M., Puranen T., Vehmaanpera J., Siika-Aho M. (2007). Thermostable enzymes in lignocellulose hydrolysis. Adv. Biochem. Eng. Biotechnol. 108, 121–145. 10.1007/10_2007_065 [DOI] [PubMed] [Google Scholar]
- Wang H., Ma R., Shi P., Xue X., Luo H., Huang H., et al. (2014). A new alpha-galactosidase from thermoacidophilic alicyclobacillus sp. A4 with wide acceptor specificity for transglycosylation. Appl. Biochem. Biotechnol. 174, 328–338. 10.1007/s12010-014-1050-8 [DOI] [PubMed] [Google Scholar]