ABSTRACT
Mycobacterium tuberculosis can persist for decades in the human host. Stringent response pathways involving inorganic polyphosphate [poly(P)], which is synthesized and hydrolyzed by polyphosphate kinase (PPK) and exopolyphosphatase (PPX), respectively, are believed to play a key regulatory role in bacterial persistence. We show here that M. tuberculosis poly(P) accumulation is temporally linked to bacillary growth restriction. We also identify M. tuberculosis Rv1026 as a novel exopolyphosphatase with hydrolytic activity against long-chain poly(P). Using a tetracycline-inducible expression system to knock down expression of Rv1026 (ppx2), we found that M. tuberculosis poly(P) accumulation leads to slowed growth and reduced susceptibility to isoniazid, increased resistance to heat and acid pH, and enhanced intracellular survival during macrophage infection. By transmission electron microscopy, the ppx2 knockdown strain exhibited increased cell wall thickness, which was associated with reduced cell wall permeability to hydrophilic drugs rather than induction of drug efflux pumps or altered biofilm formation relative to the empty vector control. Transcriptomic and metabolomic analysis revealed a metabolic downshift of the ppx2 knockdown characterized by reduced transcription and translation and a downshift of glycerol-3-phosphate levels. In summary, poly(P) plays an important role in M. tuberculosis growth restriction and metabolic downshift and contributes to antibiotic tolerance through altered cell wall permeability.
IMPORTANCE
The stringent response, involving the regulatory molecules inorganic polyphosphate [poly(P)] and (p)ppGpp, is believed to mediate Mycobacterium tuberculosis persistence. In this study, we identified a novel enzyme (Rv1026, PPX2) responsible for hydrolyzing long-chain poly(P). A genetically engineered M. tuberculosis strain deficient in the ppx2 gene showed increased poly(P) levels, which were associated with early bacterial growth arrest and reduced susceptibility to the first-line drug isoniazid, as well as increased bacterial survival during exposure to stress conditions and within macrophages. Relative to the control strain, the mutant showed increased thickness of the cell wall and reduced drug permeability. Global gene expression and metabolite analysis revealed reduced expression of the transcriptional and translational machinery and a shift in carbon source utilization. In summary, regulation of the poly(P) balance is critical for persister formation in M. tuberculosis.
INTRODUCTION
Tuberculosis (TB) remains a major global health problem (1, 2). The prolonged duration of therapy required to eradicate TB infection is due to the ability of Mycobacterium tuberculosis to persist in host tissues despite antibiotic treatment (3). The stringent response mediates bacterial adaptation to stress conditions (3, 4). Inorganic polyphosphate [poly(P)], a linear polymer of many tens or hundreds of inorganic phosphate residues linked by high-energy phosphoanhydride bonds, has been implicated in the transition to bacterial persistence (5, 6). Intracellular poly(P) content increases when bacteria encounter growth-limiting conditions, such as phosphate depletion, amino acid starvation, or osmotic stress (6, 7). Poly(P) accumulation has been shown to control various bacterial processes, including protein synthesis, nucleotide balance, lipid metabolism, energy utility, and susceptibility to antibiotics (5, 6). Regulation of bacterial poly(P) content has been linked to the stringent response alarmone (p)ppGpp, whose stochastic expression may contribute to bacterial persistence (8).
M. tuberculosis, like other bacteria, possesses two groups of enzymes, which control inorganic polyphosphate homeostasis: polyphosphate kinase (PPK1/Rv2984) (9) and exopolyphosphatase (PPX1/Rv0496) (10, 11). ppk1 plays a central role in the regulatory network controlling expression of relA (9), which encodes a dual-function enzyme responsible for synthesis and hydrolysis of (p)ppGpp (12, 13). Despite its name, M. tuberculosis Rv3232c/PPK2 catalyzes poly(P)-dependent phosphorylation of ADP to ATP at a rate >800-fold higher than that of poly(P) synthesis (14, 15), and a ppk2-deficient mutant inappropriately accumulates poly(P) (11). PPX1, which hydrolyzes short-chain poly(P) (10, 16), is inhibited by pppGpp (16), suggesting the presence of a positive-feedback regulatory loop in the M. tuberculosis stringent response.
Poly(P) content has been implicated in M. tuberculosis antibiotic tolerance. Thus, poly(P)-accumulating strains deficient in ppx1 (10) or ppk2 (11) showed reduced susceptibility to the bactericidal drug isoniazid, which targets the mycolic acid synthesis pathway (17). Conversely, an M. tuberculosis ppk1 deletion mutant was found to have enhanced susceptibility to isoniazid and fluoroquinolones (18). Furthermore, maintenance of intracellular poly(P) balance is critical for M. tuberculosis survival during host infection. The ppk1 (9), ppk2 (11, 14), and ppx1 (10) genes are each required for optimal M. tuberculosis growth and survival during macrophage infection. A mutant deficient in ppk2 showed impaired growth during acute infection in the lungs of mice (11). In addition, a poly(P)-deficient strain lacking ppk1 (18) and a poly(P)-accumulating strain lacking ppx1 (10) were found to have reduced long-term survival in guinea pig lungs. These findings suggest that M. tuberculosis poly(P) levels must be tightly regulated during different stages of animal infection.
Bioinformatic predictions have identified M. tuberculosis Rv1026 as a putative PPX belonging to the single-domain Ppx-GppA family (19). However, Rv1026 was to shown to lack PPX activity against short-chain poly(P) (16). Rv1026, which appears to be an essential gene (20, 21), was found to be significantly upregulated during chronic TB infection in two different mouse strains (21). Overexpression of Rv1026 in Mycobacterium smegmatis leads to altered sliding motility and biofilm formation (22), and the gene is present in the Mycobacterium leprae minimal genome required for slow growth in human tissues (23). In the current study, we tested the hypothesis that M. tuberculosis Rv1026 encodes a PPX capable of hydrolyzing long-chain poly(P). We studied the intracellular poly(P) content of wild-type M. tuberculosis during exposure to various stress conditions to determine whether poly(P) behaves as a transient molecular signal during bacillary growth restriction. Next, we generated recombinant strains conditionally deficient in Rv1026 to determine whether poly(P) accumulation is sufficient for inducing M. tuberculosis growth restriction and antibiotic tolerance and to test the role of poly(P) accumulation during macrophage infection. Using RNA sequencing and ultrahigh-performance liquid chromatography-tandem mass spectrometry (UHPLC/MS/MS), we characterized the regulatory and metabolic pathways controlled by poly(P)-mediated signaling. Finally, we used transmission electron microscopy (TEM) to characterize the bacillary morphology and cell wall thickness in the context of Rv1026 deficiency and poly(p) accumulation.
RESULTS
Rv1026 exhibits exopolyphosphatase activity against long-chain poly(P).
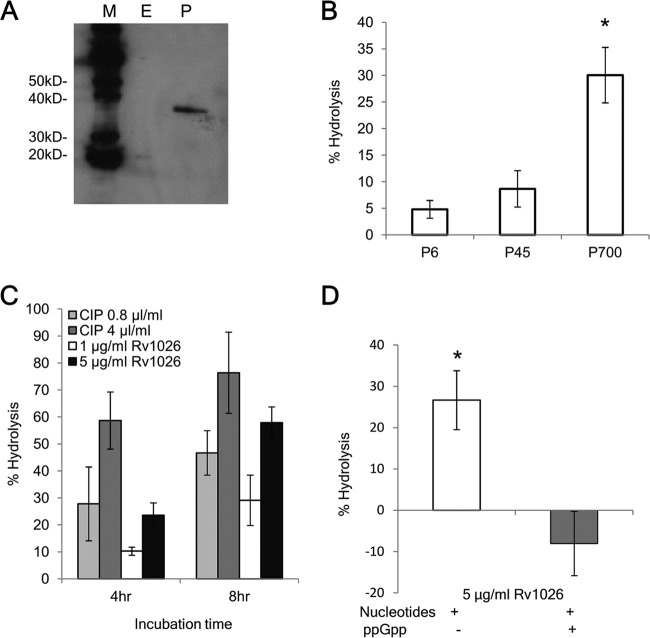
Rv1026 has been predicted to have PPX activity, but this has not been confirmed experimentally (16, 19, 22). We cloned, expressed, and purified six-histidine-tagged Rv1026 (6×His-Rv1026) from Arctic Escherichia coli, and protein lysate from E. coli transformed with the empty vector was used as the baseline control. Recombinant protein was confirmed by Western blotting using anti-His antibody (Fig. 1A). When incubated with poly(P) substrates of different lengths for 8 h, 6×His-Rv1026 showed the greatest hydrolysis activity against 700-mer poly(P) relative to calf intestinal alkaline phosphatase (CIP) (41%, 11.1%, and 6.7% for 700-mer, 45-mer, and hexametaphosphate, respectively [P < 0.05]; Fig. 1B). The hydrolytic activity of long-chain poly(P) was time and concentration dependent (Fig. 1C) and was accelerated when recombinant protein was incubated with AMP/ADP and GMP/GDP but did not favor any specific substrate (see Fig. S1 in the supplemental material). The addition of ppGpp (8 µM) inhibited the PPX activity of Rv1026 after 6-h incubation (Fig. 1D). On the basis of these findings, we concluded that recombinant Rv1026, hereafter designated PPX2, has PPX activity against long-chain poly(P), which can be inhibited by ppGpp, as in other bacteria (24).
FIG 1 .
Recombinant Rv1026 protein exhibits hydrolysis activity against long-chain polyphosphate, which is inhibited by ppGpp. (A) Western blot showing detection of 6×His-Rv1026 (34.6 kDa) using Penta-His antibody. Lanes M, protein markers; E, elution from empty vector control strain after dialysis; P, elution fraction of 6×His-Rv1026 after dialysis. The positions of molecular mass markers (in kilodaltons) are shown to the left of the blot. (B) Exopolyphosphatase activity of recombinant Rv1026. Recombinant protein (5 µg/ml) was incubated with poly(P), specifically sodium hexametaphosphate (P6) (10 µg/ml), 45-mer poly(P) (P45) (10 µg/ml), and 700-mer poly(P) (P700) (50 µM [phosphate monomer]) for 8 h. (C) PPX activity of recombinant Rv1026 as a function of concentration and incubation time. (D) Inhibition of PPX activity of recombinant Rv1026 by ppGpp. Recombinant protein was incubated with P700 (50 µM [phosphate monomer]) for 6 h with (+) or without (−) ppGpp (8 µM). Values that are significantly different (P < 0.05) are indicated by an asterisk. Values are means ± standard deviations (SD) (error bars) from three experiments.
Growth restriction is associated with transient elevation of intracellular polyphosphate content.
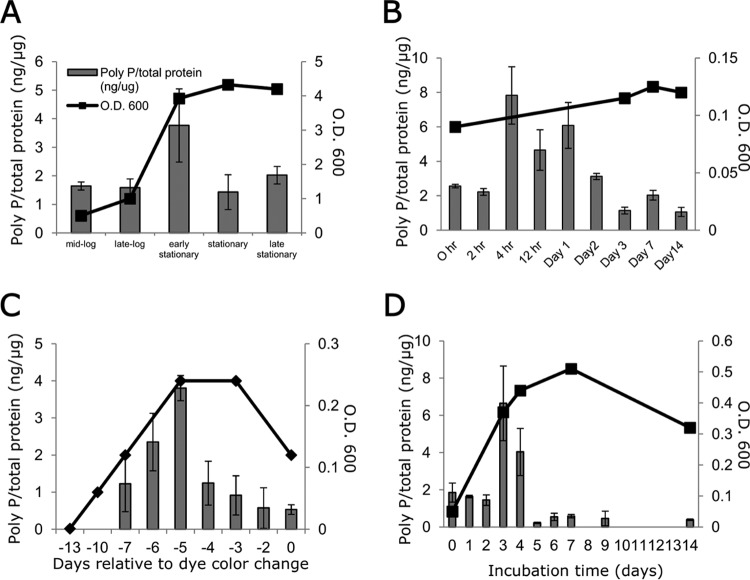
In order to gain insight into the role of poly(P) in M. tuberculosis growth arrest, we used a 4′,6-diamidino-2-phenylindole (DAPI)-based method to evaluate the intracellular poly(P) content during different growth-limiting conditions (10). During axenic growth in nutrient-rich broth, intrabacillary poly(P) levels increased during late log phase and returned to the baseline level after entry into stationary phase (Fig. 2A). Upon nutrient starvation, intrabacillary poly(P) levels peaked at 4 h and remained elevated for 24 h (Fig. 2B), coinciding with bacillary growth restriction, before decreasing to the baseline level. M. tuberculosis subjected to progressive hypoxia showed a transient elevation of poly(P) 5 to 6 days before bacterial entry into nonreplicating persistence stage 2 (Fig. 2C), which is characterized by shutdown of replication and metabolism (25). Similarly, peak M. tuberculosis poly(P) levels were observed 3 to 4 days after phosphate depletion (Fig. 2D), when bacillary growth is curbed due to exhaustion of intracellular phosphate stores (26). Under each condition, transient poly(P) accumulation closely preceded reduced M. tuberculosis growth, suggesting that poly(P) may serve as a molecular signal for M. tuberculosis growth arrest.
FIG 2 .
Intrabacillary accumulation of inorganic polyphosphate temporally coincides with M. tuberculosis growth restriction. (A to D) Intrabacillary poly(P) content was measured in wild-type M. tuberculosis during axenic growth in supplemented Middlebrook 7H9 broth (A), nutrient starvation (B), progressive hypoxia (C), and phosphate depletion (D) using a DAPI-based method and normalized to total protein content of extract lysate. Each data point represents the mean of three biological replicates. In the progressive hypoxia model in panel C, the x axis shows the days after change in color of the indicator dye, methylene blue, indicating bacterial entry into nonreplicating persistence stage 2 (mean ± SD). In panels A to D, the poly(P)/total protein ratio is shown on the left-hand x axes, and the optical density at 600 nm (O.D. 600) is shown on the right-hand x axes.
Rv1026 deficiency leads to poly(P) accumulation and slowed M. tuberculosis growth.
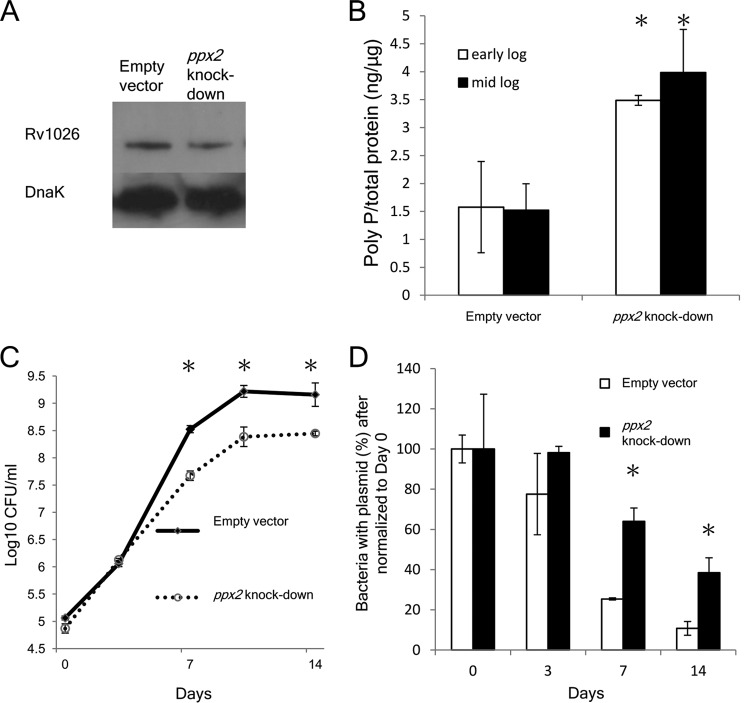
To determine whether poly(P) accumulation is sufficient for M. tuberculosis growth arrest, we used a Tet-on expression system (27) to conditionally knock down the expression of the exopolyphosphatase gene Rv1026/ppx2 in wild-type M. tuberculosis CDC1551 (see Fig. S2A in the supplemental material). The identity of the ppx2 knockdown strains was confirmed by PCR and sequencing (Fig. S2B). As shown in Fig. S2C, the growth rate of the ppx2 knockdown strain in supplemented Middlebrook 7H9 broth was inversely proportional to the concentration of the inducing agent, anhydrotetracycline (aTC). Since aTC at 250 ng/ml maximally inhibits growth of the ppx2 knockdown strain, while aTC at 1 µg/ml also inhibits growth of wild-type M. tuberculosis (28), the former concentration of inducing agent was used in all subsequent studies. The ppx2 knockdown strain displayed reduced expression of PPX2 by immunoblotting (Fig. 3A). By densitometric analysis, the ppx2 knockdown strain had 66% PPX2 expression compared to the empty vector control strain after normalization to the expression level of DnaK. The ppx2 knockdown strain showed increased intracellular poly(P) content relative to the empty vector control during logarithmic growth in nutrient-rich broth (P < 0.05; Fig. 3B), corroborating the PPX activity of Rv1026. The ppx2 knockdown strain reached a lower bacterial density during stationary phase relative to the empty vector control (Fig. 3C); this phenomenon could be attributable to reduced replication or accelerated bacterial death. To address this question, we used a “replication clock” plasmid, pBP10 (29), to transform the ppx2 knockdown strain, as the rate of plasmid loss is directly proportional to the rate of bacterial replication. The ppx2 knockdown showed reduced plasmid loss relative to the empty vector control strain during logarithmic phase, indicating that poly(P) accumulation is associated with slowed growth (Fig. 3D). Transmission electron microscopy revealed that the mean length of the ppx2 knockdown bacilli was significantly shorter than that of control bacilli (P < 0.005; Fig. S3), consistent with other studies showing decreased length of nonreplicating organisms (30). Together, these findings corroborate the hypothesis that poly(P) plays an important role in regulating M. tuberculosis growth.
FIG 3 .
Conditional knockdown of ppx2 leads to M. tuberculosis poly(P) accumulation and growth restriction. (A) Immunoblot confirmation of Rv1026 knockdown strains compared to empty vector control strain. (B) Poly(P) content of ppx2 knockdown and empty vector strains. Values that are significantly different (P < 0.05) are indicated by an asterisk. Values are means ± standard deviations (error bars) from three experiments. (C) Growth curves of ppx2 knockdown and empty vector strains in supplemented Middlebrook 7H9 broth. Values that are significantly different (P < 0.05) compared to the value for the ppx2 knockdown strain are indicated by an asterisk. Values are means ± standard deviations (error bars) from three experiments. (D) Replication rate in ppx2 knockdown strain versus empty vector control strain. The percentage of “replication clock” plasmid-containing bacteria was determined by counting the number of CFU on Middlebrook 7H10 plates containing kanamycin (50 µg/ml) and dividing this number by CFU on nonselective 7H10 plates (*, P < 0.05; n = 3).
ppx2 deficiency contributes to antibiotic tolerance.
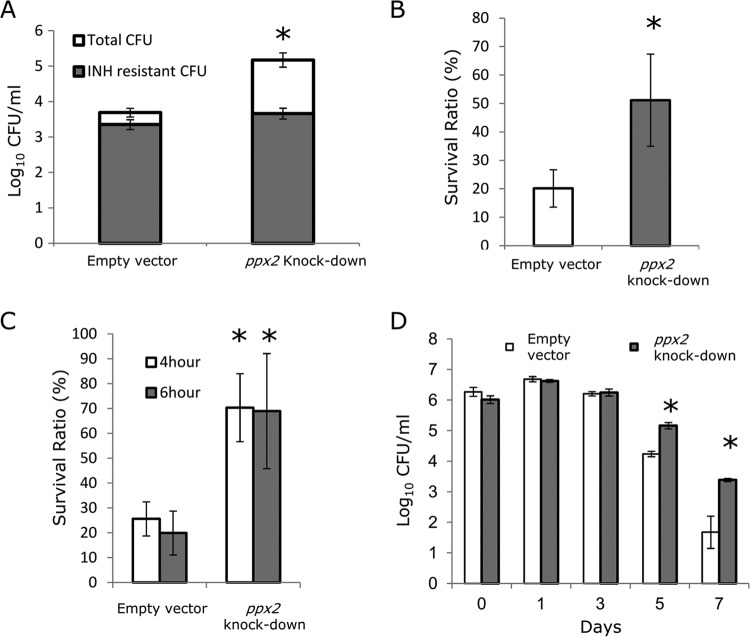
Next, we studied the activity of isoniazid, which targets actively multiplying bacilli (31, 32), against the poly(P)-accumulating ppx2 knockdown strain. Relative to the empty vector control strain, the isoniazid MIC increased 8-fold against the ppx2 knockdown (Table 1). In contrast, the MIC of rifampin was shifted only 2-fold against this recombinant strain. The ppx2 knockdown strain showed improved survival following exposure to high concentrations of isoniazid (10 µg/ml) compared to the empty vector strain (P < 0.05; Fig. 4A), confirming the importance of ppx2 in bacterial survival during exposure to cell wall synthesis inhibitors. Furthermore, the absolute numbers of isoniazid-resistant colonies were equivalent in the ppx2 knockdown and empty vector strains (Fig. 4A), accounting for 49% ± 11% and 2.5% ± 1.2% of the surviving bacilli, respectively. Together, these data are consistent with increased survival of drug-tolerant bacteria during poly(P) accumulation rather than selection of drug-resistant mutants.
TABLE 1 .
MICs of two antibiotics against the empty vector control and ppx2 knockdown strains
| Strain | MIC (µg/ml) |
|
|---|---|---|
| Isoniazid | Rifampin | |
| Empty vector control | 0.12 | 0.25 |
| ppx2 knockdown | 0.96 | 0.5 |
FIG 4 .
Polyphosphate accumulation contributes to M. tuberculosis phenotypic tolerance to isoniazid and stress resistance. (A) Logarithmically growing ppx2 knockdown and empty vector control strains were incubated with isoniazid (10 µg/ml) for 7 days. The bacteria were plated on Middlebrook 7H10 agar with or without isoniazid (INH) (1 µg/ml) to determine the number of drug-sensitive and drug-resistant CFU. Data are the means of three independent samples, and the numbers are the numbers of CFU of isoniazid-resistant (gray bar) or isoniazid-sensitive bacteria (white bar) (*, P < 0.05; n = 3). (B to D) Logarithmically growing cultures of empty vector control and ppx2 knockdown strains were incubated under various stress conditions. The stress conditions were as follows: 40°C for 24 h (B), 0.05% SDS for 4 and 6 h (C), and acidified Middlebrook 7H9 broth (pH 4.5) (D). In panels B and C, the survival ratio is the number of surviving bacteria after challenge divided by the number of bacteria prior to incubation. Values are means ± SD. Values that are significantly different (P < 0.05) compared to the value for the empty vector control strain are indicated by an asterisk.
ppx2 deficiency leads to increased M. tuberculosis resistance to various in vitro stresses.
Poly(P) regulates expression of the stringent response alarmone (p)ppGpp, and accumulation of poly(P) is believed to contribute to bacterial stress resistance (3, 8). We exposed the ppx2 knockdown strain to various growth-limiting conditions to study its phenotype in relation to the empty vector control strain. The ppx2 knockdown strain showed increased survival following exposure to 40°C for 24 h (Fig. 4B) and to 0.05% sodium dodecyl sulfate (SDS) for 4 h and 6 h (Fig. 4C) compared to the empty vector control. The recombinant strain also showed improved survival following acid shock compared to the empty vector strain (Fig. 4D).
ppx2 deficiency leads to increased M. tuberculosis growth in murine macrophages.
To determine whether ppx2 deficiency alters M. tuberculosis survival in the host, we infected J774 macrophages with the ppx2 knockdown and empty vector control strains. The ppx2 knockdown strain showed increased intracellular growth during infection of naive macrophages (Fig. 5A). The supernatant of naive macrophages infected with the ppx2 knockdown strain showed higher levels of granulocyte-macrophage colony-stimulating factor (GM-CSF) (P = 0.02), interleukin 5 (IL-5) (P = 0.003), IL-12(p40) (P = 0.001), and IL-12(p70) (P = 0.002) compared to those infected with the empty vector control (Fig. 5B). Other cytokines, including the Th1-type cytokines gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), and IL-2, were noted to be significantly different in the two strains. These data suggest that the increased growth of the ppx2 knockdown was not due to reduced macrophage production of proinflammatory cytokines.
FIG 5 .
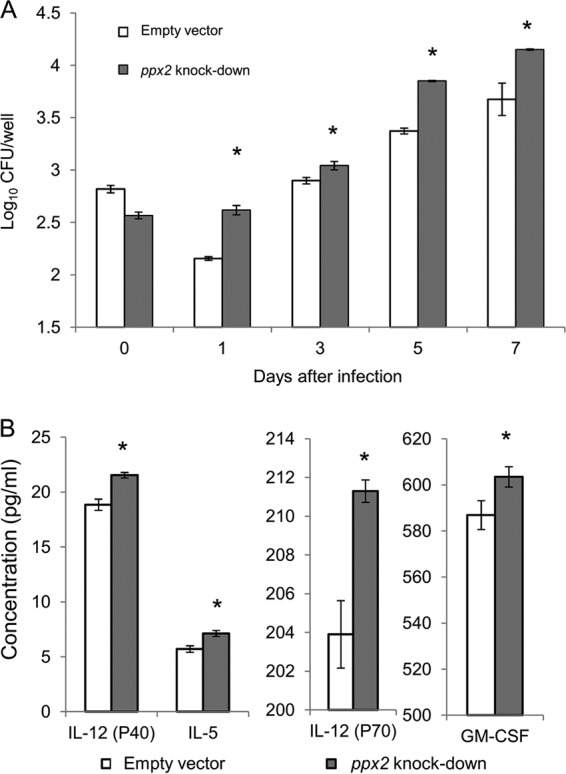
Polyphosphate accumulation contributes to enhanced M. tuberculosis survival during infection of naive macrophages. (A) Naive macrophages were infected with the empty vector control strain and ppx2 knockdown strain. The numbers of CFU were determined at days 0, 1, 3, 5 and 7 after infection (*, P < 0.05 compared to the empty vector strain; n = 3). (B) Poly(P) accumulation alters cytokine and chemokine release by naive macrophages. Cytokines and chemokines released by naive macrophages were measured after 72 h of infection with empty vector or ppx2 knockdown strain (*, P < 0.05 compared to the empty vector strain; n = 3).
Inorganic polyphosphate regulates the transcription of multiple M. tuberculosis genes.
In order to gain insight into the regulatory and metabolic changes occurring in M. tuberculosis during poly(P)-mediated growth restriction we used transcriptome sequencing (RNA-seq) to study the global gene expression of the ppx2 knockdown strain relative to the empty vector control strain during mid-log phase, when the difference in poly(P) content between the two strains is greatest and statistically significant (Fig. 3B). A total of 972 genes were significantly differentially regulated in the ppx2 knockdown strain, including 482 downregulated genes and 490 upregulated genes (see Table S1 in the supplemental material). The poly(P)-dependent stringent response signaling pathway, including mprAB and sigE (9, 33), was significantly upregulated in the ppx2 knockdown strain. In addition, the operon comprising espA (Rv3616c), espC (Rv3615c), and espD (Rv3614c) was upregulated in the ppx2 knockdown strain. This operon has been shown to play a role in M. tuberculosis virulence (34–36) and during bacillary growth when long-chain fatty acids are the carbon source (37). The cydA-cydB-cydC-cydD operon, which has been implicated in M. tuberculosis drug resistance (38), was also upregulated in the ppx2 knockdown strain. Similarly, the isoniazid-induced iniB gene, mutations of which have been implicated in M. tuberculosis resistance to isoniazid and ethambutol (39, 40), was among the most significantly upregulated genes in the ppx2 knockdown strain relative to the empty vector strain.
Based on gene ontology analysis, the ppx2 knockdown strain showed differential expression of genes involved in cell wall, growth, and protein synthesis (see Table S2 in the supplemental material). Among growth-related pathways, 46 of 460 genes were upregulated, and 138 of 460 genes were downregulated. Among cell wall-related pathways, 78 of 504 genes were upregulated, and 113 of 504 genes were downregulated. The ppx2 knockdown strain displayed significant downregulation of genes involved in translation and transcription, including rpoA, rpsB, rpsD, rpsG, rpsJ, rpsK, rpsL, rplK, rplM, rplN, rplU, and rplV, consistent with the hypothesis that ppx2 deficiency-induced poly(P) accumulation is associated with a reduction in the rate of protein synthesis.
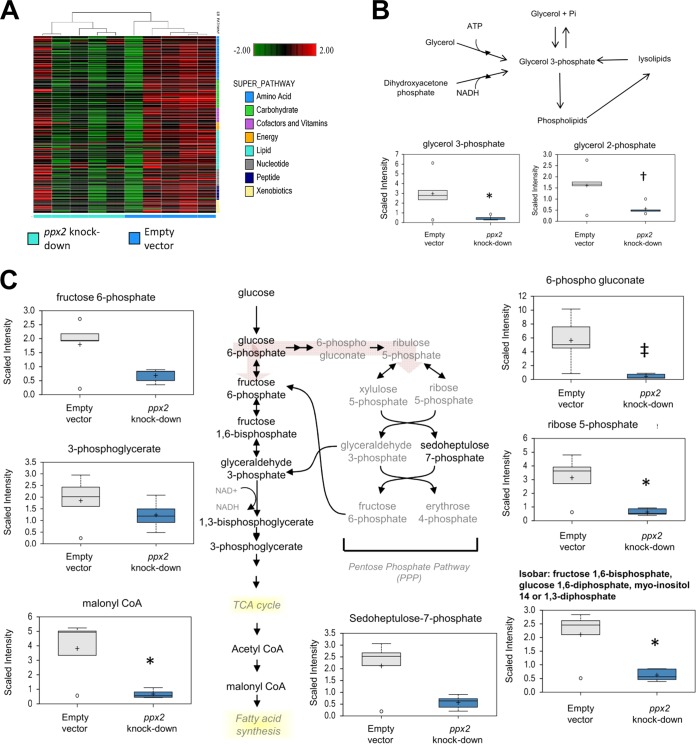
ppx2 deficiency affects global M. tuberculosis metabolism.
Metabolomics analysis using UHPLC/MS/MS revealed lower levels of the glycolytic intermediates glucose 6-phosphate (P = 0.11) and isobaric metabolites (fructose 1,6-diphosphate, glucose 1,6-diphosphate, myo-inositol 1,4- or 1,3-diphosphate) in the ppx2 knockdown strain compared to the empty vector control strain (P = 0.02; Fig. 6). Significant decreases were also observed in malonyl-coenzyme A (malonyl-CoA) levels (P = 0.017), a metabolite required for the fatty acid synthesis and pentose phosphate pathway intermediates 6-phosphogluconate (P = 0.002) and ribose-5-phosphate (P = 0.01), which could impact nucleotide biogenesis. The major phospholipid scaffold, glycerol 3-phosphate (G3P), was also lower in the ppx2 knockdown strain (P = 0.04). Statistically insignificantly lower levels of metabolites in the peptidoglycan synthesis pathway, including 1-deoxyxylose-5-phosphate (P = 0.067) and diaminopimelate (P = 0.13), were detected in the ppx2 knockdown strain. The mutant showed reduced levels of precursors in pathways responsible for glucose metabolism, fatty acid synthesis, and nucleotide biogenesis (see Table S3 in the supplemental material).
FIG 6 .
Metabolomics analysis of the ppx2 knockdown strain compared to the empty vector control strain during exponential growth. (A) Hierarchical clustering pathway analysis of ppx2 knockdown and empty vector strains. (B and C) Metabolites altered in the ppx2 knockdown strain compared to the empty vector strain include phosphate compounds (B) and components of the pentose phosphate and glucose utilization pathways (C). Values that are significantly different are indicated as follows: *, P < 0.05; + or †, 0.05 < P < 0.1; ‡, P < 0.005. TCA, tricarboxylic acid.
Previous work has highlighted the importance of alterations in lipid metabolism during M. tuberculosis dormancy (37). To gain further insight into the global metabolic changes associated with M. tuberculosis poly(P) accumulation, we calculated z scores for metabolites and gene expression values and mapped these to the Palsson’s network based on reactions (41). Significant correlations in enzyme-encoding gene expression and corresponding metabolites were observed in pathways related to fatty acid metabolism, membrane metabolism, pentose phosphate pathway, pyrimidine metabolism, and sugar metabolism (see Table S4 in the supplemental material), indicating that transcriptional changes were associated with changes at the metabolite level. For example, upregulation of the Rv1347c (mbtK) and Rv2524c (fas) genes could directly contribute to lower levels of malonyl-CoA. Conversely, expression of the fabH, inhA, and pks15 genes and the Rv2931-Rv2932-Rv2933 (ppsA-ppsB-ppsC) operon was negatively correlated with malonyl-CoA levels, suggesting the former may play a negative-feedback role in this pathway.
According to our transcriptomic and metabolomics analyses, ppx2 deficiency was associated with altered global metabolism. M. tuberculosis persistence and antibiotic tolerance have been associated with altered reduction potential (42). In order to characterize the redox potential of the ppx2 knockdown strain, we used the alamarBlue assay (Invitrogen), which measures the reduction potential of viable metabolically active cells to chemically reduce resazurin to the fluorescent molecule resorufin, and can be used as a surrogate of cellular metabolic activity (43, 44). At mid-log phase, the normalized fluorescence signal (based on CFU) in the ppx2 knockdown strain was 68.4% ± 5.1% that of the empty vector strain, consistent with reduced redox potential in the mutant strain.
ppx2 deficiency is associated with altered cell wall permeability and increased cell wall thickness but reduced biofilm formation.
Our global transcriptional analysis of the poly(P)-accumulating ppx2 knockdown strain revealed induction of several efflux genes, including iniA, iniB, mmpL10, and Rv2459 (45–47), which could contribute to the isoniazid tolerance phenotype of this strain. To further understand the potential role of altered antibiotic influx and efflux during ppx2 deficiency, we evaluated the accumulation rate of the surrogate compound, ethidium bromide (EtBr) (48), in the ppx2 knockdown and control strains by fluorescence. EtBr showed significantly reduced accumulation in the recombinant strain compared to the empty vector control strain (Fig. 7A). However, we found no difference in the fluorescence decay ratio following preincubation of each strain with EtBr, and treatment with the efflux pump inhibitors verapamil, chlorpromazine, reserpine, and carbonyl cyanide m-chlorophenyl hydrazone led to a similar increase in EtBr accumulation in each strain (data not shown). Interestingly, the ppx2 knockdown strain showed higher uptake of the Nile red dye relative to the empty vector strain (Fig. 7B), consistent with cell wall modifications in the former strain permitting greater diffusion of lipophilic molecules rather than polar molecules such as isoniazid. Transmission electron microscopy revealed no significant difference in the gross morphology of these two strains. However, the mean cell wall thickness of the ppx2 knockdown strain (20.1 nm ± 2.49 nm) was significantly greater than that of the empty vector strain (17.75 ± 2.8 nm; P < 0.001). On the other hand, the thickness of the cell capsule (49) of each strain was similar (17.25 ± 2.67 nm in the mutant versus 18.11 ± 2.76 nm in the control; Fig. 8). These data are the first to link induction of the bacterial stringent response pathway with changes in cell wall thickness, potentially contributing to antibiotic tolerance.
FIG 7 .
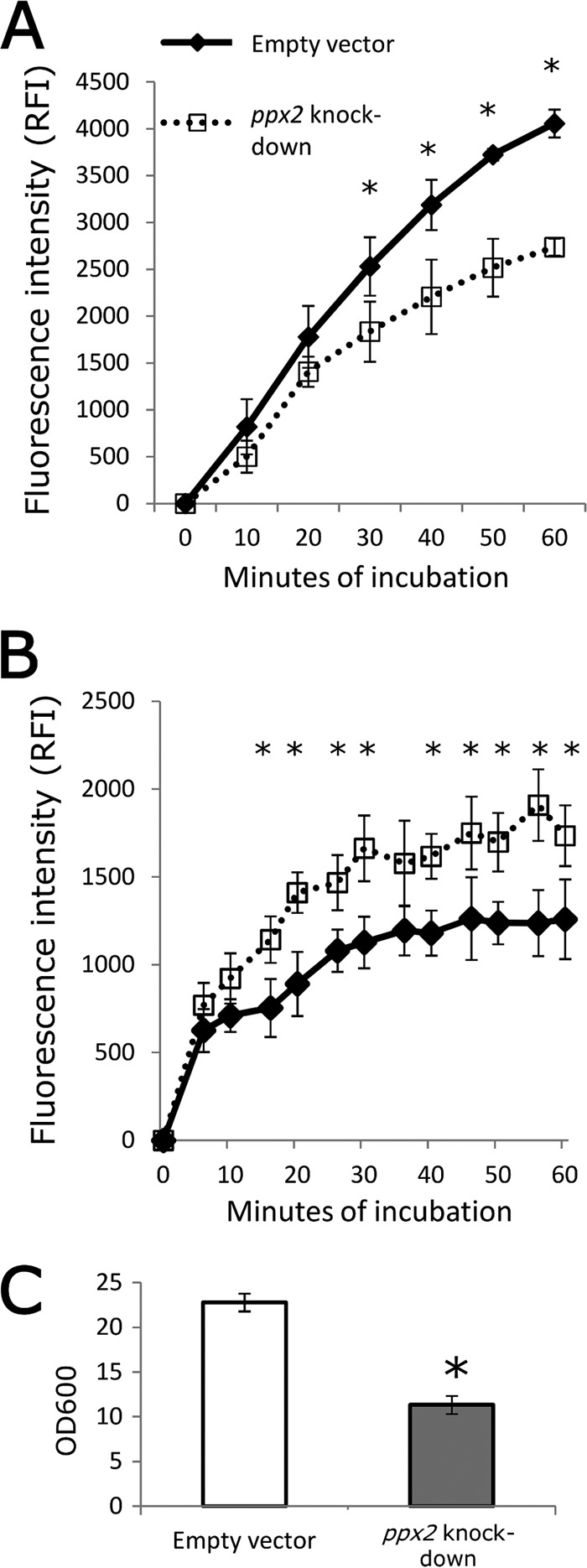
Polyphosphate accumulation results in decreased ethidium bromide accumulation, increased Nile red staining, and reduced biofilm formation. (A) Mid-log-phase cultures of empty vector control and ppx2 knockdown strains were incubated in PBST with 2 µg/ml ethidium bromide. The values at each time point are normalized to the time zero reading value (mean ± SD; *, P < 0.05; n = 3). RFI, relative fluorescence intensity. (B) Mid-log-phase cultures of each strain were incubated in PBS containing 20 µM Nile red stain (mean ± SD; *, P < 0.05; n = 3). (C) Each strain was incubated in Sauton’s medium lacking detergent for 5 weeks, and biofilms were assessed by crystal violet staining (*, P < 0.05; n = 3).
FIG 8 .

Polyphosphate accumulation is associated with increased cell wall thickness. (A to D) The empty vector control strain (A and C) and ppx2 knockdown strain (B and D) were evaluated by transmitted electronic microscopy during mid-log-phase growth. The black arrow indicates the cell wall layer. Bars, 100 nm. (E) Dot plot graph of the cell wall thickness in ppx2 knockdown and empty vector strains. The cell wall thickness in the ppx2 knockdown strain and empty vector strain were significantly different (P < 0.001).
Finally, we investigated the possibility that ppx2 deficiency and poly(P) accumulation alter formation of biofilms, which have been implicated in mycobacterial antibiotic tolerance (50). The ppx2 knockdown and empty vector control strains were grown for 5 weeks in Sauton’s medium without detergent. We found that the pellicle thickness of the ppx2 knockdown strain was reduced relative to that of the empty vector control strain. Consistent with these data, the ppx2 knockdown strain showed 40% reduced signal by crystal violet staining compared to the empty vector strain (Fig. 7C) (51). On the basis of these findings, we conclude that ppx2 deficiency-induced poly(P) accumulation was not associated with induction of drug efflux pumps or enhanced biofilm formation but was associated with increased cell wall thickness and reduced cell wall permeability to polar compounds, which may contribute to isoniazid tolerance.
DISCUSSION
The stringent response contributes to bacterial adaptation during growth-limiting conditions through the induction of specific transcriptional programs (3, 4). Poly(P) serves as a phosphate donor to activate downstream regulatory genes, including mprAB in M. tuberculosis (9, 33). Our data are consistent with the hypothesis that poly(P) serves as a transient signal to trigger the bacterial stringent response, which is required for growth arrest under nutrient-limited conditions (6). In this study, we identify a novel exopolyphosphatase (Rv1026, PPX2), and a deficiency of this enzyme leads to poly(P) accumulation, premature growth restriction, and antibiotic tolerance, likely as a result of changes in the cell wall leading to reduced permeability of polar compounds, such as isoniazid. In addition, we show for the first time that poly(P) deficiency in the context of ppx2 deficiency leads to global changes in bacterial transcriptional responses and metabolism, including a downshift in transcription and translation and a shift from utilization of carbohydrate sources of energy, as well as the induction of several important virulence factors.
Despite bioinformatic predictions (19), prior work has challenged the hypothesis that Rv1026 encodes an exopolyphosphatase in M. tuberculosis (16). Consistent with these studies, we found that recombinant Rv1026 had very weak PPX activity against short-chain poly(P) (45-mer). However, 6×His-Rv1026 was able to hydrolyze long-chain poly(P) (700-mer) in a concentration- and time-dependent manner. In addition to long-chain poly(P) as a substrate, our studies included the use of ADP, AMP, GDP, and GMP to promote hydrolysis activity, which may account for the discrepant findings between our results and those of Choi et al. (16). As in other bacteria (24), the PPX activity of recombinant Rv1026 was inhibited by the stringent response alarmone ppGpp. Further corroborating the function of Rv1026 as a PPX, we found that the ppx2 knockdown strain had higher intracellular poly(P) content than that of the empty vector control strain. M. tuberculosis PPX1 (Rv0496) has been shown to hydrolyze short-chain poly(P) (10, 16). Therefore, M. tuberculosis may employ these two enzymes to tightly regulate the number and length of intracellular poly(P) molecules, thereby quenching the transient regulatory signal during growth arrest.
Corroborating the hypothesis that poly(P) accumulation triggers M. tuberculosis growth restriction, the ppx2-deficient strain showed premature entry into stationary phase in nutrient-rich broth and reduced susceptibility to the cell wall-active agent isoniazid, which targets actively multiplying bacilli, while retaining susceptibility to the sterilizing drug rifampin which better targets nonreplicating “persisters” (52). The enhanced survival of the ppx2 knockdown strain following exposure to high-dose isoniazid was not due to the selection of isoniazid-resistant mutants, but to the presence of drug-tolerant persisters. Our findings are consistent with previous reports showing increased susceptibility of poly(P)-deficient M. tuberculosis to antibiotics (18) and reduced isoniazid susceptibility among M. tuberculosis poly(P)-accumulating strains (10, 11). We also found that poly(P) accumulation contributes to M. tuberculosis adaptation and survival during various stress challenges, including acid, heat, and detergent stresses. Although the ppx2 knockdown strain induced greater release of IL-5, IL-12, and GM-CSF, these cytokines were ineffective in controlling M. tuberculosis replication, as these strains showed enhanced intracellular survival within naive macrophages relative to the empty vector. Taken together, these findings suggest that poly(P) accumulation contributes to M. tuberculosis growth restriction, antibiotic tolerance, and survival during growth-limiting conditions.
Based on our transcriptomics analysis, a similar number of genes were found to be downregulated as were upregulated in the ppx2 knockdown strain, indicating that poly(P) accumulation does not simply lead to global transcriptional shutdown. According to previous studies, poly(P) appears to play a role in the stringent response signaling pathway, mprAB-sigE-relA (9, 33). mprAB and sigE were both upregulated in the ppx2 knockdown strain. Based on gene ontology analysis, ppx2 deficiency affected regulatory pathways related to growth, cell wall metabolism, transcription, and translation. Among the pathways related to transcription and translation, the majority of genes were downregulated, which is consistent with the putative role of poly(P) as a stringent response regulatory molecule.
Prior work has highlighted the importance of tricarboxylic acid cycle remodeling, including increased synthesis of succinate and decreased levels of α-ketoglutarate, in the metabolic adaptation of M. tuberculosis to hypoxia (53). In addition, accumulation of pyruvate and succinate and depletion of α-ketoglutarate were noted following M. tuberculosis exposure to antibiotics (54). Our metabolomics analysis revealed significantly reduced levels of glycerol 3-phosphate (G3P) and glycerol 2-phosphate (G2P) during poly(P) accumulation. G3P serves as an important scaffold for phospholipid biosynthesis (55), and reduced G3P levels are believed to be important in persister formation (56, 57). Significantly reduced intracellular G3P and G2P content has been reported in M. tuberculosis when cholesterol is the sole carbon source (58). The ppx2 knockdown strain also showed significant downregulation of the G3P dehydrogenase gene, glpD2, which has been shown to be downregulated in M. tuberculosis persisters (59). In addition, glucose phosphorylation was significantly altered in the ppx2 knockdown strain, and this process has been shown to have an important role during M. tuberculosis chronic infection in mice (60). Although several metabolites in lipid biosynthetic pathways were reduced during ppx2 deficiency, transcriptomic analysis revealed significantly increased expression of the triacylglycerol (TAG) synthesis gene, tgs1 (3-fold increase; adjusted P value of <0.0001). TAG plays an important role in M. tuberculosis persistence and antibiotic tolerance during hypoxia (61, 62). Metabolomics analysis of the ppx2 knockdown strain also showed significant decreases in glucose metabolism, fatty acid synthesis, and nucleotide biogenesis. Finally, poly(P)-induced genes include the operon comprising espA (Rv3616c), espC (MT3615c), and espD (Rv3614c), which appears to play an important role in M. tuberculosis virulence (35, 36) and may serve to promote bacillary survival within acidic environments in vitro and ex vivo (63, 64). Taken together, these data suggest that poly(P) accumulation induces changes in carbon utilization and contributes to altered lipid metabolism, which may contribute to altered cell wall properties, antibiotic tolerance, and enhanced survival during physiologically relevant stress conditions.
Previous work has linked biofilm formation with the phenomena of antibiotic tolerance and persistence in M. tuberculosis (51). Poly(P) accumulation has been associated with biofilm formation in Mycobacterium smegmatis (22), Burkholderia pseudomallei (65), Porphyromonas gingivalis (66), and Pseudomonas aeruginosa (67). However, we found that M. tuberculosis poly(P) accumulation was associated with reduced biofilm formation, which could not account for the observed tolerance to isoniazid in the ppx2-deficient strain. Our data are consistent with previous studies demonstrating that proper hydrolysis of poly(P) is required for formation of biofilms in E. coli (68) and Bacillus cereus (69).
In this study, we show for the first time that mycobacterial poly(P) accumulation leads to changes in cell wall permeability, manifested by decreased uptake of the polar compound EtBr and increased uptake of the lipophilic dye Nile red. Nonreplicating M. tuberculosis displays reduced uptake of antibiotics, and the phenotype of antibiotic tolerance is not reversed by inhibition of drug efflux pumps (70). Previous studies have shown that mycobacterial uptake of Nile red is directly related to cell wall lipid components and susceptibility to lipophilic drugs (71). The mycobacterial stringent response has been implicated previously in cell wall remodeling (72, 73), and nonreplicating bacilli exposed to hypoxia exhibit thicker cell walls (74). Cell wall remodeling and thickening have been associated with M. tuberculosis survival in the host (75), and we show here for the first time that intracellular poly(P) accumulation may contribute to this phenomenon.
In summary, we have identified Rv1026 as a novel M. tuberculosis exopolyphosphatase (PPX2), which is responsible for hydrolyzing long-chain poly(P). Transient accumulation of intracellular poly(P) serves as a trigger for M. tuberculosis growth restriction, metabolic downshift, and antibiotic tolerance, which are key features of persisters (76). Poly(P) accumulation resulting from Rv1026 deficiency is associated with changes in cell wall permeability and increased cell wall thickness, which may contribute to the observed phenotypic tolerance to isoniazid. Although poly(P) is present in all cells, the highly conserved bacterial enzymes responsible for poly(P) synthesis and hydrolysis in M. tuberculosis have not been identified in mammalian cells, thus making them potentially attractive targets for drug development.
MATERIALS AND METHODS
Bacteria and growth conditions.
Wild-type M. tuberculosis CDC1551 was grown in Middlebrook 7H9 broth (Difco, Sparks, MD) supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC) (Difco), 0.1% glycerol, and 0.05% Tween 80 at 37°C on a roller. Nutrient starvation and phosphate depletion conditions were established as previously described (26, 77). A progressive hypoxia model (Wayne model) was used to study bacterial adaptation during hypoxia, and the change in color of the methylene blue dye was used as an indicator of nonreplicating persistence stage 2 (NRP-2) (10, 25).
Expression of His-tagged Rv1026 protein.
The full-length sequence of Rv1026 was amplified from M. tuberculosis CDC1551 and cloned into plasmid pET15b with an N-terminal six-His tag (6×-His tag; Novagen) using the restriction enzymes XhoI and BamHI. The resulting plasmid was used to transform E. coli Arctic Express (DE3) RP competent cells (Stratagene). Original plasmid pET15b was transformed as a negative-control strain. The transformed bacteria were selected by ampicillin (100 µg/ml), and cloning was confirmed by DNA sequencing. Confirmed clones were grown in LB broth with gentamicin (20 µg/ml) and ampicillin (100 µg/ml), and the culture was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in 12°C for 24 h to overexpress recombinant Rv1026. The negative-control strain was induced by using the same protocol. Protein was harvested from cell pellets and stored at −20°C.
Purification of His-tagged Rv1026 protein.
Protein was purified using a nickel-nitrilotriacetic acid (Ni-NTA) slurry under native conditions using standard protocols (Qiagen). The dialyzed active protein fractions were quantified using Qubit protein assay (Invitrogen) and analyzed by SDS-PAGE. Protein specificity was confirmed by Western blotting using Penta-His antibody (Qiagen). The recombinant protein was frozen and stored at −80°C until use.
Assay of polyphosphatase activity of recombinant Rv1026.
Dialyzed recombinant Rv1026 in phosphate-buffered saline (PBS) solution was used to determine PPX activity (10). Briefly, the elution fraction of six-histidine-tagged Rv1026 (6×His-Rv1026) was prepared as previously described, and protein from the original plasmid (pET15b) served as a negative control. 6×His-Rv1026 (5 µg/ml) or eluted protein harvested from negative control was incubated with 10 µg/ml sodium hexametaphosphate (P6; Sigma), 10 µg/ml 45-mer polyphosphate [poly(P)] (P45; Sigma), or 50 µM (phosphate monomer) 700-mer poly(P) (P700; KeraFAST, Inc., Boston, MA) in 50 mM Tris-HCl reaction buffer (pH 8.0), which contained 10 mM MgCl2,1.5 mM KCl, and 50 µg/ml of ADP, AMP, GDP, and GMP (Sigma) for 8 h. Calf intestinal alkaline phosphatase (CIP) (New England BioLabs [NEB]) (40 U/ml) was used as a positive control, and buffer alone was used to determine total poly(P) content. Poly(P) levels were measured in three or four samples at each time point using a 4′,6-diamidino-2-phenylindole (DAPI)-based method (10, 11, 78). Each experiment had triplicate independent reactions and was repeated at least once. The data represent the averages of these triplicate reactions. To determine the effect of ppGpp on the hydrolytic activity of Rv1026, 8 µM ppGpp (TriLink BioTechnologies, San Diego, CA) was incubated with 6×His-Rv1026 (5 µg/ml) and 50 µM (phosphate monomer) P700, and total poly(P) content was measured after 6-h incubation.
Determination of intrabacillary inorganic polyphosphate.
A DAPI-based method was used to determine intracellular polyphosphate content (11, 78, 79). Briefly, the bacteria were lysed by bead beating in 50 mM Tris-HCl (pH 7.0) buffer supplemented with 5 M guanidinium thiocyanate (GITC) (Sigma), and the total protein levels of the lysates were determined by colorimetric protein assay (Bio-Rad). Poly(P) was harvested by glass milk from Geneclean III kit (MP Biomedicals LLC) and then treated with DNase (Ambion) and RNase (NEB) before elution with 95°C distilled water (pH 8.0). The poly(P) concentration was determined by fluorescence of the DAPI-poly(P) complex following excitation at 415 nm and emission at 525 nm on a FLUOstar OPTIMA microplate reader (BMG Labtech). Increasing concentrations of poly(P) (type 65; Sigma-Aldrich) were used to generate a standard curve to measure the poly(P) contents. All data were from three biological replicate experiments.
Conditional Rv1026 knockdown strains.
A conditional expression plasmid, pUV15tetORm, was obtained from Addgene (Addgene plasmid 17975) (27). For generation of the Rv1026 (ppx2) knockdown strain, the sequence containing the Rv1026 gene, including 123 bp upstream and 963 bp downstream of the transcription start and stop sites, respectively, was digested using AclI and PacI and cloned in reverse orientation into this plasmid. The segment containing attB and Int from pMH94 was cloned into puv15tetORm using the MfeI and AclI sites to generate a single-copy plasmid conferring hygromycin resistance (pUVatt Rv1026 knockdown; see Fig. S2A in the supplemental material) (80, 81). For generation of the empty vector, the segment containing Rv1026 in the reverse orientation was replaced by the segment containing attB and Int from pMH94 using the MfeI and PacI sites (pUVatt empty vector). Primers are listed in Table S5 in the supplemental material. The integrating plasmids pUVatt Rv1026 knockdown, pUVatt ppk1 knock-in, and pUVatt empty vector, were introduced into the wild-type M. tuberculosis CDC1551 strain by electroporation, and transformants were selected on hygromycin-containing Middlebrook 7H10 plates. Plasmid insertion was confirmed by PCR. For all experiments, the empty vector and Rv1026 knockdown and empty vector strains were diluted to an optical density (OD600) of 0.001 and allowed to grow to mid-log phase in the presence of the inducer anhydrotetracycline (aTC) at 250 ng/ml.
Mouse immunization and generation of polyclonal antisera.
Female BALB/c mice (4 to 5 weeks old) were purchased from Charles River Laboratories. Recombinant Rv1026 protein was harvested as described above and purified from an SDS-polyacrylamide gel. At day 0, five mice were immunized by subcutaneous injection with a mixed emulsion of 6×His-Rv1026 and complete Freund’s adjuvant (Sigma). At weeks 2 and 4, mice received booster immunizations with an emulsion of incomplete Freund’s adjuvant (Sigma) and 6×His-Rv1026. At weeks 6 and 8, polyclonal sera were collected from the tail veins, and immunoblotting was performed to confirm the activity of antisera. Antisera for Rv1026 were used at a 1:100 titer as the primary antibody. DnaK expression was used as a loading control in Western blots using an anti-DnaK antibody (BEI Resources). The densitometric results of Rv1026 and DnaK in the same experiment were analyzed with ImageJ. All experiments were repeated at least once, and similar results were obtained.
Determination of the strain replication rate by “replication clock” plasmid.
The Rv1026 knockdown and empty vector strains were transformed with plasmid pBP10 (kind gift of David Sherman) (29), and transformants were selected on Middlebrook 7H10 plates containing kanamycin (50 µg/ml). Individual colonies were grown to mid-log phase in kanamycin-containing Middlebrook 7H9 broth, and the bacteria were pelleted and resuspended in enriched Middlebrook 7H9 broth without kanamycin at day 0. At days 0, 3, 7 and 14, bacteria were plated on Middlebrook 7H10 plates with or without kanamycin to determine the proportion of kanamycin-resistant CFU among total bacterial CFU. The ratio of kanamycin-resistant bacteria to total bacteria is considered to be inversely related to the bacterial division rate (29).
Transmission electron microscopy.
Live mid-log-growth-phase bacteria in Middlebrook 7H9 broth were fixed with equal volumes of 2× fixative and gently rocked for 10 min. Samples were then centrifuged at 8,000 rpm, supernatant was removed, and 1× fixative was added to the pellet, which was rocked overnight in the cold room. The final fixative concentrations were 2.5% glutaraldehyde, 20 mM sodium cacodylate, and 1 mM MgCl2, pH 7.2. After the samples were rinsed three times with 20 mM sodium cacodylate and 1 mM MgCl2 buffer for 15 min each time, samples were postfixed in 1% osmium tetroxide in 20 mM sodium cacodylate containing 1 mM MgCl2 for 1.5 h on ice. After a brief water rinse, samples were dehydrated through a graded series of ethanol to 100%, then transferred to propylene oxide, and gradually infiltrated with a 1:1 resin mixture of Spurr’s resin and Eponate 12 (Polysciences) with the following propylene oxide parts (30%, 50%, and 75% [rocked overnight]). After 3 changes in pure resin (Spurr’s resin−Eponate 12, 1:1), pellets were cured in a 60°C oven for 2 days. Sections were cut on a Reichert Ultracut E ultramicrotome with a Diatome diamond knife. Eighty-nanometer-thick sections were picked up on Formvar-coated 1- by 2-mm copper slot grids and stained first with 1% tannic acid (filtered aqueous), followed by 2% uranyl acetate (filtered aqueous), and then lead citrate. Grids were viewed on a Phillips CM 120 transmission electron microscope (TEM) operating at 80 kV, and digital images were captured with an AMT 8,000 by 8,000 charge-coupled-device (CCD) camera. For cell size measurement, a fixed phosphotungstate negative stain was used (26). Cell size and wall thickness were measured by ImageJ.
MIC determination.
The MIC of anti-TB drugs was determined as previously described (11). For antibiotic tolerance studies, mid-log-phase cultures were incubated with isoniazid (10 µg/ml). At days 0, 7, and 14, surviving bacteria were plated on Middlebrook 7H10 agar with and without isoniazid (1 µg/ml) to determine the number of surviving bacteria and proportion of drug-resistant mutants.
Heat shock, SDS, and acid challenge.
Heat shock and SDS challenge were performed as previously described (82), except that samples were incubated in a water bath at 40°C for 24 h. For acid challenge studies, the M. tuberculosis strains were grown in acidified Middlebrook 7H9 broth (83), and serial dilutions were plated onto Middlebrook 7H10 agar at days 1, 3, and 7 after incubation.
Macrophage infections and cytokine assays.
The mouse macrophage-like cell line J774.1 was used for these studies, as previously described. Naive macrophages were divided 1 day before infection without any treatment. At day 0, 104 macrophages were infected with an equal number of logarithmically growing bacilli of empty vector control and ppx2 knockdown strains in RPMI 1640 medium containing 250 ng/ml aTC. Intracellular M. tuberculosis was recovered and plated on days 0, 1, 3, 5, and 7. At days 1 and 3, the supernatant of each culture was collected and frozen at −80°C until analysis. Macrophage-secreted cytokines were analyzed by immunobead cytokine assays (mouse cytokine 23-plex assay; Bio-Rad) (11).
RNA-Seq.
Following exposure of logarithmically growing cultures (OD = 0.5) to the inducer aTC for at least 1 week, RNA was harvested using Trizol-based methods (10). RNA samples were treated with DNase, and the quality of the RNA samples was assessed by using an Agilent bioanalyzer (Agilent Technologies). The samples were sent to the Next Generation Sequencing Center at the Sidney Kimmel Comprehensive Cancer Center of the Johns Hopkins University School of Medicine for library construction and sequencing using Illumina Hi Seq 2000 (Illumina). The sequence quality of the data sets was checked by fastQC software. The transcriptome sequencing (RNA-seq) data were aligned with the M. tuberculosis CDC1551 genome obtained from Ensembl (bacteria, http://bacteria.ensembl.org) using bowtie2. HTSeq-count script written in python was used to obtain the read counts of each CDC1551 gene from the significance analysis of microarray (SAM) alignment file. The GTF annotation file from the bacterial Ensembl website was used during read counting for gene annotation. Since the library was strand specific, high-throughput sequencing (HTSeq) was employed with appropriate configurations (default). Normalization and analysis of differential expression were performed using DEseq2 package of bioconductor. DESeq2 generates statistics for each of the genes, including the Wald test P value and Benjamini Hotchberg (BH) corrected P values. Standard cutoffs of corrected P values (≤0.05) were used to denote differentially expressed genes under specific conditions. Finally, the Bioconductor package UniProt.ws was used for a detailed annotation of the genes, including mapping between MT and Rv identifiers.
Gene ontology analysis.
The gene identifier to gene ontology (GO) identifier was obtained using UniProt.ws software. Gene ontology analysis was conducted using the goseq package, which is useful, because it considers the effect of variations of gene length, which is a relevant normalization criterion for RNA-Seq data. The Bioconductor package GO.db was used to obtain a detailed annotation of the GO identifiers. A BH-adjusted P value cutoff of 0.05 was used to identify differentially regulated GO terms.
Metabolomics analysis.
Sample preparation and analysis were performed as previously described (84) with minor modifications. After at least 1 week of incubation with aTC, logarithmically growing cultures (OD = 0.5) of the ppx2 knockdown and empty vector control strains were pelleted, and the samples were extracted in 1 ml of extraction buffer (chloroform-methanol, 2:1) and then concentrated by centrifugal evaporation. Samples were processed and analyzed by Metabolon, Inc. (Durham, NC). The nontargeted metabolic profiling instrumentation employed for this analysis combined three independent platforms: ultrahigh performance liquid chromatography-tandem mass spectrometry (UHPLC/MS/MS) optimized for basic species, UHPLC/MS/MS optimized for acidic species, and gas chromatography-mass spectrometry (GC-MS) (85, 86). Metabolites were identified by automated comparison of the ion features in the experimental samples to a reference library of chemical standards (87). For statistical analyses and data display purposes, any missing values were assumed to be below the limits of detection, and these values were inputted with the compound minimum. Statistical analysis of log-transformed data was performed using R (http://cran.r-project.org). Welch’s t tests were performed to compare data between experimental groups. Multiple comparisons were accounted for by estimating the false discovery rate (FDR) using q values (88). For correlation of metabolites and transcriptome data, we identified the metabolites based on Palsson’s network (41), and 70 metabolites have been mapped. The z scores of metabolite and enzymatic genes were calculated by the fold change ratio and P values. After the transcriptome was mapped to the metabolic network, the data were classified into major metabolic pathways.
Resazurin assay for intracellular NAD/NADH ratio.
Serial dilutions of mid-log-growth-phase M. tuberculosis cultures were incubated with resazurin (Invitrogen) for 16 to 18 h, and the fluorescence intensity was read by an BMG Optima microplate reader at 544-nm excitation and 590-nm emission wavelengths. The signal was corrected with the signal from negative controls (medium alone), and results yielding a linear relationship were used as representative data. The fluorescence intensity was further normalized to total CFU count.
Ethidium bromide accumulation/efflux assay and Nile red uptake assays.
The ethidium bromide accumulation and efflux assays were measured by florescence intensity (48, 89) with minor modifications. Briefly, mid-log-phase cultures were washed with PBS containing 0.05% Tween 80 (PBST) and then stained with 2 µg/ml ethidium bromide (Sigma). Ethidium bromide (1 µg/ml) was used for accumulation assays with efflux inhibitors, including chloropromazine (10 µg/ml; Sigma), verapamil (100 µg/ml; Sigma), reserpine (6 µg/ml; Sigma), or carbonyl cyanide m-chlorophenyl hydrazone (1 µg/ml; Sigma). For the ethidium bromide efflux assay, bacteria were washed with PBST and then incubated with 2 µg/ml ethidium and 100 µg/ml verapamil for 60 min. After the bacteria were washed twice with PBST, efflux activity was measured as the decay ratio of fluorescence intensity. For Nile red uptake staining, mid-log-phase cultures were washed with PBS and then stained with 20 µM Nile red (Sigma) (90). In all assays, the cells were incubated in 96-well plates, and analysis was performed at the indicated time points by excitation at 544 nm and emission at 590 nm on a FLUOstar OPTIMA microplate reader (BMG Labtech). All data were normalized to the time zero reading of each well. All experiments were repeated at least three times and similar results were obtained. Representative results are shown in Fig. 7A and B.
Biofilm formation assay and crystal violet staining.
Crystal violet staining was performed (51) with minor modifications. Briefly, mid-log-phase cultures (5 ml; density of 106/ml) were grown in 50-ml standing conical tubes containing Sauton’s medium (Himedia, India) without detergent for 5 weeks. The extracellular matrix of biofilm was measured by using crystal violet stain and a FLUOstar OPTIMA microplate reader (BMG Labtech).
Statistical analysis.
Data from at least three biological replicates were used to calculate means and standard deviation (SD) for graphing purposes. Statistical analysis employed the unpaired Student t test, and a P value of <0.05 was considered significant.
Microarray data accession number.
The RNA-seq data were deposited in the GEO database under accession no. GSE57868.
SUPPLEMENTAL MATERIAL
Recombinant Rv1026 exhibits greater poly(P) hydrolysis activity in the presence of diphosphate or monophosphate nucleotides. (A) Recombinant Rv1026 shows increased PPX activity when coincubated with ADP, AMP, GDP, and GMP (P < 0.05; n = 3 or 4). (B) The presence of ADP, AMP, GDP, or GMP promotes hydrolytic activity of recombinant Rv1026 in 8 h. Download
Confirmation of Rv1026/ppx2 knockdown strains. (A) Diagram of the Rv1026 knockdown plasmid. The backbone plasmid was pUV15tetOR, with cloning of attP and Int from pMH94. The Rv1026 gene was amplified by PCR from wild-type M. tuberculosis and then cloned in reverse orientation using the AclI and PacI sites. (B) PCR analysis of genomic DNA of empty vector control and ppx2 knockdown strains. Primer set 1 (forward primer [F], GCCGTTAATTAAGGGAGAACTCCCTATCA; reverse primer [R], GCTGACCGAACGCTGTTT) targets the Rv1026 knockdown segment, yielding a 658-bp product in the Rv1026 knockdown. Primer set 2 (F, CGCACTAACGTTCGTTTGTCAGCATCGAAAG; R, GCGCAATTGATCGGACCTACAACGACCAG) targets the plasmid, yielding a 1,694-bp product in all strains. The image was sliced for labeling. (C) Growth of the ppx2 knockdown strain in supplemented Middlebrook 7H9 broth containing different concentrations of aTC (*, P < 0.05; †, P < 0.001; n = 3). Download
ppx2 knock-down strain has a shorter longitudinal axis compared to the empty vector strain. (A) Transmission electron microscopy (TEM) of empty vector and ppx2 knock-down strains during mid-log growth phase. (B) The longitudinal axis of ppx2 knock-down was significantly shorter than that of the empty vector (P = 0.005). Download
Significantly differentially regulated genes in the ppx2 knockdown strain compared to the empty vector control strain
Gene ontology analysis of gene expression in the ppx2 knockdown strain compared to the empty vector control strain
Metabolomic analysis of the empty vector control strain and ppx2 knockdown strain
Correlation analysis of metabolomics and transcriptomics data during M. tuberculosis poly(P) accumulation
Primers used in the ppx2 knockdown study
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Heart, Lung, and Blood Institute and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health by grants R01 HL106786 to P.C.K. and J.S.B. and grant R01 AI083125 to P.C.K., respectively.
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Citation Chuang Y, Bandyopadhyay N, Rifat D, Rubin H, Bader JS, Karakousis PC. 2015. Deficiency of the novel exopolyphosphatase Rv1026/PPX2 leads to metabolic downshift and altered cell wall permeability in Mycobacterium tuberculosis. mBio 6(2):e02428-14. doi:10.1128/mBio.02428-14.
Contributor Information
Christina L. Stallings, Washington University in St Louis School of Medicine.
L. David Sibley, Washington University School of Medicine.
REFERENCES
- 1.Fauci AS, NIAID Tuberculosis Working Group . 2008. Multidrug-resistant and extensively drug-resistant tuberculosis: the National Institute of Allergy and Infectious Diseases Research agenda and recommendations for priority research. J Infect Dis 197:1493–1498. doi: 10.1086/587904. [DOI] [PubMed] [Google Scholar]
- 2.Lawn SD, Zumla AI. 2011. Tuberculosis. Lancet 378:57–72. doi: 10.1016/S0140-6736(10)62173-3. [DOI] [PubMed] [Google Scholar]
- 3.Cohen NR, Lobritz MA, Collins JJ. 2013. Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutte CC, Crosson S. 2013. Bacterial lifestyle shapes stringent response activation. Trends Microbiol 21:174–180. doi: 10.1016/j.tim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulaev I, Kulakovskaya T. 2000. Polyphosphate and phosphate pump. Annu Rev Microbiol 54:709–734. doi: 10.1146/annurev.micro.54.1.709. [DOI] [PubMed] [Google Scholar]
- 6.Rao NN, Gómez-García MR, Kornberg A. 2009. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem 78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- 7.Kornberg A, Rao NN, Ault-Riché D. 1999. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem 68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- 8.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 9.Sureka K, Dey S, Datta P, Singh AK, Dasgupta A, Rodrigue S, Basu J, Kundu M. 2007. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol Microbiol 65:261–276. doi: 10.1111/j.1365-2958.2007.05814.x. [DOI] [PubMed] [Google Scholar]
- 10.Thayil SM, Morrison N, Schechter N, Rubin H, Karakousis PC. 2011. The role of the novel exopolyphosphatase MT0516 in Mycobacterium tuberculosis drug tolerance and persistence. PLoS One 6:e28076. doi: 10.1371/journal.pone.0028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuang YM, Belchis DA, Karakousis PC. 2013. The polyphosphate kinase gene ppk2 is required for Mycobacterium tuberculosis inorganic polyphosphate regulation and virulence. mBio 4(3):e00039-13. doi: 10.1128/mBio.00039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avarbock A, Avarbock D, Teh JS, Buckstein M, Wang ZM, Rubin H. 2005. Functional regulation of the opposing (p)ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 44:9913–9923. doi: 10.1021/bi0505316. [DOI] [PubMed] [Google Scholar]
- 13.Avarbock D, Salem J, Li LS, Wang ZM, Rubin H. 1999. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene 233:261–269. doi: 10.1016/S0378-1119(99)00114-6. [DOI] [PubMed] [Google Scholar]
- 14.Sureka K, Sanyal S, Basu J, Kundu M. 2009. Polyphosphate kinase 2: a modulator of nucleoside diphosphate kinase activity in mycobacteria. Mol Microbiol 74:1187–1197. doi: 10.1111/j.1365-2958.2009.06925.x. [DOI] [PubMed] [Google Scholar]
- 15.Shum KT, Lui EL, Wong SC, Yeung P, Sam L, Wang Y, Watt RM, Tanner JA. 2011. Aptamer-mediated inhibition of Mycobacterium tuberculosis polyphosphate kinase 2. Biochemistry 50:3261–3271. doi: 10.1021/bi2001455. [DOI] [PubMed] [Google Scholar]
- 16.Choi MY, Wang Y, Wong LL, Lu BT, Chen WY, Huang JD, Tanner JA, Watt RM. 2012. The two PPX-GppA homologues from Mycobacterium tuberculosis have distinct biochemical activities. PLoS One 7:e42561. doi: 10.1371/journal.pone.0042561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsen MH, Vilchèze C, Kremer L, Besra GS, Parsons L, Salfinger M, Heifets L, Hazbon MH, Alland D, Sacchettini JC, Jacobs WR Jr.. 2002. Overexpression of inhA, but not kasA, confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis, M. bovis BCG and M. tuberculosis. Mol Microbiol 46:453–466. doi: 10.1046/j.1365-2958.2002.03162.x. [DOI] [PubMed] [Google Scholar]
- 18.Singh R, Singh M, Arora G, Kumar S, Tiwari P, Kidwai S. 2013. Polyphosphate deficiency in Mycobacterium tuberculosis is associated with enhanced drug susceptibility and impaired growth in guinea pigs. J Bacteriol 195:2839–2851. doi: 10.1128/JB.00038-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindner SN, Knebel S, Wesseling H, Schoberth SM, Wendisch VF. 2009. Exopolyphosphatases PPX1 and PPX2 from Corynebacterium glutamicum. Appl Environ Microbiol 75:3161–3170. doi: 10.1128/AEM.02705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamichhane G, Zignol M, Blades NJ, Geiman DE, Dougherty A, Grosset J, Broman KW, Bishai WR. 2003. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 100:7213–7218. doi: 10.1073/pnas.1231432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A 100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi T, Fu T, Xie J. 2011. Polyphosphate deficiency affects the sliding motility and biofilm formation of Mycobacterium smegmatis. Curr Microbiol 63:470–476. doi: 10.1007/s00284-011-0004-4. [DOI] [PubMed] [Google Scholar]
- 23.Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honoré N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies RM, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutherford KM, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward JR, Barrell BG. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 24.Kuroda A, Murphy H, Cashel M, Kornberg A. 1997. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem 272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 25.Wayne LG, Hayes LG. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun 64:2062–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rifat D, Bishai WR, Karakousis PC. 2009. Phosphate depletion: a novel trigger for Mycobacterium tuberculosis persistence. J Infect Dis 200:1126–1135. doi: 10.1086/605700. [DOI] [PubMed] [Google Scholar]
- 27.Guo XV, Monteleone M, Klotzsche M, Kamionka A, Hillen W, Braunstein M, Ehrt S, Schnappinger D. 2007. Silencing Mycobacterium smegmatis by using tetracycline repressors. J Bacteriol 189:4614–4623. doi: 10.1128/JB.00216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, Schnappinger D. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res 33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gill WP, Harik NS, Whiddon MR, Liao RP, Mittler JE, Sherman DR. 2009. A replication clock for Mycobacterium tuberculosis. Nat Med 15:211–214. doi: 10.1038/nm.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chien AC, Hill NS, Levin PA. 2012. Cell size control in bacteria. Curr Biol 22:R340–R349. doi: 10.1016/j.cub.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takayama K, Wang L, David HL. 1972. Effect of isoniazid on the in vivo mycolic acid synthesis, cell growth, and viability of Mycobacterium tuberculosis. Antimicrob Agents Chemother 2:29–35. doi: 10.1128/AAC.2.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winder FG, Collins PB. 1970. Inhibition by isoniazid of synthesis of mycolic acids in Mycobacterium tuberculosis. J Gen Microbiol 63:41–48. doi: 10.1099/00221287-63-1-41. [DOI] [PubMed] [Google Scholar]
- 33.Sanyal S, Banerjee SK, Banerjee R, Mukhopadhyay J, Kundu M. 2013. Polyphosphate kinase 1, a central node in the stress response network of Mycobacterium tuberculosis, connects the two-component systems MprAB and SenX3-RegX3 and the extracytoplasmic function sigma factor, sigma E. Microbiology 159:2074–2086. doi: 10.1099/mic.0.068452-0. [DOI] [PubMed] [Google Scholar]
- 34.Champion PA, Champion MM, Manzanillo P, Cox JS. 2009. ESX-1 secreted virulence factors are recognized by multiple cytosolic AAA ATPases in pathogenic mycobacteria. Mol Microbiol 73:950–962. doi: 10.1111/j.1365-2958.2009.06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garces A, Atmakuri K, Chase MR, Woodworth JS, Krastins B, Rothchild AC, Ramsdell TL, Lopez MF, Behar SM, Sarracino DA, Fortune SM. 2010. EspA acts as a critical mediator of ESX1-dependent virulence in Mycobacterium tuberculosis by affecting bacterial cell wall integrity. PLoS Pathog 6:e1000957. doi: 10.1371/journal.ppat.1000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Millington KA, Fortune SM, Low J, Garces A, Hingley-Wilson SM, Wickremasinghe M, Kon OM, Lalvani A. 2011. Rv3615c is a highly immunodominant RD1 (region of difference 1)-dependent secreted antigen specific for Mycobacterium tuberculosis infection. Proc Natl Acad Sci U S A 108:5730–5735. doi: 10.1073/pnas.1015153108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodríguez JG, Hernández AC, Helguera-Repetto C, Aguilar Ayala D, Guadarrama-Medina R, Anzóla JM, Bustos JR, Zambrano MM, Gonzalez YMJ, García MJ, Del Portillo P. 2014. Global adaptation to a lipid environment triggers the dormancy-related phenotype of Mycobacterium tuberculosis. mBio 5(3):e01125-14. doi: 10.1128/mBio.01125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padiadpu J, Vashisht R, Chandra N. 2010. Protein-protein interaction networks suggest different targets have different propensities for triggering drug resistance. Syst Synth Biol 4:311–322. doi: 10.1007/s11693-011-9076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramaswamy SV, Amin AG, Göksel S, Stager CE, Dou SJ, El Sahly H, Moghazeh SL, Kreiswirth BN, Musser JM. 2000. Molecular genetic analysis of nucleotide polymorphisms associated with ethambutol resistance in human isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother 44:326–336. doi: 10.1128/AAC.44.2.326-336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramaswamy SV, Reich R, Dou SJ, Jasperse L, Pan X, Wanger A, Quitugua T, Graviss EA. 2003. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 47:1241–1250. doi: 10.1128/AAC.47.4.1241-1250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jamshidi N, Palsson BØ. 2007. Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst Biol 1:26. doi: 10.1186/1752-0509-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CT, Lobritz MA, Braff D, Schwarz EG, Ye JD, Pati M, Vercruysse M, Ralifo PS, Allison KR, Khalil AS, Ting AY, Walker GC, Collins JJ. 2014. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A 111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gloeckner H, Jonuleit T, Lemke HD. 2001. Monitoring of cell viability and cell growth in a hollow-fiber bioreactor by use of the dye Alamar Blue. J Immunol Methods 252:131–138. doi: 10.1016/S0022-1759(01)00347-7. [DOI] [PubMed] [Google Scholar]
- 44.Nakayama GR, Caton MC, Nova MP, Parandoosh Z. 1997. Assessment of the Alamar Blue assay for cellular growth and viability in vitro. J Immunol Methods 204:205–208. doi: 10.1016/S0022-1759(97)00043-4. [DOI] [PubMed] [Google Scholar]
- 45.Louw GE, Warren RM, Gey van Pittius NC, McEvoy CR, Van Helden PD, Victor TC. 2009. A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob Agents Chemother 53:3181–3189. doi: 10.1128/AAC.01577-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szumowski JD, Adams KN, Edelstein PH, Ramakrishnan L. 2013. Antimicrobial efflux pumps and Mycobacterium tuberculosis drug tolerance: evolutionary considerations. Curr Top Microbiol Immunol 374:81–108. doi: 10.1007/82_2012_300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarathy JP, Dartois V, Lee EJ. 2012. The role of transport mechanisms in Mycobacterium tuberculosis drug resistance and tolerance. Pharmaceuticals (Basel) 5:1210–1235. doi: 10.3390/ph5111210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodrigues L, Machado D, Couto I, Amaral L, Viveiros M. 2012. Contribution of efflux activity to isoniazid resistance in the Mycobacterium tuberculosis complex. Infect Genet Evol 12:695–700. doi: 10.1016/j.meegid.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 49.Sani M, Houben EN, Geurtsen J, Pierson J, de Punder K, van Zon M, Wever B, Piersma SR, Jiménez CR, Daffé M, Appelmelk BJ, Bitter W, van der Wel N, Peters PJ. 2010. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog 6:e1000794. doi: 10.1371/journal.ppat.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambandan D, Dao DN, Weinrick BC, Vilchèze C, Gurcha SS, Ojha A, Kremer L, Besra GS, Hatfull GF, Jacobs WR Jr.. 2013. Keto-mycolic acid-dependent pellicle formation confers tolerance to drug-sensitive Mycobacterium tuberculosis. mBio 4(3):e00222-13. doi: 10.1128/mBio.00222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pang JM, Layre E, Sweet L, Sherrid A, Moody DB, Ojha A, Sherman DR. 2012. The polyketide Pks1 contributes to biofilm formation in Mycobacterium tuberculosis. J Bacteriol 194:715–721. doi: 10.1128/JB.06304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dutta NK, Karakousis PC. 2014. Latent tuberculosis infection: myths, models, and molecular mechanisms. Microbiol Mol Biol Rev 78:343–371. doi: 10.1128/MMBR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eoh H, Rhee KY. 2013. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 110:6554–6559. doi: 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nandakumar M, Nathan C, Rhee KY. 2014. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun 5:4306. doi: 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- 55.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amato SM, Fazen CH, Henry TC, Mok WW, Orman MA, Sandvik EL, Volzing KG, Brynildsen MP. 2014. The role of metabolism in bacterial persistence. Front Microbiol 5:70. doi: 10.3389/fmicb.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spoering AL, Vulic M, Lewis K. 2006. GlpD and PlsB participate in persister cell formation in Escherichia coli. J Bacteriol 188:5136–5144. doi: 10.1128/JB.00369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, McKinney JD, Bertozzi CR, Sassetti CM. 2012. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem Biol 19:218–227. doi: 10.1016/j.chembiol.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keren I, Minami S, Rubin E, Lewis K. 2011. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2(3):e00100-11. doi: 10.1128/mBio.00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marrero J, Trujillo C, Rhee KY, Ehrt S. 2013. Glucose phosphorylation is required for Mycobacterium tuberculosis persistence in mice. PLoS Pathog 9:e1003116. doi: 10.1371/journal.ppat.1003116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baek SH, Li AH, Sassetti CM. 2011. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol 9:e1001065. doi: 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. 2011. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog 7:e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fisher MA, Plikaytis BB, Shinnick TM. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J Bacteriol 184:4025–4032. doi: 10.1128/JB.184.14.4025-4032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rohde KH, Abramovitch RB, Russell DG. 2007. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2:352–364. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 65.Tunpiboonsak S, Mongkolrob R, Kitudomsub K, Thanwatanaying P, Kiettipirodom W, Tungboontina Y, Tungpradabkul S. 2010. Role of a Burkholderia pseudomallei polyphosphate kinase in an oxidative stress response, motilities, and biofilm formation. J Microbiol 48:63–70. doi: 10.1007/s12275-010-9138-5. [DOI] [PubMed] [Google Scholar]
- 66.Chen W, Palmer RJ, Kuramitsu HK. 2002. Role of polyphosphate kinase in biofilm formation by Porphyromonas gingivalis. Infect Immun 70:4708–4715. doi: 10.1128/IAI.70.8.4708-4715.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rashid MH, Rumbaugh K, Passador L, Davies DG, Hamood AN, Iglewski BH, Kornberg A. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97:9636–9641. doi: 10.1073/pnas.170283397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grillo-Puertas M, Villegas JM, Rintoul MR, Rapisarda VA. 2012. Polyphosphate degradation in stationary phase triggers biofilm formation via LuxS quorum sensing system in Escherichia coli. PLoS One 7:e50368. doi: 10.1371/journal.pone.0050368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi X, Rao NN, Kornberg A. 2004. Inorganic polyphosphate in Bacillus cereus: motility, biofilm formation, and sporulation. Proc Natl Acad Sci U S A 101:17061–17065. doi: 10.1073/pnas.0407787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sarathy J, Dartois V, Dick T, Gengenbacher M. 2013. Reduced drug uptake in phenotypically resistant nutrient-starved nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:1648–1653. doi: 10.1128/AAC.02202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bisson GP, Mehaffy C, Broeckling C, Prenni J, Rifat D, Lun DS, Burgos M, Weissman D, Karakousis PC, Dobos K. 2012. Upregulation of the phthiocerol dimycocerosate biosynthetic pathway by rifampin-resistant, rpoB mutant Mycobacterium tuberculosis. J Bacteriol 194:6441–6452. doi: 10.1128/JB.01013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dahl JL, Arora K, Boshoff HI, Whiteford DC, Pacheco SA, Walsh OJ, Lau-Bonilla D, Davis WB, Garza AG. 2005. The relA homolog of Mycobacterium smegmatis affects cell appearance, viability, and gene expression. J Bacteriol 187:2439–2447. doi: 10.1128/JB.187.7.2439-2447.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dahl JL, Kraus CN, Boshoff HI, Doan B, Foley K, Avarbock D, Kaplan G, Mizrahi V, Rubin H, Barry CE III. 2003. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A 100:10026-10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cunningham AF, Spreadbury CL. 1998. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton alpha-crystallin homolog. J Bacteriol 180:801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kieser KJ, Rubin EJ. 2014. How sisters grow apart: mycobacterial growth and division. Nat Rev Microbiol 12:550–562. doi: 10.1038/nrmicro3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomasz A, Albino A, Zanati E. 1970. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 227:138–140. doi: 10.1038/227138a0. [DOI] [PubMed] [Google Scholar]
- 77.Karakousis PC, Yoshimatsu T, Lamichhane G, Woolwine SC, Nuermberger EL, Grosset J, Bishai WR. 2004. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med 200:647–657. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aschar-Sobbi R, Abramov AY, Diao C, Kargacin ME, Kargacin GJ, French RJ, Pavlov E. 2008. High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J Fluoresc 18:859–866. doi: 10.1007/s10895-008-0315-4. [DOI] [PubMed] [Google Scholar]
- 79.Thayil SM, Morrison N, Schechter N, Rubin H, Karakousis PC. 2011. The role of the novel exopolyphosphatase MT0516 in Mycobacterium tuberculosis drug tolerance and persistence. PLoS One 6:e28076. doi: 10.1371/journal.pone.0028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen P, Ruiz RE, Li Q, Silver RF, Bishai WR. 2000. Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene, sigF. Infect Immun 68:5575–5580. doi: 10.1128/IAI.68.10.5575-5580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee MH, Pascopella L, Jacobs WR Jr, Hatfull GF. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and Bacille Calmette-Guerin. Proc Natl Acad Sci U S A 88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Manganelli R, Voskuil MI, Schoolnik GK, Smith I. 2001. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol Microbiol 41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- 83.Rhoades ER, Orme IM. 1997. Susceptibility of a panel of virulent strains of Mycobacterium tuberculosis to reactive nitrogen intermediates. Infect Immun 65:1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, Sweet L, Gomes A, Rustad T, Dolganov G, Glotova I, Abeel T, Mahwinney C, Kennedy AD, Allard R, Brabant W, Krueger A, Jaini S, Honda B, Yu WH, Hickey MJ, Zucker J, Garay C, Weiner B, Sisk P, Stolte C, Winkler JK, Van de Peer Y, Iazzetti P, Camacho D, Dreyfuss J, Liu Y, Dorhoi A, Mollenkopf HJ, Drogaris P, Lamontagne J, Zhou Y, Piquenot J, Park ST, Raman S, Kaufmann SH, Mohney RP, Chelsky D, Moody DB, Sherman DR, Schoolnik GK. 2013. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499:178–183. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boudonck KJ, Mitchell MW, Német L, Keresztes L, Nyska A, Shinar D, Rosenstock M. 2009. Discovery of metabolomics biomarkers for early detection of nephrotoxicity. Toxicol Pathol 37:280–292. doi: 10.1177/0192623309332992. [DOI] [PubMed] [Google Scholar]
- 86.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. 2009. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 87.Dehaven CD, Evans AM, Dai H, Lawton KA. 2010. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform 2:9. doi: 10.1186/1758-2946-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100:9440-9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rodrigues L, Ramos J, Couto I, Amaral L, Viveiros M. 2011. Ethidium bromide transport across Mycobacterium smegmatis cell-wall: correlation with antibiotic resistance. BMC Microbiol 11:35. doi: 10.1186/1471-2180-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu WX, Zhang L, Mai JT, Peng RC, Yang EZ, Peng C, Wang HH. 2014. The Wag31 protein interacts with AccA3 and coordinates cell wall lipid permeability and lipophilic drug resistance in Mycobacterium smegmatis. Biochem Biophys Res Commun 448:255–260. doi: 10.1016/j.bbrc.2014.04.116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Recombinant Rv1026 exhibits greater poly(P) hydrolysis activity in the presence of diphosphate or monophosphate nucleotides. (A) Recombinant Rv1026 shows increased PPX activity when coincubated with ADP, AMP, GDP, and GMP (P < 0.05; n = 3 or 4). (B) The presence of ADP, AMP, GDP, or GMP promotes hydrolytic activity of recombinant Rv1026 in 8 h. Download
Confirmation of Rv1026/ppx2 knockdown strains. (A) Diagram of the Rv1026 knockdown plasmid. The backbone plasmid was pUV15tetOR, with cloning of attP and Int from pMH94. The Rv1026 gene was amplified by PCR from wild-type M. tuberculosis and then cloned in reverse orientation using the AclI and PacI sites. (B) PCR analysis of genomic DNA of empty vector control and ppx2 knockdown strains. Primer set 1 (forward primer [F], GCCGTTAATTAAGGGAGAACTCCCTATCA; reverse primer [R], GCTGACCGAACGCTGTTT) targets the Rv1026 knockdown segment, yielding a 658-bp product in the Rv1026 knockdown. Primer set 2 (F, CGCACTAACGTTCGTTTGTCAGCATCGAAAG; R, GCGCAATTGATCGGACCTACAACGACCAG) targets the plasmid, yielding a 1,694-bp product in all strains. The image was sliced for labeling. (C) Growth of the ppx2 knockdown strain in supplemented Middlebrook 7H9 broth containing different concentrations of aTC (*, P < 0.05; †, P < 0.001; n = 3). Download
ppx2 knock-down strain has a shorter longitudinal axis compared to the empty vector strain. (A) Transmission electron microscopy (TEM) of empty vector and ppx2 knock-down strains during mid-log growth phase. (B) The longitudinal axis of ppx2 knock-down was significantly shorter than that of the empty vector (P = 0.005). Download
Significantly differentially regulated genes in the ppx2 knockdown strain compared to the empty vector control strain
Gene ontology analysis of gene expression in the ppx2 knockdown strain compared to the empty vector control strain
Metabolomic analysis of the empty vector control strain and ppx2 knockdown strain
Correlation analysis of metabolomics and transcriptomics data during M. tuberculosis poly(P) accumulation
Primers used in the ppx2 knockdown study