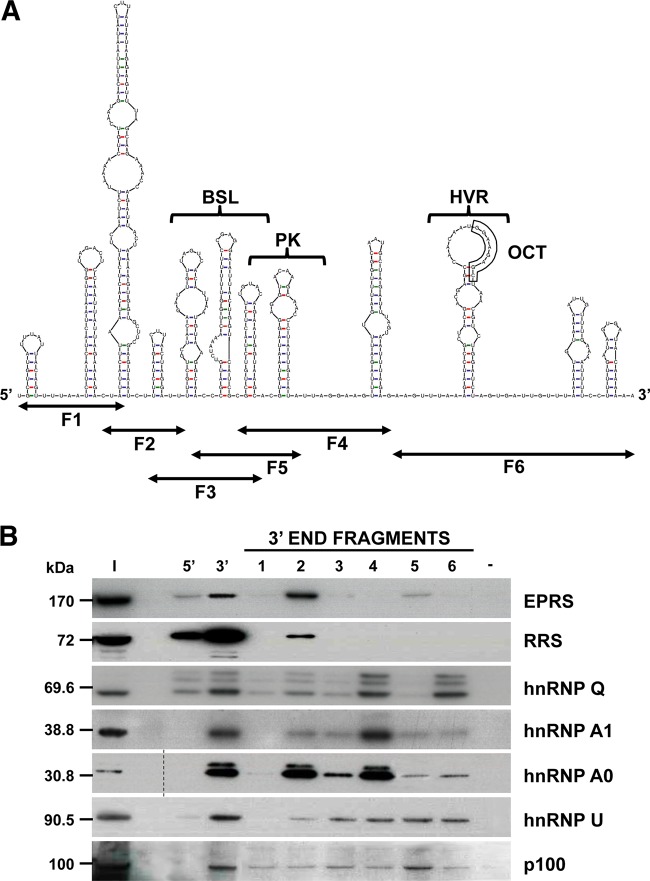
FIG 2 .
Mapping of the RNA domains interacting with the 3′-end-interacting cellular proteins. (A) Predicted secondary structure of the 493 nt from the TGEV genome 3′ end, obtained using the Mfold 3.4 software (49). The sequence covered by the six overlapping fragments (F1 to F6) and the higher-order RNA structures similar to the bulge stem-loop (BSL), the pseudoknot (PK), the hypervariable region (HVR), and the conserved octanucleotide (OCT) are indicated. (B) Identification of the 3′-end domains interacting with the selected cellular proteins. Proteins (600 µg) from cytoplasmic extracts of TGEV-infected Huh-7 cells were pulled down with the six overlapping fragments (1 to 6) covering the last 493 nt of the TGEV genome or with TGEV genome ends 5′ and 3′ without poly(A) or without RNA (−) as specificity controls. The pulled down proteins were separated in SDS-PAGE gels together with 50 µg of the initial protein extract (I) and analyzed by immunoblotting with specific antibodies against EPRS, RRS, hnRNP Q, hnRNP A1, hnRNP A0, hnRNP U, and p100. To simplify the information of the figure, an empty lane was removed in the hnRNP A0 panel, in which the dashed line indicates the splicing site. The molecular masses in kDa of the analyzed proteins are indicated at the left.