ABSTRACT
The surface capsular polysaccharide (CP) is a virulence factor that has been used as an antigen in several successful vaccines against bacterial pathogens. A vaccine has not yet been licensed against Staphylococcus aureus, although two multicomponent vaccines that contain CP antigens are in clinical trials. In this study, we evaluated CP production in USA300 methicillin-resistant S. aureus (MRSA) isolates that have become the predominant community-associated MRSA clones in the United States. We found that all 167 USA300 MRSA and 50 USA300 methicillin-susceptible S. aureus (MSSA) isolates were CP negative (CP−). Moreover, all 16 USA500 isolates, which have been postulated to be the progenitor lineage of USA300, were also CP−. Whole-genome sequence analysis of 146 CP− USA300 MRSA isolates revealed they all carry a cap5 locus with 4 conserved mutations compared with strain Newman. Genetic complementation experiments revealed that three of these mutations (in the cap5 promoter, cap5D nucleotide 994, and cap5E nucleotide 223) ablated CP production in USA300 and that Cap5E75 Asp, located in the coenzyme-binding domain, is essential for capsule production. All but three USA300 MSSA isolates had the same four cap5 mutations found in USA300 MRSA isolates. Most isolates with a USA500 pulsotype carried three of these four USA300-specific mutations, suggesting the fourth mutation occurred in the USA300 lineage. Phylogenetic analysis of the cap loci of our USA300 isolates as well as publicly available genomes from 41 other sequence types revealed that the USA300-specific cap5 mutations arose sequentially in S. aureus in a common ancestor of USA300 and USA500 isolates.
IMPORTANCE
The USA300 MRSA clone emerged as a community-associated pathogen in the United States nearly 20 years ago. Since then, it has rapidly disseminated and now causes health care-associated infections. This study shows that the CP-negative (CP−) phenotype has persisted among USA300 isolates and is a universal and characteristic trait of this highly successful MRSA lineage. It is important to note that a vaccine consisting solely of CP antigens would not likely demonstrate high efficacy in the U.S. population, where about half of MRSA isolates comprise USA300. Moreover, conversion of a USA300 strain to a CP-positive (CP+) phenotype is unlikely in vivo or in vitro since it would require the reversion of 3 mutations. We have also established that USA300 MSSA isolates and USA500 isolates are CP− and provide new insight into the evolution of the USA300 and USA500 lineages.
INTRODUCTION
Staphylococcus aureus is an important pathogen that causes a wide range of infections in health care and community settings. Methicillin-resistant S. aureus (MRSA) isolates in particular, which have become increasingly prevalent in the last decade, are resistant to nearly all β-lactams and are often multiply resistant to several classes of antibiotics. A vaccine that could protect against S. aureus infection would be important for public health, although development of an effective vaccine has remained elusive (1, 2).
Capsular polysaccharides (CPs) envelope the surface of many bacterial pathogens and have been the primary or sole protective antigen used in vaccines that are effective against certain serotypes of Streptococcus pneumoniae, Haemophilus influenzae, and Neisseria meningitidis (serotypes A, CW, and Y) (3). Among encapsulated S. aureus isolates, serotypes 5 and 8 prevail (4–9). Capsular polysaccharides 5 (CP5) and 8 (CP8) have similar trisaccharide repeating units but differ in the linkages between the sugars and the sites of O-acetylation of the monosaccharide residues (10). Previous reports have indicated that ~20% of S. aureus isolates fail to produce CP5 or CP8 (4–9).
The S. aureus CP protects the bacterium from host phagocytes (2); however, this protection can be overcome by CP-specific antibodies that enhance opsonophagocytic killing by human neutrophils (10, 11). Vaccines that contained CP5 and CP8 antigens conjugated to Pseudomonas aeruginosa exotoxoid A were tested for efficacy in patients with end-stage renal disease. In phase III clinical trials, these CP-based vaccines failed to protect against S. aureus bacteremia (12–14).
In the United States, the community-associated (CA)-MRSA clonal type USA300 has been the most frequently isolated S. aureus genotype from all body sites, including blood, surpassing the USA100 health care-associated MRSA strain type as a cause of nosocomial infection in some locales (15). USA300 MRSA strains are most often associated with CA skin and soft tissue infection (SSTI) (16) and belong to multilocus sequence type 8 (ST8). USA300 MRSA isolates characteristically contain the SCCmec type IV element (17), a phage carrying the genes encoding Panton-Valentine leukocidin (PVL) (18) and the arginine catabolic mobile genetic element (ACME) carrying the arginine deiminase cluster and the gene encoding the spermidine resistance factor, SpeG, which promotes skin colonization (19–21). Although they usually lack SCCmec IV, ACME, and PVL, USA500 MRSA isolates are the closest relative of USA300 among the members of the ST8 lineage and have been postulated to be the progenitor of USA300 (21).
We previously demonstrated that neither CP5 nor CP8 was produced by several USA300 MRSA clinical isolates obtained in 2004 and 2005 from our center in Chicago, IL (22). A subsequent study of isolates obtained during the same time period (2004 to 2005) from Washington, DC, also reported CP-negative (CP−) USA300 MRSA strains (23). These studies suggested that the failure to produce a CP was a common trait among USA300 MRSA isolates circulating in 2004 through 2005, but this was not investigated subsequently.
A decade has passed since the CP− USA300 MRSA isolates were described. Since then, USA300 MRSA strains have widely disseminated and increased in prevalence in the United States (15). It is not known if there are CP5-positive (CP5+) USA300 variants in circulation or if the CP− USA300 MRSA isolates identified in 2004 have become the sole or dominant USA300 clone.
Because CP5 and CP8 are components in two S. aureus vaccines under development (1, 24, 25), the aim of this study was to determine if currently circulating USA300 MRSA isolates produce a CP. Further, we evaluated whether the CP− phenotype was shared by USA300 methicillin-susceptible S. aureus (MSSA) isolates and the closely related USA500 lineage (21, 26). Since we found that all USA300 and USA500 isolates were uniformly CP−, we analyzed the evolutionary relationship between the cap5 mutations in USA300 and USA500 isolates.
(This work was presented in part at the International Symposium on Staphylococci and Staphylococcal Infections, Chicago, IL, 2014.)
RESULTS
CP serotyping.
We chose for CP serotyping 233 clinical S. aureus isolates from our collection that were identified as either USA300 MRSA, USA300 MSSA, or USA500. Isolates were obtained from either colonized or infected body sites from subjects in Chicago, IL, Los Angeles, CA, San Francisco, CA, or Connecticut between 1995 and 2011, as described in Table 1. Clonal types consisted of (i) 167 USA300 MRSA isolates, (ii) 50 USA300 MSSA isolates, (iii) 14 USA500 MRSA isolates, and (iv) 2 USA500 MSSA isolates. The USA300 sample included 16 ACME arcA-negative USA300 MRSA isolates and 38 ACME arcA-negative USA300 MSSA isolates (as detailed in Table S1 in the supplemental material), since the absence of ACME arcA is atypical compared with other USA300 strains, and these isolates might exhibit altered CP phenotypes. Table S1 provides the typing characteristics of USA300 MSSA isolates, USA500 isolates, and ACME arcA-negative USA300 isolates.
TABLE 1.
Demographics of USA300 and USA500 clinical isolates
| Parameter | No. of isolates |
|||||
|---|---|---|---|---|---|---|
| USA300 |
USA500 |
|||||
| MRSA | MSSA | Total | MRSA | MSSA | Total | |
| Colonization vs infection | ||||||
| Colonization | 134 | 20 | 154 | 0 | 0 | 0 |
| Infection | 33 | 30 | 63 | 14 | 2 | 16 |
| Total | 167 | 50 | 217 | 14 | 2 | 16 |
| Geographic source | ||||||
| Chicago | 83 | 45 | 128 | 12 | 0 | 12 |
| Los Angeles | 84 | 3 | 87 | 1 | 0 | 1 |
| San Francisco | 0 | 2 | 2 | 0 | 2 | 2 |
| Connecticut | 0 | 0 | 0 | 1a | 0 | 1 |
| Total | 167 | 50 | 217 | 14 | 2 | 16 |
| Yr of isolation | ||||||
| 1995 | 1 | 0 | 1 | 0 | 0 | 0 |
| 1996 | 0 | 0 | 0 | 5 | 0 | 5 |
| 1997 | 0 | 0 | 0 | 2 | 0 | 2 |
| 2004 | 1 | 0 | 1 | 0 | 0 | 0 |
| 2008 | 37 | 0 | 37 | 5 | 0 | 5 |
| 2009 | 92 | 17 | 109 | 0 | 2 | 2 |
| 2010 | 33 | 21 | 54 | 0 | 0 | 0 |
| 2011 | 3 | 12 | 15 | 0 | 0 | 0 |
| Unknown | 0 | 0 | 0 | 2 | 0 | 1 |
| Total | 167 | 50 | 217 | 14 | 2 | 16 |
This isolate is the USA500 reference strain NRS385.
USA300 MRSA and MSSA isolates.
Capsule production was evaluated in 167 USA300 MRSA isolates. Although all USA300 MRSA strains tested carried a cap5 locus, all were nonreactive to CP5 polyclonal antiserum, whereas the CP5+ control strains Reynolds and Newman produced a signal (Fig. 1). Figure 1 also shows the nonreactivity to CP5 antiserum of the CP8+ strains, ST80 and MN8, as well as the CP− strains NCTC 8325-4 (27) and USA300 strain LAC. All 9 MRSA isolates that had a pulsotype consistent with USA300 but lacked ACME arcA were also CP−. USA300 MRSA isolates were nonreactive with CP8 polyclonal antiserum, as expected, since they do not encode a cap8 locus (data not shown). All 50 USA300 MSSA isolates tested were CP−.
FIG 1 .
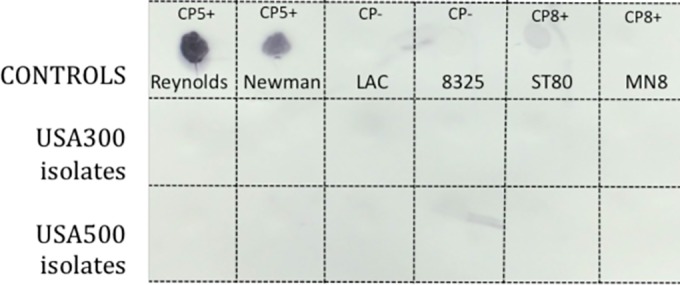
CP immunoblots of USA300 and USA500 clinical isolates. Capsule serotyping was performed by a colony immunoblot method with the use of CP5-specific antibodies as described previously (43). The first row shows the results from the CP5-positive (CP5+) control strains Reynolds and Newman, the CP5- and CP8-negative (CP−) control strains LAC and 8325, and the CP8+ control strains ST80 and MN8. The second and third rows include six representative USA300 and USA500 clinical isolates, respectively.
To address the possibility that USA300 strains might produce CP5 in vivo but not in vitro, we performed experiments similar to those described previously (28, 29). Both strain Newman and the USA300 MRSA strain CDC3 reacted with antibodies raised to killed acapsular S. aureus cells (see Fig. S1 in the supplemental material). The control CP5+ strain Newman produced CP5 as visualized by the blue-staining Alexa Fluor 660-tagged secondary antibodies reacting with CP5 antibodies. In contrast, neither the inoculum of USA300 MRSA strain CDC3 nor the CDC3 bacteria isolated from mouse blood 6 h after bacterial challenge showed detectable reactivity with the CP5 antibodies. Similar results were obtained with samples of blood from mice challenged with USA300 strain LAC. Thus, our results show no evidence that USA300 MRSA isolates produce CP5 in vivo.
USA500 isolates.
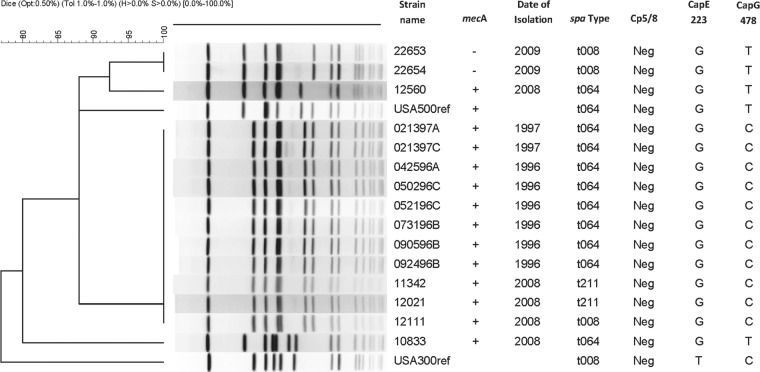
Due to the close phylogenetic relationship between USA500 and USA300 isolates (21, 30), we hypothesized that USA500 isolates might also be CP5−. Figure 2 documents the pulsed-field gel electrophoresis (PFGE) patterns and relevant typing characteristics of the 16 USA500 isolates we tested for CP production. The USA500 isolates belonged to ST8, carried SCCmec type IV, and lacked ACME arcA (26, 30, 31) (see Table S1 in the supplemental material). However, most of these USA500 isolates carried pvl genes, which is atypical for USA500. The USA500 isolates belonged to three spa types: t064 (11 of 16 isolates), t008 (n = 3), or t211 (n = 2) (see Table S1). All 16 USA500 isolates were CP− as represented in a panel in Fig. 1.
FIG 2 .
Pulsed-field gel electrophoresis (PFGE) documenting the SmaI digestion patterns of USA500 isolates relative to the USA500 reference strain NRS385 (USA500ref) and relevant typing characteristics. The USA300 reference strain (USA300ref) has PFGE pattern USA300-0114 and was obtained from the Network for Antimicrobial Resistance in Staphylococcus aureus (NARSA). “Cp5/8” refers to capsule polysaccharide types 5 and 8, “CapE 223” refers to the nucleotide at position 223 in cap5E, and “CapG 478” refers to the nucleotide at position 478 in cap5G.
DNA sequence analysis of cap loci of USA300 MRSA isolates.
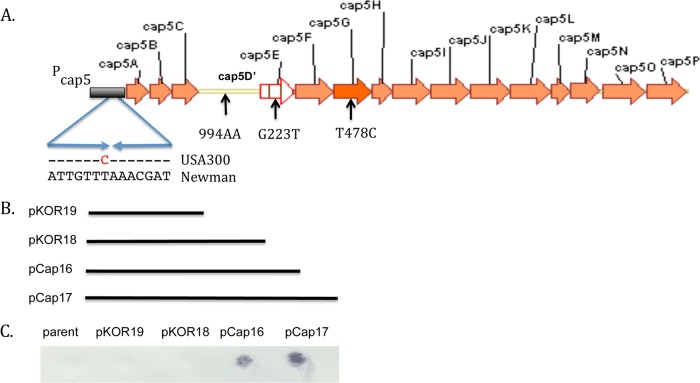
To evaluate the genetic basis for the CP5− phenotype, we evaluated the cap loci from assembled whole-genome shotgun data from 146 CP− USA300 MRSA isolates. Each strain carried an intact cap5 locus but had 4 identical point mutations relative to the CP5+ reference strain Newman (Fig. 3A).
FIG 3 .
USA300 cap5 locus showing universal mutations in USA300 relative to the cap5 reference strain Newman. Orange arrows depict open reading frames (ORFs). “Pcap5” refers to the cap5 promoter region. The inverted repeat sequence, as reported by Ouyang et al. (32), is shown as opposing arrows beneath Pcap5 indicating the T→C mutation in strain USA300 relative to strain Newman. The yellow line indicates a disruption of the cap5D ORF due to insertion of an A at nt 994 in USA300 (994AA). The cap5E G→T mutation at nt 223 (G223T) and the cap5G T→C mutation at nt 478 (T478C) are indicated beneath each ORF. (B) Lines beneath the map indicate the cap5 regions from strain Newman that are cloned in the complementation plasmids pKOR19 (cap5ABC plus truncated cap5D), pKOR18 (cap5ABCD plus a portion of cap5E), pCap16 (cap5ABCDE), and pCap17 (which contains cap5ABCDEF). (C) CP5 serotyping results from USA300 MRSA strain 923 transformants harboring the cap5 complementation plasmids depicted in panel B.
The cap5 promoter had a T→C replacement 73 bp upstream (−73) from the ATG translation start codon of cap5A (Pcap5 −73). This was located in the cap5 promoter in an inverted repeat (Fig. 3) known to be essential for transcription of capABCDEF (27, 32). We also identified a frameshift mutation within a polyadenine (AA) tract that begins at nucleotide (nt) 1006 of cap5D in strain Newman. This corresponds to nt 994 in cap5D of the USA300 reference strain because the annotations for the start codon of strain Newman and USA300 differ by four codons. This segment contains six A’s in the wild-type (wt) cap5D gene (strain Newman) and seven A’s in USA300 MRSA isolates, resulting in premature termination during translation (after 338 residues of the full-length Newman protein and 334 residues in USA300). There was also a G→T mutation at position 223 of cap5E (cap5E-223), which converts Asp to Tyr in the encoded protein at codon 75 (Asp75Tyr) and a T→C mutation in cap5G-478, resulting in a Phe160Leu conversion in the encoded protein. Additional nonconserved mutations were found in various cap5 genes among USA300 isolates (data not shown) (33). The mutations in USA300 at cap5E-223 and cap5G-478 have not been previously recognized in USA300 isolates, whereas the mutations in Pcap5 and cap5D were identified previously in three USA300 MRSA isolates from 2004 to 2005 (22).
cap5 loci in publicly available USA300 S. aureus genome sequences.
To confirm that the USA300-specific mutations were not confined to the Chicago and Los Angeles locales, we also analyzed 319 previously published USA300 isolate sequences from San Diego, CA (n = 35), and New York (n = 277). (Annotations of the included isolates are outlined in Table S2 in the supplemental material.) All but one isolate had all 4 cap5 mutations identical to those in our USA300 collection.
cap5 mutations among USA300 arcA-negative MRSA and USA300 MSSA isolates.
We performed Sanger sequencing of PCR products amplified from the cap5 locus to determine if the four cap5 mutations identified in USA300 MRSA isolates were also conserved among 9 atypical USA300 MRSA isolates that were ACME arcA negative and 19 USA300 MSSA isolates (Table 2). All 4 USA300-specific mutations were conserved among 8 of 9 (89%) ACME arcA-negative USA300 MRSA isolates tested (Table 2). One USA300 MRSA outlier (isolate 111395F) had only 2 of the 4 cap mutations (Pcap5 −73 and cap5D-994). However, this isolate was obtained in 1995 and is not typical of recent USA300 MRSA isolates. Although isolate 111395F had a SmaI digestion pattern similar to that of a USA300 isolate, it had features consistent with USA500 isolates, including spa type t064, ACME arcA, and PVL. However, strain 111395F also had features atypical for USA500, including carriage of SCCmec II.
TABLE 2 .
Summary of conserved mutations in cap5 loci detected in USA300 and USA500 isolates in our collectiona
| Gene | nt position | nt in: |
Codon (aa) in: |
Codon position | nt (no./total) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain Newman | USA300 | Strain Newman | USA300 | USA300 |
USA500 |
||||||
| MRSA ACME arcA+ (n = 155)b | MRSA ACME arcA− (n = 9)c | MSSA (n = 19)c | MRSA (n = 14)c | MSSA (n = 2)c | |||||||
| Pcap5d | −73e | T | C | NAf | NA | NA | C (155/155) | C (9/9) | C (19/19) | C (14/14) | C (2/2) |
| cap5D | 994g | A | AA | AAA (Lys) | STOP | 338 | AA (155/155) | AA (9/9) | AA (19/19) | AA (14/14) | AA (2/2) |
| cap5E | 223 | G | T | GAT (Asp) | TAT (Tyr) | 75 | T (155/155) | T (8/9) | T (16/19) | G (14/14) | G (2/2) |
| G (1/9) | G (3/19) | ||||||||||
| cap5G | 478 | T | C | TTC (Phe) | CTC (Leu) | 160 | C (155/155) | C (8/9) | C (10/19) | C (11/14) | T (2/2) |
| T (1/9) | T (3/14) | ||||||||||
SNPs of clinical isolates subjected to whole-genome sequencing or Sanger sequencing of PCR products as described in the text.
USA300 ACME arcA+ MRSA isolates were characterized by PFGE or by molecular typing as described in Materials and Methods.
Details of the molecular typing characteristics are provided in Table S1 in the supplemental material.
Pcap5, promoter region of the cap5 operon.
Seventy-three nucleotides upstream of the cap5A ATG translation initiation codon.
NA, not applicable since the polymorphism is in an intergenic region.
cap5D nt 994 in the USA300 reference strain corresponds to cap5D nt 1006 in strain Newman.
Among the USA300 MSSA isolates tested, 16 of 19 (84.2%) had all 4 USA300-specific mutations in Pcap5, cap5D-994, cap5E-223, and cap5G-478 (Table 2; see Table S1 in the supplemental material). Three outlier MSSA isolates had just three of the USA300-specific mutations (Pcap5, cap5D-994, and cap5G-478).
USA500 isolates.
Among USA500 isolates, the four USA300 cap5 mutations described above were common but not universal. By Sanger sequencing, we found that 11 of the 14 (78%) USA500 MRSA isolates had 3 of the 4 USA300-specific mutations (Pcap5 −73, cap5D-994, and capG-478), and none had the G→T mutation at cap5E-223. Three outlier USA500 MRSA isolates and both USA500 MSSA isolates had only 2 of the 4 mutations (Pcap5 −73 and cap5D-993) (Table 2), but these are each sufficient to yield a CP− phenotype (27).
Analysis of USA300-specific cap5 mutations in S. aureus phylogenies.
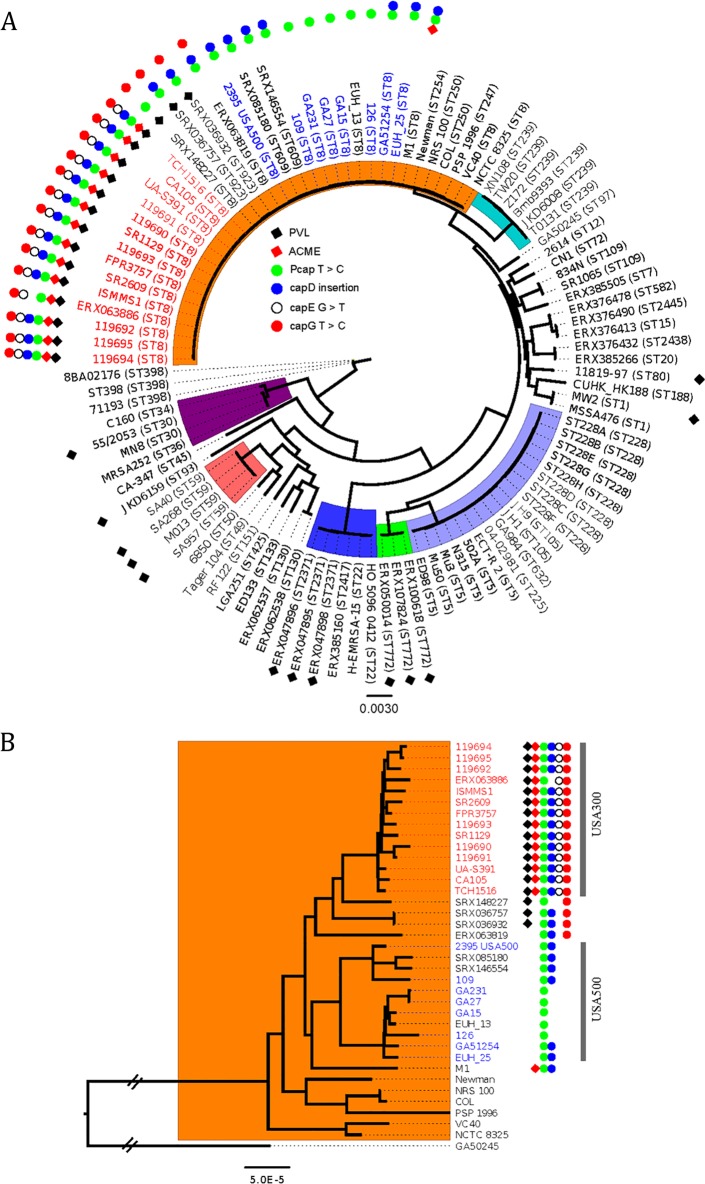
To evaluate the appearance of the cap5 mutations on the core phylogeny of S. aureus, we analyzed the USA300-specific cap5 mutations in 319 publicly available USA300 S. aureus genome sequences, 146 USA300 MRSA genomes in this study, and 90 non-USA300 genomes from 41 STs (Fig. 4). This allowed us to propose that the USA300 cap5 mutations evolved in a stepwise fashion. Moreover, these data suggest that USA300 and USA500 appear to have emerged from a common ancestor, and the Pcap5 T→C mutation likely occurred in the last common ancestor (LCA) of all USA300 and USA500 strains. The cap5D insertion may also have occurred in the LCA of both USA300 and USA500 but may have reverted back to wt in a recently emerged branch within USA500 (exemplified by strains GA231 and GA27). The cap5G T→C mutation had its origins in an ancestor of all USA300 strains that was also a progenitor of a clade of strains falling between the classical USA300 and USA500 patterns.
FIG 4 .
A maximum likelihood phylogeny based on whole-genome alignments of publicly available S. aureus genomes. (A) Phylogenetic analysis of 104 S. aureus strains from 41 STs showing the distribution of conserved mutations in cap5 in USA300 (Table 2). There are 14 USA300 strains in the tree that are representative of 146 strains from our study and 319 publicly available USA300 genomes. The remaining 90 genomes are from non-USA300 isolates. Strain names are shown as either the common name or the accession number followed by the sequence type (ST) in parentheses. Strains with red and blue text, respectively, are previously confirmed USA300 and USA500 isolates. The clade in orange is clonal complex 8 (CC8). Other common CCs are also highlighted. For simplicity, only a representative strain from each ST with the same pattern of cap5 mutations was included. Carriage of PVL and ACME is indicated with black and red diamonds, respectively, due to their association with the USA300 subclone. cap5 mutations are shown as solid or open circles as indicated. (B) High-resolution phylogeny of CC8 strains (a subset from panel A). USA300 and USA500 strains are annotated as in panel A.
Our analysis also suggests that the isolates with ST609 and ST623 are intermediate between USA300 and USA500 and might have a common ancestry. The conserved cap5 mutations that we describe were not present in any of the other 20 clonal complexes and were not in all ST8 strains. Thus, these mutations are unique to USA300 and its close relatives USA500, ST609, and ST623.
Complementation studies to test the phenotypic effect of cap5 gene mutations.
To determine the phenotypic relevance of the cap5 single nucleotide polymorphisms (SNPs) in strain USA300, we tested whether expression of wild-type cap5 genes in trans on a complementation plasmid could restore CP5 production to USA300 MRSA strain 923. The cap5 genes included in each complementation plasmid are depicted in Fig. 3B. Transformation of USA300 strain 923 with pKOR19 (expressing wt cap5ABC) or pKOR18 (expressing wt capABCD) was not sufficient to restore the CP5+ phenotype (Fig. 3C). This was not due to mutations in the cloned cap5 promoter or cap5D, as confirmed by DNA sequence analysis. In contrast, transformation of USA300 strain 923 with pCap16 (expressing wt cap5ABCDE) or pCap17 (wt cap5ABCDEF genes) restored CP5 production (Fig. 3C). These data indicate that the mutations in cap5D and Pcap5 in USA300 and USA500 strains ablate CP production. These data also reveal a vital role for the Asp→Tyr substitution at Cap5E-75 in ablating CP production. In contrast, Cap5G tolerates the Phe160Leu substitution in USA300 because complementation of CP5 production in a USA300 strain was achieved by a plasmid lacking cap5G.
DISCUSSION
The fact that CP antigens have been used successfully in vaccines against several bacterial pathogens has encouraged a similar vaccine strategy for S. aureus. Several vaccines that have been designed for use against S. aureus have included CP5 and CP8 because they are the most common CP types produced by S. aureus clinical isolates. However, this study shows that USA300 MRSA isolates, as well as USA300 MSSA and closely related USA500 isolates, are universally CP− negative. We also showed no evidence that USA300 isolates produce CP5 in vivo during infection in mice. In contrast, Timofeyeva et al. demonstrated that CP5 was detected in vivo on USA300 MRSA after a 6-h infection (28). We cannot explain the discrepancy between the two studies. However, it is unlikely that a strain could revert all 3 mutations that would be required for USA300 to revert to a CP+ phenotype.
USA300 isolates have increased in prevalence in the United States to become one of the most common S. aureus genotypes (15), especially among SSTIs, where they have accounted for 98% of MRSA isolates (16). Because USA300 isolates comprise a substantial portion of the S. aureus disease burden in the United States, a vaccine based solely on CP would likely demonstrate low efficacy in a U.S. population.
These results extend the observations reported a decade ago when USA300 MRSA isolates were first reported to be acapsular (22, 23). We now show that the CP− phenotype has persisted among USA300 MRSA isolates and is a universal and characteristic trait of this highly successful MRSA lineage. Moreover, we have now established that USA300 MSSA isolates and USA500 isolates are also CP−.
Our prior understanding of the genetic basis of the CP− phenotype among USA300 isolates was limited to our identification of two mutations in the cap5 locus in three USA300 isolates (22) that had been shown previously to ablate CP production in other S. aureus strains (27, 32). The comprehensive genetic analyses of USA300 isolates in this study reveals that the 4 cap5 mutations are present in all but 1 USA300 MRSA isolate and in all but 3 USA300 MSSA isolates. Importantly, 3 of those 4 mutations are alone sufficient to ablate CP production. It would therefore require reversion of all 3 mutations in USA300 MRSA to produce a strain that produces a CP. Also, 3 of the 4 same mutations are common in the USA500 lineage. It has been shown previously that the mutations in Pcap5 −73 and the frameshift mutation in cap5D ablate CP production in S. aureus (22, 27). Whereas the promoter mutation attenuates expression of capABCDEF, the cap5D and cap5E mutations ablate production or activity of enzymes responsible for producing key soluble precursors UDP-d-FucNAc and UDP-l-FucNAc, respectively, that are needed for the biosynthesis of CP (2, 34). This is the first report to show that Asp75 is a critical residue in Cap5E, which is an essential enzyme for CP5 production (35). This enzyme converts the precursor (UDP-GlcNAc) to a keto-intermediate that is further reduced (by Cap5F) and epimerized at C-2 (by Cap5G) to yield UDP-N-acetyl l-fucosamine (36). Thus, USA300 is unable to produce UDP-N-acetyl l-fucosamine, which is required for CP5 and CP8 production. When the Cap5E Asp75Tyr conversion is overlaid on the Cap5E three-dimensional crystal structure (37), it lies within the coenzyme-binding domain. This information could be useful in developing inhibitors of CP biosynthesis.
Our analysis of S. aureus whole-genome sequences (WGSs) in the public databases from 41 STs also provides insight into the evolution of the USA500 and USA300 S. aureus lineages not previously appreciated. All USA500 isolates that we evaluated had at least 2 of the 4 cap5 mutations found in USA300 MRSA (in Pcap5 and in cap5D—both essential for CP production [36]). Moreover, most USA500 MRSA isolates had the cap5G−478 T→C mutation. Notably, because none of the USA500 isolates had the cap5E-223 G→T mutation, this is a feature in the core genome that might be used to distinguish USA500 from USA300 isolates in future epidemiological studies. By overlaying the USA300 mutations on the phylogeny of S. aureus genomes (Fig. 4), it is apparent that the 4 USA300 cap5-specific mutations only exist in clonal cluster 8, and they arose sequentially, starting with Pcap5, in a common ancestor of USA300 and USA500 isolates. Importantly, this is the first study to reveal that USA300 and USA500 lineages inherited the cap mutations from a common ancestor rather than USA300 inheriting them from USA500, as suggested previously (21).
MRSA isolates arise by horizontal acquisition of a mobile genetic element called SCCmec that carries the methicillin resistance gene, mec (18). Interestingly, only a limited number of genetic backgrounds have become successful MRSA clones (18). Despite the close genetic similarity between the USA300 and USA500 lineages (21, 26, 30), USA300 has disseminated more widely in the United States. Comparisons of virulence factors produced by USA500 versus USA300 may provide insight into their relative successes. USA500 and USA300 MRSA isolates both exhibit high-level virulence in animal infection (22, 26, 30), and both express high levels of global regulators and exotoxins encoded in the core genome (22, 26). CP is another core virulence determinant that we have ruled out as a key player in the relative success of USA300 over USA500, because CP is not expressed in either subclone. The success of USA300 may lie in the fact that it carries ACME, which is thought to enhance transmissibility and competitiveness of USA300, although not its virulence (19, 20, 22).
Our findings shed light on the roles of CP5 and CP8 in staphylococcal virulence of USA300 and USA500 strains. CP has been shown to protect S. aureus from opsonophagocytic killing by human neutrophils (11, 29) and thus protects S. aureus from host immune killing. Moreover, the capsule has been shown to enhance virulence in animal models of bacteremia, subcutaneous and renal abscess formation, surgical wound infection, septic arthritis, and lethality (2). Yet its absence in USA300 and USA500 strains underscores the fact that a CP is not necessary for virulence of these strains. This is consistent with studies that have shown that CP production can attenuate staphylococcal virulence in situations in which bacterial adherence is critical, as in endocarditis and murine mastitis, because it masks the adhesins on the cell surface (2). USA300 is best known for causing SSTIs, so we can speculate that perhaps this niche favors a CP− phenotype by exposing adhesins on the surface of the bacteria. Because the pathogenesis of staphylococcal infections is multifactorial, it is likely that other virulence factors produced by USA300 and USA500 compensate for lack of CP during infection. Overexpression of agr, sae, α-hemolysin, and a variety of leukocidins (22), may even have been selected for in the absence of a CP.
Our study has limitations. Our sample of isolates was obtained mainly from patients in Chicago and Los Angeles. This was offset by analyzing public genome sequences, which supported and extended the findings from our isolates.
We conclude that a vaccine designed for a U.S. population should not be solely based on CP antigens because USA300 and USA500 CP− isolates cause a large proportion of S. aureus infections.
MATERIALS AND METHODS
Determination of USA300 and USA500 genetic backgrounds.
S. aureus isolates were confirmed with a catalase test and by agglutination using Staphaurex Plus (Remel) and underwent genotyping by multilocus sequence typing (MLST) (38), Ridom spa typing (39), SCCmec typing (40), and PCR detection of mecA, the ACME-borne arcA gene (ACME arcA) (19), and genes encoding PVL (41) as described previously (39). USA300 MRSA isolates were classified by SmaI digestion patterns using pulsed-field gel electrophoresis (PFGE) as described previously (39). In the absence of PFGE data, the USA300 genotype was inferred in MRSA isolates that belonged to ST8 and carried SCCmec type IV and PVL genes (39). All USA500 isolates and USA300 MSSA isolates were identified solely by PFGE using SmaI digestion patterns. PFGE patterns were assigned to a given clonal group by comparison to a reference strain using an 80% similarity cutoff using the Dice coefficient in BioNumerics software (Applied Maths, TX) (42). Isolates were obtained from consenting patients as approved by the Institutional Review Board of the participating institutions.
CP5 and CP8 serotyping.
Capsule serotyping was performed by a colony immunoblot method with the use of CP5- and CP8-specific polyclonal antibodies as described (43). Briefly, tryptic soy agar plates were spot inoculated in a grid pattern with up to 60 S. aureus isolates and incubated overnight at 37°C. The colonies were blotted onto nitrocellulose filter membranes (diameter, 82.5 mm) for 5 min at ambient temperature. Adherent colonies were fixed to the membranes by being heated at 60°C for 15 min. After being washed twice in phosphate-buffered saline (PBS) to remove excess cells, the filters were immersed in a solution of trypsin (1 mg/ml) for 60 min at 37°C to remove protein A. After two washes in PBS, the filters were blocked with 0.05% skim milk for 1 h and washed in PBS containing 0.05% Tween 20 (PBST). Capsule type-specific polyclonal antiserum (diluted 1:1,000) was incubated with each filter at 37°C for 1 h. After being washed in PBST, horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin was incubated with each filter for 1 h at 37°C. After three washes in PBST and two washes in PBS, substrate (3 mg 4-chloro-1-naphthol per ml of methanol diluted 1:5 in PBS and containing 0.1% H2O2) was added to the filters. A purple color developed within 15 min and was scored visually from 0 to 4+. Positive reactions were scored as 2+ to 4+. The reactivity of the isolates was evaluated by comparison to those of control S. aureus strains (CP5+, CP8+, and CP− isolates) included on each filter membrane. Isolates with no reaction to CP-specific antibodies were defined as CP−.
USA300 strains subjected to whole-genome sequencing.
To determine the basis for the CP5− phenotype, we analyzed the entire cap5 locus from the WGS assemblies we produced (33) from 146 USA300 MRSA isolates that were included in the sample tested for CP production. These isolates were obtained between 2008 to 2010 from the University of Chicago Medical Center (n = 75) and Harbor-UCLA Medical Center in Los Angeles (n= 71) (44).
WGS and SNP calling.
Genomic DNA was sequenced on an Illumina HiSeq 2000 (Illumina Inc., San Diego, CA) at ~215× the median depth of coverage per strain as described in reference 33 and the supplemental material. Mutations in the cap5 locus were retrieved from the assemblies by comparison to the cap5 locus of the CP5+ reference strain Newman using Jalview (45). All nucleotide positions are reported relative to the USA300 reference strain TCH1516.
Confirmation of cap5 mutations.
To evaluate whether the conserved mutations we identified in the WGS analysis were present in S. aureus isolates of interest, the regions surrounding the mutations were PCR amplified from the indicated strains using primers listed in Table 3. The amplification products were subjected to Sanger sequencing in the DNA sequencing and genotyping facility (University of Chicago) using the amplification primers. Mutations were identified using Vector NTI software (Invitrogen) by alignment to the cap5 locus of the CP5+ reference strain Newman.
TABLE 3 .
Sequences of the primers used for PCR amplification and sequencing of the cap5 gene fragments
| Primer (expected size [bp])a | Sequence |
|---|---|
| cap5A promoter, Pcap5 | |
| Forward | 5′ GAATCATTAGCTAAAGCTGTCTAC 3′ |
| Reverse | 5′ GTCACCCTTAGTTTGATTCA 3′ |
| cap5D (803) | |
| Forward | 5′ GTAAAATTGCGGATATTCCAGAAC 3′ |
| Reverse | 5′ AGTGGAATCACAGATCCTCT 3′ |
| cap5E (381) | |
| Forward | 5′ GCACAGGATCATTCGGTAAT 3′ |
| Reverse | 5′ CTTTTGAAATACCCATAGCA 3′ |
| cap5G (543) | |
| Forward | 5′ TGGAAGCGGGTAATAGATGC 3′ |
| Reverse | 5′ GGACACCAGGGAACTTCAAA 3′ |
Forward primers have a sense orientation, and reverse primers have an antisense orientation.
cap5 mutations in publicly available USA300 and non-USA300 S. aureus genomes.
To examine whether the four cap5 locus mutations found in the WGSs of our sample of 146 USA300 MRSA isolates were universal in this clonal lineage, we also interrogated cap5 gene cluster sequences from 319 publicly available USA300 MRSA genomes. These included 35 isolates from San Diego (46), 277 isolates from New York City (47), the completed genome sequences of strains TCH1516 (48), FPR3757 (49), UA-S391 (50), and ISMMS1 (51), and three isolates from Alam et al. (52). Also, the phylogenetic distribution and origin of the cap mutations among 90 publicly available non-USA300 S. aureus genomes from 41 STs were studied using whole-genome alignment-based maximum likelihood phylogeny of publicly constructed genomes using PhyML, as implemented in REALPHY (53). Further details of the public sequence annotations are provided in Table S2 in the supplemental material.
Complementation studies of the cap locus.
In order to test the effect of the cap5 mutations, we electrotransformed (54) a USA300 MRSA strain, 923, with plasmids containing various cap5 genes that were isolated from strain Newman and cloned into the vector pCU1 as summarized in Table 4. Briefly we used pKOR18 (which contains cap5ABCD and a portion of cap5E), pKOR19 (which contains cap5ABC and truncated cap5D) (27), pCap16 (which contains intact cap5A-E) (35), and pCap17 (which contains intact cap5A-F) (35). Following transformation, strain 923 was cultured in the presence of chloramphenicol at 5 mg/liter to maintain the plasmid. CP5 serotyping was performed as described above.
TABLE 4 .
Plasmid constructs used in cap5 genetic complementation experiments
| Plasmida | cap5 genes contained in plasmid | Reference |
|---|---|---|
| pKOR19 | Pcap5 + capABC | 27 |
| pKOR18 | Pcap5 + capABCD | 27 |
| pCap16 | Pcap5 + capABCDE | 35 |
| pCap17 | Pcap5 + cap5ABCDEF | 35 |
pCap16 and pCap17 were kindly provided by Timothy Foster.
Nucleotide sequence accession number.
The raw sequence reads from the project have been deposited into the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) database under accession no. SRP039020. These sequences were also analyzed in reference 33.
SUPPLEMENTAL MATERIAL
Supplemental methods. Download
Analysis of CP5 production in vivo by USA300 MRSA strain CDC3. Mice were challenged by the intraperitoneal route with strain CDC3. Cardiac blood collected at 6 h was sonicated and treated with DNase and trypsin, and the pellet was incubated with a monoclonal antibody specific for CP5 or anti-S. aureus polyclonal antiserum followed by reaction with secondary antibodies labeled with Alexa Fluor 660 (blue [anti-CP5]) or Alexa Fluor 568 (red [anti-S. aureus]), as detailed in the supplemental material. Samples were viewed on a Zeiss Pascal scanning confocal microscope, and images were analyzed with Zeiss LSM AxioVision software. The bar represents 10 µm. (A) CP5+ S. aureus strain Newman (grown in vitro); (B) USA300 MRSA inoculum from strain CDC3 (grown in vitro); (C) blood sample from a mouse infected with USA300 MRSA strain CDC3. These data show that USA300 MRSA does not produce CP5 in vivo. Download
Characteristics of USA300 MSSA and USA500 isolates tested for CP production. col, colonization; arcA, encoded in the arginine catabolic mobile element ACME; shading signifies a difference compared with USA300.
Annotations of public genome sequences included in the tree in Figure 4. The bases highlighted in yellow are mutations compared to Newman strain (accession number NC_009641). ND, Not determined.
ACKNOWLEDGMENTS
We acknowledge that pCap16 and pCap17 were the kind gift of Timothy Foster. The USA500 and USA300-0114 reference strains were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) under a contract from the National Institute of Allergy and Infectious Diseases.
This study was funded by National Institutes of Health grants awarded to R.S.D. (grant no. R01AI103342), J.C.L. (grant no. R01 AI088754), and T.D.R. (grant no. AI091827).
Footnotes
Citation Boyle-Vavra S, Li X, Alam MT, Read TD, Sieth J, Cywes-Bentley C, Dobbins G, David MZ, Kumar N, Eells SJ, Miller LG, Boxrud DJ, Chambers HF, Lynfield R, Lee JC, Daum RS. 2015. USA300 and USA500 clonal lineages of Staphylococcus aureus do not produce a capsular polysaccharide due to conserved mutations in the cap5 locus. mBio 6(2):e02585-14. doi:10.1128/mBio.02585-14.
REFERENCES
- 1.Daum RS, Spellberg B. 2012. Progress toward a Staphylococcus aureus vaccine. Clin Infect Dis 54:560–567. doi: 10.1093/cid/cir828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Riordan K, Lee JC. 2004. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev 17:218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trotter CL, McVernon J, Ramsay ME, Whitney CG, Mulholland EK, Goldblatt D, Hombach J, Kieny M-P, SAGE Subgroup . 2008. Optimising the use of conjugate vaccines to prevent disease caused by Haemophilus influenzae type b, Neisseria meningitidis and Streptococcus pneumoniae. Vaccine 26:4434–4445. doi: 10.1016/j.vaccine.2008.05.073. [DOI] [PubMed] [Google Scholar]
- 4.Hochkeppel HK, Braun DG, Vischer W, Imm A, Sutter S, Staeubli U, Guggenheim R, Kaplan EL, Boutonnier A, Fournier JM. 1987. Serotyping and electron microscopy studies of Staphylococcus aureus clinical isolates with monoclonal antibodies to capsular polysaccharide types 5 and 8. J Clin Microbiol 25:526–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arbeit RD, Karakawa WW, Vann WF, Robbins JB. 1984. Predominance of two newly described capsular polysaccharide types among clinical isolates of Staphylococcus aureus. Diagn Microbiol Infect Dis 2:85–91. doi: 10.1016/0732-8893(84)90002-6. [DOI] [PubMed] [Google Scholar]
- 6.Karakawa WW, Vann WF. 1982. Capsular polysaccharides of Staphylococcus aureus. Semin Infect Dis 4:285–293. [Google Scholar]
- 7.Sompolinsky D, Samra Z, Karakawa WW, Vann WF, Schneerson R, Malik Z. 1985. Encapsulation and capsular types in isolates of Staphylococcus aureus from different sources and relationship to phage types. J Clin Microbiol 22:828–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JC, Liu MJ, Parsonnet J, Arbeit RD. 1990. Expression of type-8 capsular polysaccharide and production of toxic shock syndrome toxin-1 are associated among vaginal isolates of Staphylococcus aureus. J Clin Microbiol 28:2612–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roghmann M, Taylor KL, Gupte A, Zhan M, Johnson JA, Cross A, Edelman R, Fattom AI. 2005. Epidemiology of capsular and surface polysaccharide in Staphylococcus aureus infections complicated by bacteraemia. J Hosp Infect 59:27–32. doi: 10.1016/j.jhin.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Nanra JS, Buitrago SM, Crawford S, Ng J, Fink PS, Hawkins J, Scully IL, McNeil LK, Aste-Amézaga JM, Cooper D, Jansen KU, Anderson AS. 2013. Capsular polysaccharides are an important immune evasion mechanism for Staphylococcus aureus. Hum Vaccin Immunother 9:480–487. doi: 10.4161/hv.23223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thakker M, Park JS, Carey V, Lee JC. 1998. Staphylococcus aureus serotype 5 capsular polysaccharide is antiphagocytic and enhances bacterial virulence in a murine bacteremia model. Infect Immun 66:5183–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinefield H, Black S, Fattom A, Horwith G, Rasgon S, Ordonez J, Yeoh H, Law D, Robbins JB, Schneerson R, Muenz L, Fuller S, Johnson J, Fireman B, Alcorn H, Naso R. 2002. Use of a Staphylococcus aureus conjugate vaccine in patients receiving hemodialysis. N Engl J Med 346:491–496. doi: 10.1056/NEJMoa011297. [DOI] [PubMed] [Google Scholar]
- 13.Fattom A, Matalon A, Buerkert J, Taylor K, Damaso S, Boutriau D. 2014. Efficacy profile of a bivalent Staphylococcus aureus glycoconjugated vaccine in adults on hemodialysis: phase III randomized study. Hum Vaccin Immunother e34414. doi: 10.4161/hv.34414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagnoli F, Bertholet S, Grandi G. 2012. Inferring reasons for the failure of Staphylococcus aureus vaccines in clinical trials. Front Cell Infect Microbiol 2:16. doi: 10.3389/fcimb.2012.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diekema DJ, Richter SS, Heilmann KP, Dohrn CL, Riahi F, Tendolkar S, McDanel JS, Doern GV. 2014. Continued emergence of USA300 methicillin-resistant Staphylococcus aureus in the United States: results from a nationwide surveillance study. Infect Control Hosp Epidemiol 35:285–292. doi: 10.1086/675283. [DOI] [PubMed] [Google Scholar]
- 16.Talan DA, Krishnadasan A, Gorwitz RJ, Fosheim GE, Limbago B, Albrecht V, Moran GJ, EMERGEncy ID Net Study Group . 2011. Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients, 2004 and 2008. Clin Infect Dis 53:144–149. doi: 10.1093/cid/cir308. [DOI] [PubMed] [Google Scholar]
- 17.Ma XX, Ito T, Tiensasitorn C, Jamklang M, Chongtrakool P, Boyle-Vavra S, Daum RS, Hiramatsu K. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother 46:1147–1152. doi: 10.1128/AAC.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyle-Vavra S, Daum RS. 2007. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin. Lab Invest 87:3–9. doi: 10.1038/labinvest.3700501. [DOI] [PubMed] [Google Scholar]
- 19.Diep BA, Stone GG, Basuino L, Graber CJ, Miller A, des Etages SA, Jones A, Palazzolo-Ballance AM, Perdreau-Remington F, Sensabaugh GF, DeLeo FR, Chambers HF. 2008. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis 197:1523–1530. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- 20.Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, Richardson AR. 2013. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13:100–107. doi: 10.1016/j.chom.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Planet PJ, LaRussa SJ, Dana A, Smith H, Xu A, Ryan C, Uhlemann AC, Boundy S, Goldberg J, Narechania A, Kulkarni R, Ratner AJ, Geoghegan JA, Kolokotronis SO, Prince A. 2013. Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin. mBio 4(6):e00889-13. doi: 10.1128/mBio.00889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montgomery CP, Boyle-Vavra S, Adem PV, Lee JC, Husain AN, Clasen J, Daum RS. 2008. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J Infect Dis 198:561–570. doi: 10.1086/590157. [DOI] [PubMed] [Google Scholar]
- 23.Sutter DE, Summers AM, Keys CE, Taylor KL, Frasch CE, Braun LE, Fattom AI, Bash MC. 2011. Capsular serotype of Staphylococcus aureus in the era of community-acquired MRSA. FEMS Immunol Med Microbiol 63:16–24. doi: 10.1111/j.1574-695X.2011.00822.x. [DOI] [PubMed] [Google Scholar]
- 24.Anderson AS, Miller AA, Donald RG, Scully IL, Nanra JS, Cooper D, Jansen KU. 2012. Development of a multicomponent Staphylococcus aureus vaccine designed to counter multiple bacterial virulence factors. Hum Vaccin Immunother 8:1585–1594. doi: 10.4161/hv.21872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy J, Licini L, Haelterman E, Moris P, Lestrate PR, Damaso S, Boutriau D. 2014. Evaluation of an investigational 4-component Staphylococcus aureus vaccine with or without ASO3B adjuvant in healthy adults: results from a phase I trial, abstr 145. Int Symp Staphylococci Staphylococcal Infect, Chicago, IL. [Google Scholar]
- 26.Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF, Lu Y, Otto M. 2009. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A 106:5883–5888. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cocchiaro JL, Gomez MI, Risley A, Solinga R, Sordelli DO, Lee JC. 2006. Molecular characterization of the capsule locus from non-typeable Staphylococcus aureus. Mol Microbiol 59:948–960. doi: 10.1111/j.1365-2958.2005.04978.x. [DOI] [PubMed] [Google Scholar]
- 28.Timofeyeva Y, Scully IL, Anderson AS. 2014. Immunofluorescence microscopy for the detection of surface antigens in methicillin-resistant Staphylococcus aureus (MRSA). Methods Mol Biol 1085:85–95. doi: 10.1007/978-1-62703-664-1_4. [DOI] [PubMed] [Google Scholar]
- 29.Wacker M, Wang L, Kowarik M, Dowd M, Lipowsky G, Faridmoayer A, Shields K, Park S, Alaimo C, Kelley KA, Braun M, Quebatte J, Gambillara V, Carranza P, Steffen M, Lee JC. 2014. Prevention of Staphylococcus aureus infections by glycoprotein vaccines synthesized in Escherichia coli. J Infect Dis 209:1551–1561. doi: 10.1093/infdis/jit800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benson MA, Ohneck EA, Ryan C, Alonzo F III, Smith H, Narechania A, Kolokotronis SO, Satola SW, Uhlemann AC, Sebra R, Deikus G, Shopsin B, Planet PJ, Torres VJ. 2014. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol Microbiol 93:664–681. doi: 10.1111/mmi.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diep BA, Carleton HA, Chang RF, Sensabaugh GF, Perdreau-Remington F. 2006. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. J Infect Dis 193:1495–1503. doi: 10.1086/503777. [DOI] [PubMed] [Google Scholar]
- 32.Ouyang S, Sau S, Lee CY. 1999. Promoter analysis of the cap8 operon, involved in type 8 capsular polysaccharide production in Staphylococcus aureus. J Bacteriol 181:2492–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alam MT, Read TD, Petit RA III, Boyle-Vavra S, Miller LG, Eells SJ, Daum RS, David MZ. 2015. Transmission and microevolution of USA300 MRSA in U.S. households: evidence from whole-genome sequencing. mBio 6(2):e00054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, Ulm H, Rausch M, Li X, O’Riordan K, Lee JC, Schneider T, Müller CE. 2014. Analysis of the Staphylococcus aureus capsule biosynthesis pathway in vitro: characterization of the UDP-GlcNAc C6 dehydratases CapD and CapE and identification of enzyme inhibitors. Int J Med Microbiol 304:958–969. doi: 10.1016/j.ijmm.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Wann ER, Dassy B, Fournier JM, Foster TJ. 1999. Genetic analysis of the cap5 locus of Staphylococcus aureus. FEMS Microbiol Lett 170:97–103. doi: 10.1111/j.1574-6968.1999.tb13360.x. [DOI] [PubMed] [Google Scholar]
- 36.Kneidinger B, O’Riordan K, Li J, Brisson JR, Lee JC, Lam JS. 2003. Three highly conserved proteins catalyze the conversion of UDP-N-acetyl-d-glucosamine to precursors for the biosynthesis of O antigen in Pseudomonas aeruginosa O11 and capsule in Staphylococcus aureus type 5. Implications for the UDP-N-acetyl-l-fucosamine biosynthetic pathway. J Biol Chem 278:3615–3627. doi: 10.1074/jbc.M203867200. [DOI] [PubMed] [Google Scholar]
- 37.Miyafusa T, Caaveiro JM, Tanaka Y, Tanner ME, Tsumoto K. 2013. Crystal structure of the capsular polysaccharide synthesizing protein CapE of Staphylococcus aureus. Biosci Rep 33:e0043. doi: 10.1042/BSR20130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.David MZ, Taylor A, Lynfield R, Boxrud DJ, Short G, Zychowski D, Boyle-Vavra S, Daum RS. 2013. Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for Panton-Valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a U.S. medical center. J Clin Microbiol 51:814–819. doi: 10.1128/JCM.02429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 42.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol 41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JC, Liu MJ, Parsonnet J, Arbeit RD. 1990. Expression of type 8 capsular polysaccharide and production of toxic shock syndrome toxin 1 are associated among vaginal isolates of Staphylococcus aureus. J Clin Microbiol 28:2612–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller LG, Eells SJ, Taylor AR, David MZ, Ortiz N, Zychowski D, Kumar N, Cruz D, Boyle-Vavra S, Daum RS. 2012. Staphylococcus aureus colonization among household contacts of patients with skin infections: risk factors, strain discordance, and complex ecology. Clin Infect Dis 54:1523–1535. doi: 10.1093/cid/cis213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tewhey R, Cannavino CR, Leake JA, Bansal V, Topol EJ, Torkamani A, Bradley JS, Schork NJ. 2012. Genetic structure of community acquired methicillin-resistant Staphylococcus aureus USA300. BMC Genomics 13:508. doi: 10.1186/1471-2164-13-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uhlemann A-, Dordel J, Knox JR, Raven KE, Parkhill J, Holden MTG, Peacock SJ, Lowy FD. 2014. Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community. Proc Natl Acad Sci U S A 111:6738–6743. doi: 10.1073/pnas.1401006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Highlander SK, Hultén KG, Qin X, Jiang H, Yerrapragada S, Mason EO Jr, Shang Y, Williams TM, Fortunov RM, Liu Y, Igboeli O, Petrosino J, Tirumalai M, Uzman A, Fox GE, Cardenas AM, Muzny DM, Hemphill L, Ding Y, Dugan S, Blyth PR, Buhay CJ, Dinh HH, Hawes AC, Holder M, Kovar CL, Lee SL, Liu W, Nazareth LV, Wang Q, Zhou J, Kaplan SL, Weinstock GM. 2007. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol 7:99. doi: 10.1186/1471-2180-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 50.Sabirova JS, Xavier BB, Hernalsteens JP, De Greve H, Ieven M, Goossens H, Malhotra-Kumar S. 2014. Complete genome sequences of two prolific biofilm-forming Staphylococcus aureus isolates belonging to USA300 and EMRSA-15 clonal lineages. Genome Announc 2:e00610-14. doi: 10.1128/genomeA.00610-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altman DR, Sebra R, Hand J, Attie O, Deikus G, Carpini KW, Patel G, Rana M, Arvelakis A, Grewal P, Dutta J, Rose H, Shopsin B, Daefler S, Schadt E, Kasarskis A, van Bakel H, Bashir A, Huprikar S. 2014. Transmission of methicillin-resistant Staphylococcus aureus via deceased donor liver transplantation confirmed by whole genome sequencing. Am J Transplant 14:2640–2644. doi: 10.1111/ajt.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alam MT, Petit RA III, Crispell EK, Thornton TA, Conneely KN, Jiang Y, Satola SW, Read TD. 2014. Dissecting vancomycin-intermediate resistance in Staphylococcus aureus using genome-wide association. Genome Biol Evol 6:1174–1185. doi: 10.1093/gbe/evu092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bertels F, Silander OK, Pachkov M, Rainey PB, van Nimwegen E. 2014. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol 31:1077–1088. doi: 10.1093/molbev/msu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Augustin J, Götz F. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol Lett 54:203–207. doi: 10.1016/0378-1097(90)90283-V. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download
Analysis of CP5 production in vivo by USA300 MRSA strain CDC3. Mice were challenged by the intraperitoneal route with strain CDC3. Cardiac blood collected at 6 h was sonicated and treated with DNase and trypsin, and the pellet was incubated with a monoclonal antibody specific for CP5 or anti-S. aureus polyclonal antiserum followed by reaction with secondary antibodies labeled with Alexa Fluor 660 (blue [anti-CP5]) or Alexa Fluor 568 (red [anti-S. aureus]), as detailed in the supplemental material. Samples were viewed on a Zeiss Pascal scanning confocal microscope, and images were analyzed with Zeiss LSM AxioVision software. The bar represents 10 µm. (A) CP5+ S. aureus strain Newman (grown in vitro); (B) USA300 MRSA inoculum from strain CDC3 (grown in vitro); (C) blood sample from a mouse infected with USA300 MRSA strain CDC3. These data show that USA300 MRSA does not produce CP5 in vivo. Download
Characteristics of USA300 MSSA and USA500 isolates tested for CP production. col, colonization; arcA, encoded in the arginine catabolic mobile element ACME; shading signifies a difference compared with USA300.
Annotations of public genome sequences included in the tree in Figure 4. The bases highlighted in yellow are mutations compared to Newman strain (accession number NC_009641). ND, Not determined.