ABSTRACT
Endophytes are microbes that inhabit plant tissues without any apparent signs of infection, often fundamentally altering plant phenotypes. While endophytes are typically studied in plant roots, where they colonize the apoplast or dead cells, Methylobacterium extorquens strain DSM13060 is a facultatively intracellular symbiont of the meristematic cells of Scots pine (Pinus sylvestris L.) shoot tips. The bacterium promotes host growth and development without the production of known plant growth-stimulating factors. Our objective was to examine intracellular colonization by M. extorquens DSM13060 of Scots pine and sequence its genome to identify novel molecular mechanisms potentially involved in intracellular colonization and plant growth promotion. Reporter construct analysis of known growth promotion genes demonstrated that these were only weakly active inside the plant or not expressed at all. We found that bacterial cells accumulate near the nucleus in intact, living pine cells, pointing to host nuclear processes as the target of the symbiont’s activity. Genome analysis identified a set of eukaryote-like functions that are common as effectors in intracellular bacterial pathogens, supporting the notion of intracellular bacterial activity. These include ankyrin repeats, transcription factors, and host-defense silencing functions and may be secreted by a recently imported type IV secretion system. Potential factors involved in host growth include three copies of phospholipase A2, an enzyme that is rare in bacteria but implicated in a range of plant cellular processes, and proteins putatively involved in gibberellin biosynthesis. Our results describe a novel endophytic niche and create a foundation for postgenomic studies of a symbiosis with potential applications in forestry and agriculture.
IMPORTANCE
All multicellular eukaryotes host communities of essential microbes, but most of these interactions are still poorly understood. In plants, bacterial endophytes are found inside all tissues. M. extorquens DSM13060 occupies an unusual niche inside cells of the dividing shoot tissues of a pine and stimulates seedling growth without producing cytokinin, auxin, or other plant hormones commonly synthesized by plant-associated bacteria. Here, we tracked the bacteria using a fluorescent tag and confocal laser scanning microscopy and found that they localize near the nucleus of the plant cell. This prompted us to sequence the genome and identify proteins that may affect host growth by targeting processes in the host cytoplasm and nucleus. We found many novel genes whose products may modulate plant processes from within the plant cell. Our results open up new avenues to better understand how bacteria assist in plant growth, with broad implications for plant science, forestry, and agriculture.
INTRODUCTION
Bacterial endophytes are increasingly recognized for their ability to stimulate plant growth, assist in nutrient acquisition, and protect against stress and pathogens (1, 2), yet our understanding of the mechanisms by which they interact with and colonize the host is limited. The general view of endophyte transmission posits that bacterial endophytes enter the plant through the root from soil and colonize intercellular spaces (2). However, a number of endophytes have been detected in seeds (3, 4), suggesting vertical transmission. In addition, plant tissue culture, which is started from shoot meristem or seed embryos, is not axenic (5), pointing toward common associations between plants and bacteria in reproductive tissues.
Intracellular colonization is either unusual or understudied in endophytes. A few exceptions include Gluconacetobacter diazotrophicus in Arabidopsis thaliana, rice, and maize (6) and the rare intracellular invasion of Bradyrhizobium in rice roots (7). Typically, intracellular colonization occurs in dead plant cells or results in the death of the plant cell (8). Intracellular colonization occurs in living Scots pine (Pinus sylvestris L.) shoot meristematic cells by endophytic bacteria and yeasts, shown by in situ hybridization (9, 10) and transmission electron microscopy (Fig. 1). The strain Methylobacterium extorquens DSM13060, which was originally isolated from shoot tip-derived tissue cultures of P. sylvestris, is consistently associated with P. sylvestris buds across seasons and geographic locations (9, 11). The endophyte is most abundant just prior to bud elongation and differentiation (11) and produces compounds that induce cell divisions in host tissue (12). Inoculation of P. sylvestris seedlings with DSM13060 in vitro significantly increases root and needle growth compared to their growth in control seedlings (13), demonstrating a prominent role for M. extorquens DSM13060 in the growth and development of the plant host.
FIG 1 .
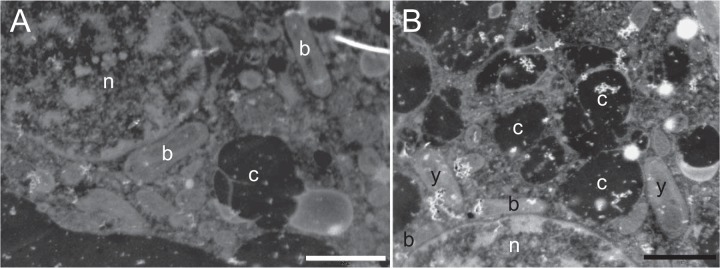
Transmission electron micrographs of bud cells of Scots pine displaying intracellular bacteria (b) and yeasts (y) residing near the nucleus (n). The cells are rich with dark chromoplasts (c). Scale bar, 2 µm.
Unlike a number of epiphytic plant hormone-producing Methylobacterium strains (14), strain DSM13060 does not produce any common phytohormones (12). Clues to the mechanisms by which DSM13060 affects host growth may be found in its unique life style within the host cells, where it may be better positioned to directly affect host processes in the cytoplasm or nucleus. To explore mechanisms underlying the intracellular lifestyle and host growth promotion by M. extorquens DSM13060, we investigated its colonization in Scots pine and sequenced and analyzed its genome.
RESULTS AND DISCUSSION
M. extorquens DSM13060 colonizes host cells in association with the nucleus.
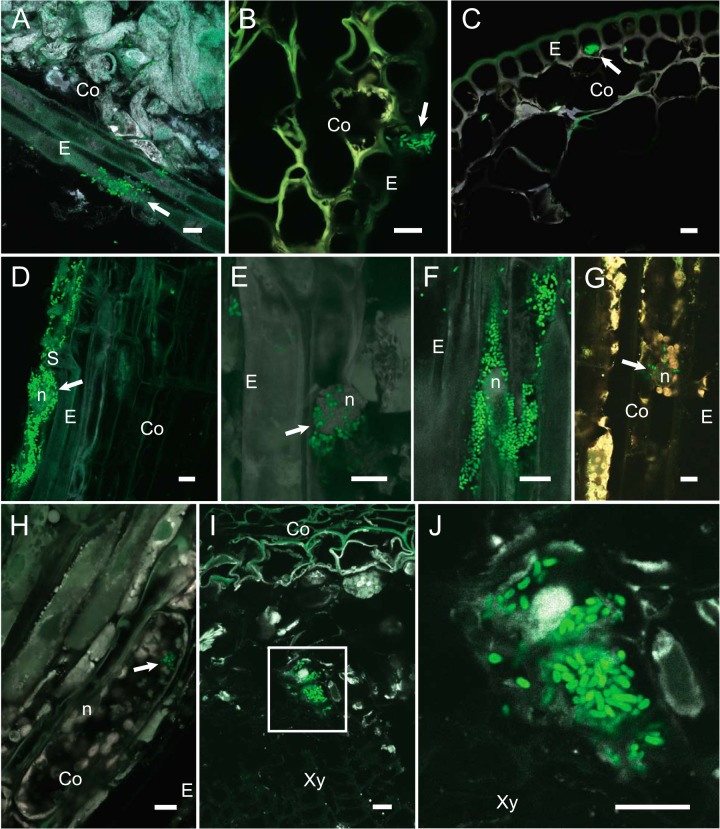
Confocal microscopic studies using M. extorquens DSM13060 tagged with green fluorescent protein (GFP) (strains 13061 and 13062) were used to follow the colonization of P. sylvestris seedlings, from the inoculation of bacteria onto emergent seedlings to the establishment and intracellular colonization of roots and aerial tissues. To distinguish GFP from endogenous autofluorescence, which significantly complicates the identification of intracellular bacteria in plants, especially in conifer tissues, we used two consecutive copies of GFP in combination with the mCherry fluorescent reporter, along with a moderate fixation and cryosectioning process. DSM13060 cells from microcolonies formed on the surface actively penetrated the epidermal cells of the root (Fig. 2A) and shoot (Fig. 2B) of pine seedlings. Once intracellular, the bacterial cells regrouped and eventually fully colonized pine epidermal cells (Fig. 2C and D) without any detectable adverse effects on the plant host. Inside cells, bacteria were frequently observed to congregate around the nucleus (Fig. 2E to G). Intracellular colonies were most abundant in the parenchymal cells of the transition zone (Fig. 2H; see also Movie S1 in the supplemental material) and in the stem parenchyma (Fig. 2I and J). To verify colonization of live plant cells, we used acridine orange-ethidium bromide (AOEB) staining with cell morphological analysis to assess the viability of the pine tissues colonized by M. extorquens DSM13060 (15).
FIG 2 .
Laser scanning confocal microscopy of Scots pine seedlings colonized by M. extorquens 13061. Bacterial cells carrying a fluorescent GFP reporter under the control of a constitutive promoter are visualized in green. Cryosections of the pine seedlings colonized by M. extorquens 13061 were analyzed 15 to 90 days after inoculation. (A) Lateral section of a pine root. Arrow indicates a microcolony of 13061 actively penetrating through the root epidermis. Individual bacterial cells are visible in the root cortex. (B) Cross section of a pine stem where a 13061 colony is penetrating an epidermal cell. (C) Cross section of a pine shoot with an intracellular 13061 microcolony inside an epidermal cell. (D) Lateral section of a pine root where epidermal cells are colonized by 13061, with bacteria aggregated around the nucleus. (E) Lateral section of a root, where the arrow indicates 13061 cells congregated around the nucleus of an epidermal cell. (F) Lateral section of a pine root where epidermal cells are completely colonized by 13061 and bacteria surround the nucleus. (G) Lateral section of a pine stem where individual 13061 cells reside close to the nucleus of a cortical cell. (H) Lateral section of a lower-stem (transition zone between root and stem) cortex, where the arrow indicates 13061 cells within a parenchymatic cell. (I) Cross section of a pine shoot where intracellular 13061 cells are visible inside parenchymatic cells. (J) Magnification of the boxed area from panel I, displaying the intracellular 13061 cells near the nucleus. Co, cortex; E, epiderm; n, nucleus; S, cylindrical sheath; Xy, xylem; scale bars, 10 µm.
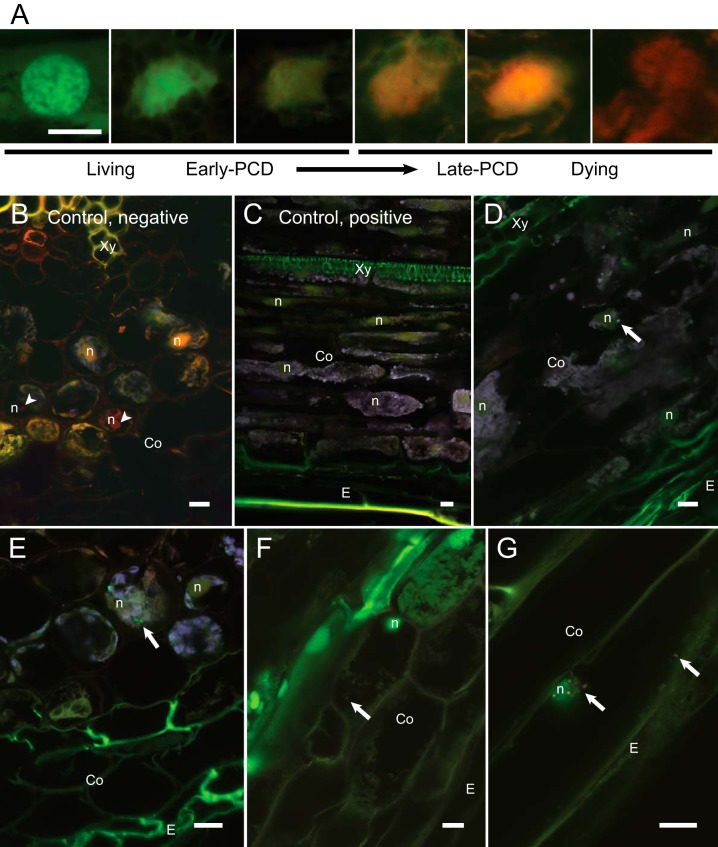
Negative controls treated with hydrogen peroxide frequently exhibited nuclear characteristics of Programmed Cell Death (PCD) or necrosis (Fig. 3A and B). The majority of nuclei were stained predominantly green and were without visible fragmentation or condensation of chromatin in positive untreated controls (Fig. 3C) and in M. extorquens DSM13060-colonized samples (Fig. 3D to G), which, overall, displayed identical tissue morphologies. M. extorquens DSM13060 was observed only in plant cells with healthy nuclear morphology, and the bacteria were localized inside plant cells near nuclei (Fig. 3D and G; see also Movies S2 and S3 in the supplemental material). Although the AOEB staining increased the overall fluorescence and made the detection of intracellular bacteria a challenge, the method clearly demonstrated that DSM13060 colonizes intact, living pine cells. A few cases of endonuclear bacteria have been observed in animals, amoeba, and protozoa (16–18). To our knowledge, aggregation near the nucleus has not been reported for plant-associated bacteria, but the nucleus is a common effector target of pathogenic plant bacteria that manipulate host transcription.
FIG 3 .
Morphological identification of cell viability in M. extorquens 13061- and 13062-colonized tissues of Scots pine seedlings. (A) Confocal micrographs representing the spectrum of nuclei observed in the acridine orange (AO)-ethidium bromide (EB)-stained tissue. Morphology of nuclei and increased incorporation of EB (red) over AO (green) correspond with advancing programmed cell death (PCD). (B to G) Representative merged confocal micrographs of Scots pine seedling tissues. In addition to AOEB stains, bacterial cells carrying a fluorescent GFP reporter are visualized in bright green and endogenous autofluorescence of plant cells is shown in purple. (B) Cross section of an uninoculated pine root incubated in 20 mM H2O2 for 2 h at room temperature. (C) Lateral section of an uninoculated root incubated in PBS for 2 h at room temperature. (D) Lateral section of a root where an individual bacterial cell resides close to the healthy nucleus of a cortical cell. (E) Cross section of a root where intracellular 13061 cells are visible inside a parenchymatic cell. (F) Lateral section of a root where a single bacterial cell is localized in the cytoplasm of a viable cortical cell. (G) Lateral section of a root where 13062 cells are congregated around the nucleus of a cortical cell. Bacterial cells carrying fluorescent GFP and mCherry reporters under the control of constitutive promoters are visualized in white-red to distinguish them from the green nucleus. Bacteria are indicated with white arrows, and arrowheads highlight cells with dead nuclei. Co, cortex; E, epiderm; n, nucleus; Xy, xylem; scale bars, 10 µm.
Genome sequence and structure.
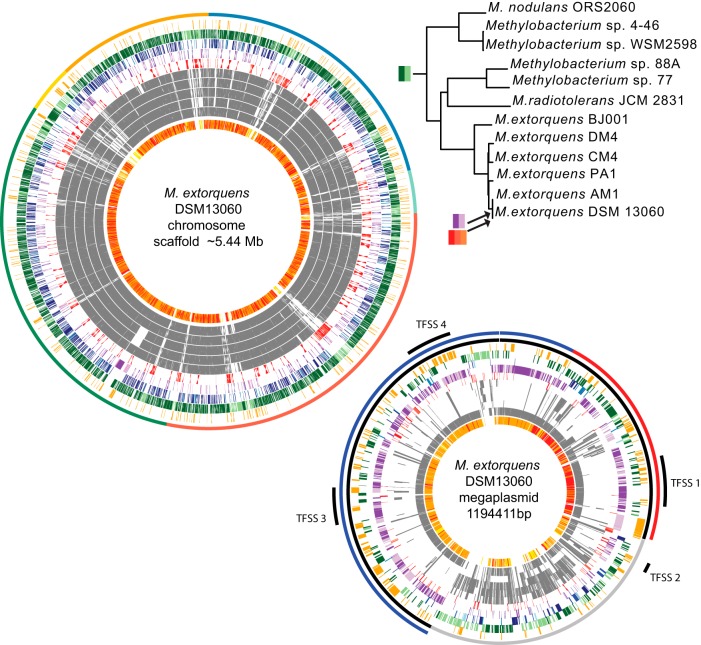
The draft genome of M. extorquens DSM13060 encompassed 12 scaffolds totaling approximately 6.7 Mb, and its general features were compared with those of 11 previously sequenced Methylobacterium genomes (19, 20). The phylogenetic relationship among the Methylobacterium strains included and their key characteristics are shown in Fig. S1 in the supplemental material. DSM13060 is very closely related to M. extorquens AM1 (e.g., the 16S rRNA gene sequences are 100% identical). All M. extorquens genomes described to date have highly syntenic chromosomes and strain-specific plasmids (19, 20). The DSM13060 genome includes a putative replicon that is largely syntenic with the M. extorquens AM1 megaplasmid (19). The DSM13060 draft chromosome (approximately 5.44 Mb) is made up almost entirely by a backbone of orthologs found in the other Methylobacterium genomes (Fig. 4). In contrast, 37% of the genes on the megaplasmid are unique to DSM13060 and AM1 (Fig. 4; see also Table S1 in the supplemental material). These genes may have been introduced into the ancestor of AM1 and DSM13060 from a distantly related species. In support of this, the phylogenetic relationship of the plasmid replication RepA protein (Fig. S2A) and its homologs in other alphaproteobacterial plasmids suggests that the plasmid is only distantly related to other RepABC plasmids (21). Following its introduction into the AM1/DSM13060 ancestor, the megaplasmid appears to have undergone some recombination. The DSM13060 megaplasmid is divided into distinct sections that differ in their degree of similarity to the AM1 megaplasmid (Fig. 4; Table S1). Approximately 75% of the megaplasmid (447 genes) is enriched in hypothetical genes that are unique to DSM13060/AM1 and potentially horizontally transferred genes (HGTs) (Fig. 4, black in outer circle). This region is perfectly syntenic with the AM1 megaplasmid (Fig. S2B). The remaining 25% of the megaplasmid consists mostly of genes that lack homologs in AM1. Instead, this region has more orthologs in other M. extorquens strains (Fig. 4, grey in outer circle). This could be the result of recombination between the DSM13060 megaplasmid and the main chromosome or smaller plasmids. In support of this, the 328-kb region is flanked by site-specific recombinases at its tail end (Fig. 4, breakpoint between grey and black in the outer circle). The first, larger segment can be further divided into two sections that differ in their percentages of protein identity between AM1 and DSM13060 orthologs, an identical section (Fig. 4, red in outer circle) versus a section with somewhat lower identity (Fig. 4, blue in outer circle). This structure in ortholog similarity could be the result of recombination between one of the megaplasmids and that of an intermediate strain. We identified genes putatively imported from outside the Alphaproteobacteria to DSM13060 after the split with AM1 (79 genes) and to the DSM13060/AM1 ancestor after the split with strain PA1 (294 genes) (Table S1). Many of the HGTs are similar to proteins from plant- and soil-associated Gammaproteobacteria and Betaproteobacteria and encode functions potentially relevant to the endophytic lifestyle, including polyketide synthases, secretion systems, exopolysaccharide biosynthesis, catalases, and glycosyltransferases (22, 23).
FIG 4 .
Structure of the M. extorquens DSM13060 genome and origins of genes. Large circle, DSM13060 chromosome; small circle, DSM13060 megaplasmid (not to scale). Successive circles from inside to outside are as follows for both chromosome and megaplasmid. First circle, AM1 BLAST hits (E < 1e−30); colors denote percentages of identity between orthologous proteins, with red denoting 100%, orange <100% and >85%, and yellow <85%. Second to 6th circles, OrthoMCL orthologs in M. extorquens strains AM1, PA1, CM4, DM4, and BJ001. Seventh to 10th circles, inferred gene origins (see Table S1 in the supplemental material). Eleventh circle, HGT to Methylobacterium spp., M. extorquens spp., or DSM13060. The outermost circle on the chromosome shows the arrangement of the 5 largest contigs in the DSM13060 scaffold, assuming colinearity with strain AM1. The outermost circles on the megaplasmid show regions that differ in their degrees of similarity to the AM1 megaplasmid (see “Genome sequence and structure,” in the text). The color coding keyed to the phylogenic tree shows when genes in different categories may have been imported, as follows: red denotes genes of the DSM13060 lineage, purple genes of the DSM13060/AM1 lineages, and green genes present before the diversification of the genus. TFSS, type IV secretion system.
Purifying selection and lifestyle.
Intracellularity, when combined with obligate host association and vertical transmission, often leads to massive gene loss, small genome size, and relaxed purifying selection on genes, as commonly observed in, e.g., insect symbionts (24). While common in animal symbionts and pathogens, only a few examples of genome reduction in plant symbionts and pathogens exist (25–27). Although DSM13060 colonizes the interior of host cells, its genome is not reduced relative to those of other Methylobacterium species (see Fig. S1 in the supplemental material). On the contrary, the presence of the shared megaplasmid makes the DSM13060 and AM1 genomes the largest among all M. extorquens strains. We found that the average ω ratio (ratio of nonsynonymous [dN] to synonymous [dS] substitutions) between DSM13060 and AM1 (ω = 0.12 ± 0.03 [mean ± standard deviation]) was higher than is typical for free-living bacteria (0.02 to 0.09), but this was largely due to weaker purifying selection on the megaplasmid (ω = 0.16 ± 0.03) than on the chromosome (ω = 0.09 ± 0.03). A similar pattern has been shown in other bacteria and has been proposed to result from the sorting of less frequently used and therefore faster-evolving genes to megaplasmids (28).
M. extorquens DSM13060’s ability to colonize the bud meristem cells (9) and flower and seed tissues (A. M. Pirttilä, H. Pospiech, H. Laukkanen, R. Myllylä, A. Hohtola, unpublished data) raises the intriguing possibility that it is vertically transmitted via the shoot meristem to seeds, but the strain’s culturability in vitro and ability to colonize epidermal cells suggests a facultative host association. Indeed, the large genome size, large amount of HGTs, strong purifying selection, and distribution of functional clusters of orthologous groups (COG) categories are all characteristic of a free-living bacterium with large population size, suggesting that between hosts, DSM13060 spends time in the environment, e.g., the soil, water, phyllosphere, and endosphere of other plant species. It is possible that obligate association with large and long-lived plants is different than obligate association with animals and does not limit bacterial population size. While the occurrence of DSM13060 has not been studied in other hosts or environments, the bacterium could conceivably spend most of its time throughout the tissues of Scots pine and, over hundreds to thousands of years, encounter other endophytic bacteria and experience extensive gene transfer. The close relative M. extorquens AM1 was isolated as an airborne contaminant. It is an efficient phyllosphere colonizer (29), but to our knowledge, its ability to colonize internal tissues and cells has not been evaluated.
Genes involved in intracellular colonization.
The mechanism behind host cell entry in DSM13060 is unknown but likely involves active degradation of the plant cell wall using glycoside hydrolases (GH), such as cellulases/endoglucanases, lichenases, and xylanases. Rhizobial intracellular plant cell colonization involves local degradation of host cell walls by the bacterial endoglucanase CelC2 (G8) (30) or a pectate lyase produced by the plant host after local induction by rhizobium-produced Nod factors (31). DSM13060 encodes two putative exoglucanases (GH3) and an endoglucanase (G8). The latter is a homolog of CelC2, which also plays a role in rhizobial cellulose biosynthesis and biofilm formation (32) and, in most species, is located next to cellulose synthase genes. DSM13060 lacks these cellulose synthase genes, and cell wall degradation may be the sole function of the DSM13060 celC2 homolog. In addition, a prophage unique to DSM13060 harbors two consecutive pectate lyase genes. Finally, DSM13060 encodes a putative glyoxal oxidase, an enzyme that is rare in bacteria but is involved in lignin degradation in fungi (33). Our experiments demonstrate that DSM13060 expresses and secretes cellulases (see Text S1 in the supplemental material).
Known endophytic and plant interaction traits.
A search for traits commonly associated with endophytes (23) showed that DSM13060 is comparable to other endophytes in all aspects except plant growth promotion. Our genome analysis confirmed previous observations that DSM13060 lacks the ability to synthesize auxin and other known plant hormones (12). Our experimental and bioinformatic analysis showed that DSM13060 does not encode the protein aminocyclopropane-1-carboxylate (ACC) deaminase (AcdS), which in many plant-associated bacteria promotes growth by lowering ethylene levels (34). The genome encodes a homolog of the acdS gene, but a 132-day colonization experiment with a reporter construct controlled by the promoter of this gene showed that it was not active during colonization, and DSM13060 was unable to utilize ACC as the sole N source in an in vitro plate assay (see Text S1). In accordance with these results, the amino acid residues in the active center of the DSM13060 acdS homolog suggest that it encodes a d-cysteine desulfhydrase. The two functions are interconvertible through a change in only 2 amino acids (35). Finally, no genes for synthesis of the plant growth-promoting volatiles acetoin and 2,3-butanediol (36) were found.
In an earlier study, DSM13060 was found to secrete adenine derivatives that are precursors of cytokinin, but not the phytohormone itself (12). Compared to most bacteria, the DSM13060/AM1 megaplasmid harbors two additional copies of adenylosuccinate synthase, which has a role in purine biosynthesis; this explains the demonstrated secretion of adenine derivatives.
Vitamin B12 production has been considered a plant growth-promoting trait in epiphytes (37) and endophytes (38). The DSM13060 genome encodes the complete pathway for vitamin B12 synthesis. However, our reporter gene construct for the cobalamin synthase (cobS) promoter of DSM13060 did not demonstrate activity inside the plant within 132 days (see Fig. S3 in the supplemental material), suggesting that bacterial vitamin B12 synthesis is not a main mechanism of host growth promotion in this endophyte.
In stem-nodulating Bradyrhizobium strains, photosynthesis is activated in response to far-red light by the bacteriophytochrome (BphP) (39) and is associated with higher infection rates (40). We constructed an mCherry fluorescent protein controlled by the BphP promoter and found it to be activated during the infection of plant cells in P. sylvestris seedling roots (see Fig. S3A in the supplemental material) but not shoots (Fig. S3B). When the bacteria penetrated deeper pine tissues, no promoter activity was seen. This suggests that, similar to Bradyrhizobium, the DSM13060 BphP is synthesized in the dark (39) and is important during the first steps of infection.
Secretion systems.
Genes for a type IV secretion system (TFSS), a type II secretion system, and a type I secretion system were identified on the megaplasmid and were shared only with strain AM1. The TFSS was scattered in four locations, TFSS1 to -4 (Fig. 4), and was closely related to the Legionella and dot/icm systems (see Fig. S4 in the supplemental material), which play a role in Legionella intracellular replication (41). The three secretion systems appear to have been imported together, along with a suite of replication- and recombination-associated genes, all most closely related to and often syntenic with genes from the halophilic alphaproteobacterium Salipiger mucescens (42).
Putative host effectors.
Other than the genes for synthesis of cytokinin precursors, no genes previously known to be involved in endophytic plant growth promotion were identified. Given the bacterium’s intracellular location and association with the host nucleus, we hypothesized that DSM13060 encodes protein effectors that are active in the host cytoplasm or nucleus. The nucleus has recently emerged as a key target for so called “nucleomodulins,” effectors produced by intracellular bacteria to modulate host transcription and other nuclear processes in order to subvert the host defense (18) or inhibit or promote cellular proliferation (43). Effectors that target the host cytoplasm or nucleus often display distinctive eukaryotic domains or functions (44).
We used a simple chi-square test to identify 233 DSM13060 Pfam domains (45) that were present more often than expected in eukaryotes and less often than expected in bacteria (i.e., eukaryote-like). By excluding mitochondrial sequences (some of which are present in members of the Alphaproteobacteria but are overall rare in bacteria), and Pfam families with established bacterial functions unrelated to host infection, we obtained 35 candidate host effectors (Table 1), which are discussed below. Although most were found in all Methylobacterium species under comparison, a few were unique to DSM13060/AM1 and located on the megaplasmid or were unique to DSM13060 and located on the chromosome.
TABLE 1 .
Proteins with eukaryote-like domains
| Eukaryote-like protein # | Pfam identifier | Pfam symbol | Percentage of sequences belonging to this Pfam family that are bacterial | Proposed function in host cell |
|---|---|---|---|---|
| 1 | pfam00777 | Glyco_transf_29 | 1.6 | Sialyltransferase; host immune evasion |
| 2, 3, 4 | pfam00068 | Phospholip_A2_1 | 2.1 | Host growth promotion |
| 5 | pfam06839 | zf-GRF | 2.8 | DNA binding; host transcription |
| 6 | pfam05686 | Glyco_transf_90 | 4.4 | None/various |
| 7 | pfam02229 | PC4 | 4.6 | DNA binding; host transcription |
| 8, 9 | pfam00400 | WD40 | 4.8 | Various (possibly host growth and development) |
| 10 | pfam13964 | Kelch_6 | 4.8 | None/various |
| 11 | pfam13202 | EF_hand_3 | 5.4 | Signaling; cell division, cell elongation, cell differentiation, and plant defense and stress responses |
| 12 | pfam13202 | EF_hand_3 | 5.4 | Signaling; cell division, cell elongation, cell differentiation, and plant defense and stress responses |
| 13 | pfam03098 | An_peroxidase | 5.8 | Systemic resistance |
| 14, 15 | pfam00069 | Pkinase | 6.5 | Protein kinase; host signal transduction |
| 16 | pfam12796 | Ank_2 | 7.4 | Protein or DNA binding; possibly host transcription |
| 17, 18 | pfam02201 | SWIB | 8.8 | Host chromatin remodeling |
| 19 | pfam00782 | DSPc | 9.3 | Tyrosine phosphatase; host signal transduction |
| 20 | pfam00450 | Peptidase_S10 | 9.5 | Defense |
| 21, 22 | pfam14226 | DIOX_N | 10.4 | Gibberellin biosynthesis |
| 23 | pfam11721 | Malectin | 10.6 | None/various |
| 24 | pfam11523 | DUF3223 | 10.9 | None/various |
| 25 | pfam07250 | Glyoxal_oxid_N | 12.3 | Lignin degradation |
| 26, 27, 28 | pfam04577 | DUF563 | 13.1 | None/various |
| 29–34 | pfam00067 | p450 | 13.7 | Gibberellin biosynthesis |
| 35 | pfam12799 | LRR_4 | 17.3 | Host defense suppression |
Plant growth and development.
Among the most interesting putative effectors identified were three copies of a putative phospholipase A2 (PLA2) (Table 1, eukaryote-like protein #2-4). The PLA2 domain is extremely rare in bacteria (less than 2% of the sequences in the Pfam database with this domain are bacterial) but ubiquitous in the genus Methylobacterium, where most strains have 2 to 3 copies (Fig. 5). PLA2 sequences were also found in some Firmicutes (Streptococcus, Lactobacillus, and Bacillus). All three proteins in DSM13060 were predicted to possess a signal peptide for secretion. PLA2 enzymes and their enzymatic products—free fatty acids and lysophospholipids (46)—are implicated in a range of cellular processes in plants (e.g., growth, development, stress responses, and defense signaling [46, 47]) and, therefore, are candidates for contributing to the growth-promoting effect of DSM13060 on P. sylvestris seedlings.
FIG 5 .
Phylogeny of a 73-bp conserved region of an alignment of the M. extorquens DSM13060 PLA2 protein, orthologs in the Methylobacterium spp. under study (Fig. S1), and all 60 similar sequences in the BLAST nr database (E < 0.001), excluding other Methylobacterium hits. Only bootstrap values above 50 are shown in the tree.
Gibberellins control diverse aspects of plant growth and development, including seed germination, stem elongation, leaf expansion, and flower and seed development (48). We identified two genes encoding putative oxoglutarate-dependent dioxygenases (2OGDs) (Table 1, eukaryote-like protein #21-22) in the DOXC class from the Pfam family DIOX_N, members of which are required for the biosynthesis of gibberellin in plants, along with terpene synthases and cytochrome P450 monooxygenases (48). Five putative cytochrome P450 monooxygenase proteins were identified in the Pfam-based search for eukaryote-like proteins (Table 1). However, the genome does not have any homologs of terpene synthases, which can explain why M. extorquens DSM13060 does not synthesize gibberellins (12).
Putative eukaryotic transcription factors.
Several of the identified proteins with eukaryote-like Pfam domains were putative eukaryotic transcription factors (TFs) (Table 1). Two were located in the TFSS3 region and could be secreted by the TFSS. The first consisted entirely of the SWIB domain (Table 1, eukaryote-like protein #17), which in eukaryotes is involved in chromatin remodeling. This domain is also found in a limited number of proteobacteria and is widespread in Chlamydia spp. (49). The second had two copies of the GRF zinc finger (zf-GRF) domain (Table 1, eukaryote-like protein #5), which is extremely unusual in bacteria (there were 854 eukaryotic sequences and 23 bacterial sequences in the Pfam database with this domain). Interestingly, in the DSM13060 protein and the AM1 orthologs, the two zf-GRFs appeared to be fused to the C-terminal end of a bacterial topoisomerase. There are many bacterial homologs in the GenBank nr (nonredundant) database corresponding to the topoisomerase part that, instead of the zf-GRF domain, have a topoisomerase DNA binding C4 zinc finger, a domain common in bacteria. The closest homolog of the N-terminal topoisomerase domain part is found in S. mucesens, which shares synteny with DSM13060/AM1 in this region and the other TFSS regions on the megaplasmid. This suggests that the domain fusion happened relatively recently. A separate BLASTP search using only the C terminus (with the zf-GRF domain) as a query revealed homology with eukaryotic proteins, suggesting that the gene fusion event occurred between genes for a bacterial and a eukaryotic protein. The top hits are from the obligate biotroph white rust pathogen (Albugo laibachii, Oomycota) of Arabidopsis (E = 1e−09) and from Phytophthora sojae (E = 1e−07), which both share domain architecture with the zf-GRF protein in DSM13060 (Topoisom_bac, Toprim, and zf-GRF). Given the role of topoisomerases in the transcriptional activation of genes (50) and the likely implication of zf-GRF in DNA binding, the gene may encode an effector with a role in host transcriptional activation.
Host signaling.
Another protein located in the TFSS3 region has two Ca2+-binding EF-hand motifs (Table 1, eukaryote-like protein #11). In plants, Ca2+ is an important intracellular messenger involved in a range of processes, e.g., cell division, cell elongation, cell differentiation, and plant defense and stress responses (51), and diverse plant proteins possess the Ca2+-binding EF-hand motif. A homolog of this protein was only found in AM1. Several pathogenic bacteria translocate protein kinases into the host cell cytoplasm to modulate eukaryotic signal transduction (52, 53). DSM13060 contains genes encoding two eukaryote-like serine/threonine protein kinases, one an HGT on the chromosome (Table 1, eukaryote-like protein #14) and the other with homologs in other Methylobacterium spp. (Table 1, eukaryote-like protein #15).
Ankyrin repeats.
Ankyrin repeat domains (Anks) mediate protein-protein interactions in eukaryotes and affect many cellular processes, including cell cycle progression, transcription, and cytoskeletal organization, and are most common in bacteria that colonize eukaryote cells (54). An Ank protein (Table 1, eukaryote-like protein #17) is widespread in the genus Methylobacterium and has homologs in plant-associated gammaproteobacteria, such as Pseudomonas species.
Defense and host immune evasion.
A eukaryote-like putative serine carboxypeptidase (Table 1, eukaryote-like protein #21) is found in M. extorquens DSM13060 and other Methylobacterium species, as well as in Bradyrhizobium, Xylella, Xanthomonas, and Burkholderia species. The wide distribution in plant-associated bacteria suggests a role in interaction with the host. Plant carboxypeptidases indirectly affect plant growth and development (55). Carboxypeptidases could also protect the bacteria against host-encoded defense-related proteases, as shown in oomycete plant pathogens (56).
DSM13060 contains genes encoding a leucine-rich repeat (LRR) protein kinase (Table 1, eukaryote-like protein #35) with homologs in other Methylobacterium species and plant-associated bacteria. The most similar sequences are found in the pathogenic Pseudomonas syringae strains and in the endophyte Pseudomonas synxantha DSM13080, isolated from P. sylvestris buds (9). LRR genes are well-known resistance genes involved in the plant defense system (57) but also fairly widespread in bacteria, where they are common effectors interacting with host cells and suppressing the host defense (58).
Plant defense and systemic resistance.
A putative eukaryote-like peroxidase with the domain An_peroxidase (Table 1, eukaryote-like protein #14) was identified. Overall, only a few homologs are found in other alphaproteobacteria; instead, they are widespread in Pseudomonas species. PP2561, a homolog in the rhizosphere colonizer Pseudomonas putida, has been shown to be essential for the induction of plant systemic resistance against foliar pathogens (59). In both DSM13060 and Pseudomonas spp., the gene is flanked by a type I secretion system, which may secrete the peroxidase into the host cytoplasm (59).
Exopolysaccharides.
Polysaccharides have been found to play a role in plant-microbe interactions in several systems (60). The M. extorquens DSM13060 genome includes two that are eukaryote-like and extremely rare in bacteria: a glycosyltransferase 29 (sialyltransferase), and a glycosyltransferase 90. In bacterial pathogens, sialylation (by sialyltransferases) is a molecular mimicking mechanism extensively used to evade the host immune system (61).
Conclusion.
Traditionally, research on bacterial endophytes is focused on agricultural crops, and the majority of endophytes studied to date colonize the apoplast or intracellular spaces of grass roots. Strain DSM13060 is different from previously sequenced endophyte strains in multiple ways; it was isolated from a gymnosperm, it colonizes shoot tissue, and it occupies the intracellular niche. The shoot meristem, a rapidly dividing tissue that gives rise to leaves and reproductive organs, hosts bacteria different from those in the roots (62). In animal symbionts and pathogens, intracellularity, even when facultative, can affect the molecular interaction and evolutionary trajectory of the association (63). Probably for these reasons, the molecular mechanisms underlying host growth stimulation appear different in strain DSM13060 than in previously studied endophytes.
We showed that M. extorquens DSM13060 colonizes host cells in association with the nucleus, while it is also capable of replicating in and moving through intercellular spaces. A bacterial nonpathogenic association with the host nucleus has, to our knowledge not been demonstrated in plants before. The discovery prompted us to search for nucleomodulins and effector proteins that could target nuclear functions. We identified a number of eukaryote-like proteins with potential roles in host-endophyte interaction. To our knowledge, there is no previous report of endophytic effectors. Effector proteins are most often described in plant (64, 65) and animal (66, 67) pathogens, with prominent exceptions like mycorrhizal fungi and rhizobia, which both use effectors to establish symbiosis with plants (68). At present, it is unknown whether M. extorquens DSM13060 translocates effectors to the nucleus. However, the nuclear aggregation and genomic evidence of eukaryote-like Pfam domains with nuclear functions (transcription and chromatin modeling) suggest a link between intracellular colonization and mediation of plant phenotypes, such as enhancement of seedling growth. We found eukaryote-like proteins with unusual domain architectures that may be the result of recent innovations, such as the fusion between a bacterial megaplasmid-encoded topoisomerase and a domain of a eukaryotic zinc finger domain.
Future research will determine whether M. extorquens DSM13060 secretes the predicted effectors, whether they modulate host nuclear processes, and whether such interactions underlie the strain’s remarkable ability to stimulate host growth and development. Intracellular endophytes may confer a more persistent advantage to plants than apoplast colonizers do. Therefore, a better understanding of this symbiosis can have broad implications for plant biotechnology, forestry, and agriculture.
MATERIALS AND METHODS
Methods for manipulations of bacterial and plant material, isolation of genomic DNA, genome sequencing and annotation, and gene activity assays are provided in the supplemental material (see Text S1 and Table S1).
Transmission electron microscopy (TEM).
A total of 10 bud samples of adult Scots pine were taken in April and May 2008 from a test site in the Botanical Gardens, University of Oulu. The buds were surface sterilized for 1 min in 70% ethanol and for 20 min in 6% calcium hypochlorite. After rinsing, bud scales were removed aseptically. Buds longer than 2 mm were dissected longitudinally. The samples were fixed overnight in a cold solution of 3% glutaraldehyde in 0.05 M sodium phosphate buffer, pH 7.0, postfixed in 1% OsO4 for 3 h, and embedded in Eponate resin. Thin sections were cut with a Nova Ultratome microtome and stained with uranyl acetate and lead citrate. Sections were visualized under a JEOL Jem 100B electron microscope.
Confocal laser scanning microscopy.
Seedlings were harvested at various time points to study the progress of pine colonization by M. extorquens DSM13060, using the GFP-tagged strain 13061 as described in reference 13. Roots, shoots, needles, and buds were cut into pieces 2 to 3 mm in diameter and fixed in 4% paraformaldehyde (wt/vol), 0.1 % glutaraldehyde (vol/vol), 20% glycerol (vol/vol), and 0.1 M sodium phosphate buffer (pH 7.4) at 4°C under vacuum. The root samples were fixed for 4 h, and the rest of the tissues were fixed for 9 h under vacuum, followed by an overnight incubation at 4°C. The fixed tissues were cut into 20- to 30-µm sections with a cryomicrotome (Reichert-Jung 2800 Frigocut with 2040 microtome) and mounted on microscopy slides with ProLong gold antifade reagent (Invitrogen). The pine tissue sections were studied with confocal laser scanning microscopy (lsm 5 Pascal; Carl Zeiss, Germany) using Plan-Neofluor 40×/1.3 and Plan-Apochromat 63×/1.4 oil objectives. The GFP fluorophore was excited at a wavelength of 488 nm by an argon ion laser, and emissions were detected using a 505-to-530-nm band-pass (BP) filter. The background autofluorescence of the plant tissues was detected using a 650-nm long-pass (LP) filter. A helium neon (HeNe) laser was used for excitation of the mCherry reporter at 543 nm, and emission was detected through a 560-to-615-nm BP filter. For multichannel images of GFP and mCherry, an HFT (HauptFarbTeiler) 488/543/633-nm beam splitter was used with a secondary NFT (NebenFarbTeiler) 545 dichromic mirror to discriminate between the emissions. The projections of all channels were analyzed and merged using Zeiss LSM Image Browser (version 4.2.0.121; Carl Zeiss, Germany). For the videos, image stacks were processed and analyzed using ZEN lite 2012 (Blue edition; Carl Zeiss).
Morphological identification of cell viability by AOEB staining.
Double staining with acridine orange (AO) and ethidium bromide (EB) allows differentiation between live and necrotic cells and enables different stages of PCD to be determined (15, 69). AO stains both live and dead cells, emitting green fluorescence when binding to double-stranded nuclear chromatin. EB will only penetrate cells that have lost cytoplasmic membrane integrity and can be detected as red fluorescence. Early-stage PCD cells have green-yellow irregular nuclei with slightly condensed or fragmented chromatin, visible in the form of bright green patches. Cells in late-stage PCD with disrupted cytoplasmic membranes incorporate EB into the chromatin, which dominates the AO and stains orange-red. Necrotic cells have bright orange nuclei with structurally normal morphology, similar to the nuclear morphology of viable cells.
Roots of in vitro-cultured Scots pine seedlings inoculated with M. extorquens 13061 and 13062 were sectioned as described above. Samples were washed twice with phosphate-buffered saline (PBS), pH 7.4, and treated with 25 µg/ml acridine orange and 25 µg/ml ethidium bromide in PBS buffer for 15 min at room temperature in the dark. Root sections of uninoculated pine seedlings were used as controls. Negative controls were incubated in 20 mM H2O2 for 2 h while positive controls were incubated in PBS for the same time before AOEB staining. After staining, the samples were washed three times with PBS. Fixation, cryosectioning, and confocal microscopic analysis were performed as described above, using appropriate excitation and emission wavelengths (488 nm/505-to-530-nm BP filter [excitation/emission] for AO, 514 nm/560-to-615-nm BP filter for EB, and 488 nm/650-nm LP filter for plant cell autofluorescence). The laser power and channel settings were kept equivalent for all samples for consistent results.
Genome analysis.
Methylobacterium orthologs were identified by using OrthoMCL. Alignments were made using MUSCLE (70). Maximum-likelihood (ML) phylogenies were inferred using RAxML-VI-HPC (71), with the GTRGAMMA model for nucleotide sequences, the predicted model of ProtTest2.4 (72) for protein sequences, and 1,000 bootstrap replicates. The species ML phylogeny presented in Fig. S1 was inferred using a concatenated nucleotide alignment from 447 single-copy orthologs (in the Methylobacterium spp. and the two outgroup species, Agrobacterium tumefaciens strain c58 and Bradyrhizobium japonicum strain USDA 110).
To search for proteins with eukaryote-like Pfam domains, we downloaded the Pfam database (version 27.0) and the NCBI Taxonomy database (used to classify sequences in each family as bacterial, eukaryotic, or other). The expected frequency of bacterial and eukaryote sequences in each family was calculated, and a chi-square test was used to compare the observed frequencies of bacterial and eukaryotic sequences to the expected frequencies. We identified 233 DSM13060 Pfam domains that were present more often than expected in eukaryotes (P < 0.05) and less often than expected in bacteria (P < 0.05) and for which 20% or less of the total sequences in the Pfam database for that family were bacterial sequences. We excluded any Pfam domains that were mitochondrial (see Text S1 in the supplemental material) or had established bacterial functions unrelated to host infection. Homologs of the PLA2 genes in DSM13060 were retrieved through a BLASTP search against the nonredundant database using the DSM13060 proteins as queries. All three proteins had the same 60 non-Methylobacterium matches. All matching sequences were downloaded (E ≥ 0.001). The PLA2 orthologs from Methylobacterium spp. under study (Fig. S1) were added, and the protein sequences were aligned using a gap penalty of −2 and manually inspected. The determination of sequence similarity across all sequences was limited to the area corresponding to the phospholipase_A2 Pfam motif. A phylogeny was inferred using this segment of the alignment only. For the DotC phylogeny shown in Fig. S4, we used a selected set of protein sequences, representing all genera with the most similar homologs of the DSM13060 dotC protein (BLASTP against the nr database, E < 1e−40). Codeml from the PAML package was used to calculate the ω ratio from alignments of DSM1360 and AM1 ortholog pairs. The alignments were made using protein sequences and then translated to nucleotide sequences.
Nucleotide sequence accession number.
The whole-genome shotgun sequences have been deposited in GenBank under accession number AGJK00000000.
SUPPLEMENTAL MATERIAL
Supplemental methods. Download
BLAST-based assignments of gene origins, showing the number of hits in each of the following taxonomic categories: non-extorquens Methylobacterium (m), non-Methylobacterium Rhizobiales (r), non-Rhizobiales Alphaproteobacteria, and non-Alphaproteobacteria (n), excluding hits classified as “unclassified sequences” and “other sequences.”
Strains, plasmids, and primers used in the study.
Maximum-likelihood genome phylogeny of DSM13060 and related species. Download
(A) Phylogeny of the RepA protein carried on the strain DSM13060 megaplasmid and homologs in members of the alphaproteobacteria. Red font shows Methylobacterium spp. (B) Synteny between the DSM13060 and AM1 megaplasmids. Colors denote the percentage of identity between orthologous proteins as follows: red, 100%; orange, <100% and >85%; yellow, <85%. Download
Reporter gene analysis of bacteriophytochrome (bphP), ACC deaminase (acdS), and cobalamin synthase (cobS) promoter activity in M. extorquens 13061 during colonization of Scots pine seedlings. M. extorquens strain 13061 with an mCherry reporter controlled by the promoter of the bphP, acdS, or cobS gene was used for laser scanning confocal microscopic analysis. The left image in each panel is a merged image of a pine tissue section infected by bacteria with both tags, and the middle and right images show bacteria carrying the mCherry (white) or GFP tag (green), respectively. In panels A and B, the middle and right images are magnifications of the boxed area in the left image. Cryosections of pine seedlings colonized by M. extorquens 13061 were analyzed 20 to 60 days after inoculation. (A) Cross section of a root displaying activated bphP in M. extorquens 13061 cells during active colonization of the outer cortex. (B) Lateral section of the lower part of a Scots pine shoot with a microcolony of M. extorquens 13061 on the epidermis without clear bphP activation. (C) Cross section of a Scots pine root. Arrows indicate M. extorquens 13061 cells without acdS promoter activity in the cortex and on the epidermis. (D) Cross section of a pine root where arrow shows an M. extorquens 13061 colony penetrating through the root epidermis without visible activity of the cobS promoter. (E) Cross section of a pine stem where arrows depict advancing 13061 colonies in the cortex without cobS promoter activity. Co, cortex; E, epiderm; En, endoderm; n, nucleus; Xy, xylem; scale bars, 10 µm. Download
Maximum-likelihood phylogeny of the DotC protein in DSM13060 and homologs from various genera, showing relationship to the Legionella DotC protein from the dot/icm Legionella TFSS. Download
Merged confocal z-stack images showing intracellular M. extorquens 13061 cells in the parenchymal cells of a shoot of Scots pine. The red dashed line highlights the GFP-tagged bacterial cells (bright green), and the plant tissue morphology visible in the green and red channels is due to autofluorescence. Images were collected at intervals of 1 µm (z axis), depicted by the numbering in the top left corner. Co, cortex; n, nucleus. Images were processed and prepared with Zen lite 2012 software (Carl Zeiss). Download
Merged confocal z-stack images of AOEB-stained Scots pine roots showing M. extorquens 13061 inside healthy parenchymal cells. Nuclear morphology of the plant cells with uniform incorporation of AO (green) over EB (red) and without condensation of chromatin corresponds with cellular viability. Bacterial cells carrying a fluorescent GFP reporter are visualized in bright green, and endogenous autofluorescence of plant cells is shown in gray/purple. The white arrows indicate intracellular bacteria in living cortical cells of the pine root. Images were collected at intervals of 1 µm (z axis), depicted by the numbering in the top left corner. Co, cortex; E, epiderm; n, nucleus; Xy, xylem. Images were processed and prepared with Zen lite 2012 software (Carl Zeiss). Download
Merged confocal z-stack images of AOEB-stained pine roots showing M. extorquens 13062 residing close to the nucleus of a cortical cell. Nuclear morphology of the plant cells with uniform incorporation of AO (green) over EB (red) and without condensation of chromatin corresponds with cellular viability. Bacterial cells carrying fluorescent GFP and mCherry reporters under the control of constitutive promoters are visualized in white-red to differentiate them from the green nucleus. The white arrow indicates intracellular bacteria congregated around the nucleus of a living cortical cell. Images were collected at intervals of 1 µm (z axis), depicted by the numbering in the top left corner. Co, cortex; E, epiderm; n, nucleus. Images were processed and prepared with Zen lite 2012 software (Carl Zeiss). Download
ACKNOWLEDGMENTS
The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. This work was supported by UC Merced startup funds, Academy of Finland (grants 129852 and 113607), Niemi Foundation, and Tauno Tönning Foundation.
We thank E. L. Lagendijk, T. Woyke, J. Pohjanen, and T. Uusitalo.
Footnotes
Citation Koskimäki JJ, Pirttilä AM, Ihantola E-L, Halonen O, Frank AC. 2015. The intracellular Scots pine shoot symbiont Methylobacterium extorquens DSM13060 aggregates around the host nucleus and encodes eukaryote-like proteins. mBio 6(2):e00039-15. doi:10.1128/mBio.00039-15.
Contributor Information
Angela Sessitsch, AIT Austrian Institute of Technology GmbH.
Nicole Dubilier, Max Planck Institute for Marine Microbiology.
REFERENCES
- 1.Rosenblueth M, Martínez-Romero E. 2006. Bacterial endophytes and their interactions with hosts. Mol Plant Microbe Interact 19:827–837. doi: 10.1094/MPMI-19-0827. [DOI] [PubMed] [Google Scholar]
- 2.Hardoim PR, van Overbeek LS, Elsas JD. 2008. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16:463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Cankar K, Kraigher H, Ravnikar M, Rupnik M. 2005. Bacterial endophytes from seeds of Norway spruce (Picea abies L. Karst). FEMS Microbiol Lett 244:341–345. doi: 10.1016/j.femsle.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Johnston-Monje D, Raizada MN. 2011. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS One 6:e20396. doi: 10.1371/journal.pone.0020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quambusch M, Pirttilä AM, Tejesvi MV, Winkelmann T, Bartsch M. 2014. Endophytic bacteria in plant tissue culture: differences between easy- and difficult-to-propagate Prunus avium genotypes. Tree Physiol 34:524–533. doi: 10.1093/treephys/tpu027. [DOI] [PubMed] [Google Scholar]
- 6.Cocking EC, Stone PJ, Davey MR. 2006. Intracellular colonization of roots of Arabidopsis and crop plants by Gluconacetobacter diazotrophicus. In Vitro Cell Dev Biol Plant 42:74–82. doi: 10.1079/IVP2005716. [DOI] [Google Scholar]
- 7.Chaintreuil C, Giraud E, Prin Y, Lorquin J, Bâ A, Gillis M, de Lajudie P, Dreyfus B. 2000. Photosynthetic bradyrhizobia are natural endophytes of the African wild rice Oryza breviligulata. Appl Environ Microbiol 66:5437–5447. doi: 10.1128/AEM.66.12.5437-5447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James E, Olivares FL. 1997. Infection and colonization of sugar cane and other graminaceous plants by endophytic diazotrophs. Crit Rev Plant Sci 17:77–119. doi: 10.1016/S0735-2689(98)00357-8. [DOI] [Google Scholar]
- 9.Pirttilä AM, Laukkanen H, Pospiech H, Myllylä R, Hohtola A. 2000. Detection of intracellular bacteria in the buds of Scotch pine (Pinus sylvestris L.) by in situ hybridization. Appl Environ Microbiol 66:3073–3077. doi: 10.1128/AEM.66.7.3073-3077.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pirttilä AM, Pospiech H, Laukkanen H, Myllylä R, Hohtola A. 2003. Two endophytic fungi in different tissues of Scots pine buds (Pinus sylvestris L.). Microb Ecol 45:53–62. doi: 10.1007/s00248-002-1038-8. [DOI] [PubMed] [Google Scholar]
- 11.Pirttilä AM, Pospiech H, Laukkanen H, Myllylä R, Hohtola A. 2005. Seasonal variations in location and population structure of endophytes in buds of Scots pine. Tree Physiol 25:289–297. doi: 10.1093/treephys/25.3.289. [DOI] [PubMed] [Google Scholar]
- 12.Pirttilä AM, Joensuu P, Pospiech H, Jalonen J, Hohtola A. 2004. Bud endophytes of Scots pine produce adenine derivatives and other compounds that affect morphology and mitigate browning of callus cultures. Physiol Plant 121:305–312. doi: 10.1111/j.0031-9317.2004.00330.x. [DOI] [PubMed] [Google Scholar]
- 13.Pohjanen J, Koskimäki JJ, Sutela S, Ardanov P, Suorsa M, Niemi K, Sarjala T, Häggman H, Pirttilä AM. 2014. Interaction with ectomycorrhizal fungi and endophytic Methylobacterium affects the nutrient uptake and growth of pine seedlings in vitro. Tree Physiol 34:993–1005. doi: 10.1093/treephys/tpu062. [DOI] [PubMed] [Google Scholar]
- 14.Koenig RL, Morris RO, Polacco JC. 2002. tRNA is the source of low-level trans-zeatin production in Methylobacterium spp. J Bacteriol 184:1832–1842. doi: 10.1128/JB.184.7.1832-1842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renvoizé C, Biola A, Pallardy M, Bréard J. 1998. Apoptosis: identification of dying cells. Cell Biol Toxicol 14:111–120. doi: 10.1023/A:1007429904664. [DOI] [PubMed] [Google Scholar]
- 16.Schulz F, Lagkouvardos I, Wascher F, Aistleitner K, Kostanjšek R, Horn M. 2014. Life in an unusual intracellular niche: a bacterial symbiont infecting the nucleus of amoebae. ISME J 8:1634–1644. doi: 10.1038/ismej.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zielinski FU, Pernthaler A, Duperron S, Raggi L, Giere O, Borowski C, Dubilier N. 2009. Widespread occurrence of an intranuclear bacterial parasite in vent and seep bathymodiolin mussels. Environ Microbiol 11:1150–1167. doi: 10.1111/j.1462-2920.2008.01847.x. [DOI] [PubMed] [Google Scholar]
- 18.Bierne H, Cossart P. 2012. When bacteria target the nucleus: the emerging family of nucleomodulins. Cell Microbiol 14:622–633. doi: 10.1111/j.1462-5822.2012.01758.x. [DOI] [PubMed] [Google Scholar]
- 19.Vuilleumier S, Chistoserdova L, Lee M-C, Bringel F, Lajus A, Zhou Y, Gourion B, Barbe V, Chang J, Cruveiller S, Dossat C, Gillett W, Gruffaz C, Haugen E, Hourcade E, Levy R, Mangenot S, Muller E, Nadalig T, Pagni M, Penny C, Peyraud R, Robinson DG, Roche D, Rouy Z, Saenampechek C, Salvignol G, Vallenet D, Wu Z, Marx CJ, Vorholt JA, Olson MV, Kaul R, Weissenbach J, Médigue C, Lidstrom ME. 2009. Methylobacterium genome sequences: a reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources. PLoS One 4:e5584. doi: 10.1371/journal.pone.0005584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marx CJ, Bringel F, Chistoserdova L, Moulin L, Farhan Ul Haque M, Fleischman DE, Gruffaz C, Jourand P, Knief C, Lee M-C, Muller EE, Nadalig T, Peyraud R, Roselli S, Russ L, Goodwin LA, Ivanova N, Kyrpides N, Lajus A, Land ML, Medigue C, Mikhailova N, Nolan M, Woyke T, Stolyar S, Vorholt JA, Vuilleumier S. 2012. Complete genome sequences of six strains of the genus Methylobacterium. J Bacteriol 194:4746–4748. doi: 10.1128/JB.01009-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinto UM, Pappas KM, Winans SC. 2012. The ABCs of plasmid replication and segregation. Nat Rev Microbiol 10:755–765. doi: 10.1038/nrmicro2882. [DOI] [PubMed] [Google Scholar]
- 22.Frank AC. 2011. The genomes of endophytic bacteria, p 107–137. In Pirttila A, Frank AC (ed), Endophytes of forest trees: biology and applications. Springer Verlag, Dordrecht, Netherlands. [Google Scholar]
- 23.Mitter B, Petric A, Shin MW, Chain PS, Hauberg-Lotte L, Reinhold-Hurek B, Nowak J, Sessitsch A. 2013. Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front Plant Sci 4:120. doi: 10.3389/fpls.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 25.Ran L, Larsson J, Vigil-Stenman T, Nylander JA, Ininbergs K, Zheng WW, Lapidus A, Lowry S, Haselkorn R, Bergman B. 2010. Genome erosion in a nitrogen-fixing vertically transmitted endosymbiotic multicellular cyanobacterium. PLoS One 5:e11486. doi: 10.1371/journal.pone.0011486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlier AL, Eberl L. 2012. The eroded genome of a Psychotria leaf symbiont: hypotheses about lifestyle and interactions with its plant host. Environ Microbiol 14:2757–2769. doi: 10.1111/j.1462-2920.2012.02763.x. [DOI] [PubMed] [Google Scholar]
- 27.Hartung JS, Shao J, Kuykendall LD. 2011. Comparison of the “Ca. Liberibacter asiaticus” genome adapted for an intracellular lifestyle with other members of the Rhizobiales. PLoS One 6:e23289. doi: 10.1371/journal.pone.0023289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper VS, Vohr SH, Wrocklage SC, Hatcher PJ. 2010. Why genes evolve faster on secondary chromosomes in bacteria. PLoS Comput Biol 6:e1000732. doi: 10.1371/journal.pcbi.1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gourion B, Rossignol M, Vorholt JA. 2006. A proteomic study of Methylobacterium extorquens reveals a response regulator essential for epiphytic growth. Proc Natl Acad Sci U S A 103:13186–13191. doi: 10.1073/pnas.0603530103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robledo M, Jiménez-Zurdo JI, Velázquez E, Trujillo ME, Zurdo-Piñeiro JL, Ramírez-Bahena MH, Ramos B, Díaz-Mínguez JM, Dazzo F, Martínez-Molina E, Mateos PF. 2008. Rhizobium cellulase CelC2 is essential for primary symbiotic infection of legume host roots. Proc Natl Acad Sci U S A 105:7064–7069. doi: 10.1073/pnas.0802547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie F, Murray JD, Kim J, Heckmann AB, Edwards A, Oldroyd GE, Downie JA. 2012. Legume pectate lyase required for root infection by rhizobia. Proc Natl Acad Sci U S A 109:633–638. doi: 10.1073/pnas.1113992109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robledo M, Rivera L, Jiménez-Zurdo JI, Rivas R, Dazzo F, Velázquez E, Martínez-Molina E, Hirsch AM, Mateos PF. 2012. Role of Rhizobium endoglucanase CelC2 in cellulose biosynthesis and biofilm formation on plant roots and abiotic surfaces. Microb Cell Fact 11:125. doi: 10.1186/1475-2859-11-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whittaker MM, Kersten PJ, Cullen D, Whittaker JW. 1999. Identification of catalytic residues in glyoxal oxidase by targeted mutagenesis. J Biol Chem 274:36226–36232. doi: 10.1074/jbc.274.51.36226. [DOI] [PubMed] [Google Scholar]
- 34.Glick BR. 2005. Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol Lett 251:1–7. doi: 10.1016/j.femsle.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 35.Todorovic B, Glick BR. 2008. The interconversion of ACC deaminase and d-cysteine desulfhydrase by directed mutagenesis. Planta 229:193–205. doi: 10.1007/s00425-008-0820-3. [DOI] [PubMed] [Google Scholar]
- 36.Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Paré PW, Kloepper JW. 2003. Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci U S A 100:4927–4932. doi: 10.1073/pnas.0730845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toraya T, Yongsmith B, Tanaka A, Fukui S. 1975. Vitamin B12 production by a methanol-utilizing bacterium. Appl Microbiol 30:477–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivanova EG, Pirttilä AM, Fedorov DNF, Doronina NV, Trotsenko YA. 2008. Association of methylotrophic bacteria with plants: Metabolic aspects, p 225–231. In Sorvari S, Pirttilä AM (ed), Prospects and applications for plant associated microbes. A laboratory manual, part A: Bacteria. Biobien Innovations, Turku, Finland. [Google Scholar]
- 39.Giraud E, Fardoux J, Fourrier N, Hannibal L, Genty B, Bouyer P, Dreyfus B, Verméglio A. 2002. Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature 417:202–205. doi: 10.1038/417202a. [DOI] [PubMed] [Google Scholar]
- 40.Giraud E, Hannibal L, Fardoux J, Verméglio A, Dreyfus B. 2000. Effect of Bradyrhizobium photosynthesis on stem nodulation of Aeschynomene sensitiva. Proc Natl Acad Sci U S A 97:14795–14800. doi: 10.1073/pnas.250484097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brand BC, Sadosky AB, Shuman HA. 1994. The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol Microbiol 14:797–808. doi: 10.1111/j.1365-2958.1994.tb01316.x. [DOI] [PubMed] [Google Scholar]
- 42.Martínez-Cánovas MJ, Quesada E, Martínez-Checa F, del Moral A, Béjar V. 2004. Salipiger mucescens gen. nov., sp. nov., a moderately halophilic, exopolysaccharide-producing bacterium isolated from hypersaline soil, belonging to the alpha-Proteobacteria. Int J Syst Evol Microbiol 54:1735–1740. doi: 10.1099/ijs.0.63166-0. [DOI] [PubMed] [Google Scholar]
- 43.Nougayrède J-P, Taieb F, De Rycke J, Oswald E. 2005. Cyclomodulins: bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol 13:103–110. doi: 10.1016/j.tim.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Hubber A, Roy CR. 2010. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 45.Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. 2000. The Pfam protein families database. Nucleic Acids Res 28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen G, Snyder CL, Greer MS, Weselake RJ. 2011. Biology and biochemistry of plant phospholipases. Crit Rev Plant Sci 30:239–258. doi: 10.1080/07352689.2011.572033. [DOI] [Google Scholar]
- 47.Chapman KD. 1998. Phospholipase activity during plant growth and development and in response to environmental stress. Trends Plant Sci 3:419–426. doi: 10.1016/S1360-1385(98)01326-0. [DOI] [Google Scholar]
- 48.Yamaguchi S. 2008. Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 49.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 50.Pedersen JM, Fredsoe J, Roedgaard M, Andreasen L, Mundbjerg K, Kruhøffer M, Brinch M, Schierup MH, Bjergbaek L, Andersen AH. 2012. DNA topoisomerases maintain promoters in a state competent for transcriptional activation in Saccharomyces cerevisiae. PLoS Genet 8:e1003128. doi: 10.1371/journal.pgen.1003128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poovaiah BW, Reddy AS. 1993. Calcium and signal transduction in plants. Crit Rev Plant Sci 12:185–211. doi: 10.1080/07352689309701901. [DOI] [PubMed] [Google Scholar]
- 52.Cozzone AJ. 2005. Role of protein phosphorylation on serine/threonine and tyrosine in the virulence of bacterial pathogens. J Mol Microbiol Biotechnol 9:198–213. doi: 10.1159/000089648. [DOI] [PubMed] [Google Scholar]
- 53.Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, Klebl B, Thompson C, Bacher G, Pieters J. 2004. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science 304:1800–1804. doi: 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- 54.Mosavi LK, Cammett TJ, Desrosiers DC, Peng Z-Y. 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci 13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li J, Lease KA, Tax FE, Walker JC. 2001. BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc Natl Acad Sci U S A 98:5916–5921. doi: 10.1073/pnas.091065998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adhikari BN, Hamilton JP, Zerillo MM, Tisserat N, Lévesque CA, Buell CR. 2013. Comparative genomics reveals Insight into virulence strategies of plant pathogenic oomycetes. PLoS One 8:e75072. doi: 10.1371/journal.pone.0075072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Belkhadir Y, Wang X, Chory J. 2006. Brassinosteroid signaling pathway. Sci STKE 2006:cm4. doi: 10.1126/stke.3642006cm4. [DOI] [PubMed] [Google Scholar]
- 58.Kedzierski Ł, Montgomery J, Curtis J, Handman E. 2004. Leucine-rich repeats in host-pathogen interactions. Arch Immunol Ther Exp (Warsz) 52:104–112. [PubMed] [Google Scholar]
- 59.Matilla MA, Ramos JL, Bakker PA, Doornbos R, Badri DV, Vivanco JM, Ramos-González MI. 2010. Pseudomonas putida KT2440 causes induced systemic resistance and changes in Arabidopsis root exudation: phytoprotection by Pseudomonas putida. Environ Microbiol Rep 2:381–388. doi: 10.1111/j.1758-2229.2009.00091.x. [DOI] [PubMed] [Google Scholar]
- 60.Jones KM, Sharopova N, Lohar DP, Zhang JQ, VandenBosch KA, Walker GC. 2008. Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proc Natl Acad Sci U S A 105:704–709. doi: 10.1073/pnas.0709338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martínez-Moliner V, Soler-Llorens P, Moleres J, Garmendia J, Aragon V. 2012. Distribution of genes involved in sialic acid utilization in strains of Haemophilus parasuis. Microbiology 158:2117–2124. doi: 10.1099/mic.0.056994-0. [DOI] [PubMed] [Google Scholar]
- 62.Pirttilä AM. 2011. Endophytic bacteria in tree shoot tissues and their effects on host, p 139–149. In Pirttila AM, Frank AC (ed), Endophytes of forest trees: biology and applications. Springer Verlag. [Google Scholar]
- 63.Toft C, Andersson SG. 2010. Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet 11:465–475. doi: 10.1038/nrg2798. [DOI] [PubMed] [Google Scholar]
- 64.Canonne J, Rivas S. 2012. Bacterial effectors target the plant cell nucleus to subvert host transcription. Plant Signal Behav 7:217–221. doi: 10.4161/psb.18885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang S, Yao J, Ma K-W, Zhou H, Song J, He SY, Ma W. 2013. Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog 9:e1003715. doi: 10.1371/journal.ppat.1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alvarez-Venegas R. 2014. Bacterial SET domain proteins and their role in eukaryotic chromatin modification. Front Genet 5:65. doi: 10.3389/fgene.2014.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deakin WJ, Broughton WJ. 2009. Symbiotic use of pathogenic strategies: rhizobial protein secretion systems. Nat Rev Microbiol 7:312–320. doi: 10.1038/nrmicro2091. [DOI] [PubMed] [Google Scholar]
- 68.Plett JM, Martin F. 2011. Mutualistic effectors: architects of symbiosis, p 295–326. In Martin F, Kamoun S (ed), Effectors in plant-microbe interactions. Wiley-Blackwell, Oxford, United Kingdom. [Google Scholar]
- 69.Byczkowska A, Kunikowska A, Kaźmierczak A. 2013. Determination of ACC-induced cell-programmed death in roots of Vicia faba ssp. minor seedlings by acridine orange and ethidium bromide staining. Protoplasma 250:121–128. doi: 10.1007/s00709-012-0383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. BioInformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 72.Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. BioInformatics 21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download
BLAST-based assignments of gene origins, showing the number of hits in each of the following taxonomic categories: non-extorquens Methylobacterium (m), non-Methylobacterium Rhizobiales (r), non-Rhizobiales Alphaproteobacteria, and non-Alphaproteobacteria (n), excluding hits classified as “unclassified sequences” and “other sequences.”
Strains, plasmids, and primers used in the study.
Maximum-likelihood genome phylogeny of DSM13060 and related species. Download
(A) Phylogeny of the RepA protein carried on the strain DSM13060 megaplasmid and homologs in members of the alphaproteobacteria. Red font shows Methylobacterium spp. (B) Synteny between the DSM13060 and AM1 megaplasmids. Colors denote the percentage of identity between orthologous proteins as follows: red, 100%; orange, <100% and >85%; yellow, <85%. Download
Reporter gene analysis of bacteriophytochrome (bphP), ACC deaminase (acdS), and cobalamin synthase (cobS) promoter activity in M. extorquens 13061 during colonization of Scots pine seedlings. M. extorquens strain 13061 with an mCherry reporter controlled by the promoter of the bphP, acdS, or cobS gene was used for laser scanning confocal microscopic analysis. The left image in each panel is a merged image of a pine tissue section infected by bacteria with both tags, and the middle and right images show bacteria carrying the mCherry (white) or GFP tag (green), respectively. In panels A and B, the middle and right images are magnifications of the boxed area in the left image. Cryosections of pine seedlings colonized by M. extorquens 13061 were analyzed 20 to 60 days after inoculation. (A) Cross section of a root displaying activated bphP in M. extorquens 13061 cells during active colonization of the outer cortex. (B) Lateral section of the lower part of a Scots pine shoot with a microcolony of M. extorquens 13061 on the epidermis without clear bphP activation. (C) Cross section of a Scots pine root. Arrows indicate M. extorquens 13061 cells without acdS promoter activity in the cortex and on the epidermis. (D) Cross section of a pine root where arrow shows an M. extorquens 13061 colony penetrating through the root epidermis without visible activity of the cobS promoter. (E) Cross section of a pine stem where arrows depict advancing 13061 colonies in the cortex without cobS promoter activity. Co, cortex; E, epiderm; En, endoderm; n, nucleus; Xy, xylem; scale bars, 10 µm. Download
Maximum-likelihood phylogeny of the DotC protein in DSM13060 and homologs from various genera, showing relationship to the Legionella DotC protein from the dot/icm Legionella TFSS. Download
Merged confocal z-stack images showing intracellular M. extorquens 13061 cells in the parenchymal cells of a shoot of Scots pine. The red dashed line highlights the GFP-tagged bacterial cells (bright green), and the plant tissue morphology visible in the green and red channels is due to autofluorescence. Images were collected at intervals of 1 µm (z axis), depicted by the numbering in the top left corner. Co, cortex; n, nucleus. Images were processed and prepared with Zen lite 2012 software (Carl Zeiss). Download
Merged confocal z-stack images of AOEB-stained Scots pine roots showing M. extorquens 13061 inside healthy parenchymal cells. Nuclear morphology of the plant cells with uniform incorporation of AO (green) over EB (red) and without condensation of chromatin corresponds with cellular viability. Bacterial cells carrying a fluorescent GFP reporter are visualized in bright green, and endogenous autofluorescence of plant cells is shown in gray/purple. The white arrows indicate intracellular bacteria in living cortical cells of the pine root. Images were collected at intervals of 1 µm (z axis), depicted by the numbering in the top left corner. Co, cortex; E, epiderm; n, nucleus; Xy, xylem. Images were processed and prepared with Zen lite 2012 software (Carl Zeiss). Download
Merged confocal z-stack images of AOEB-stained pine roots showing M. extorquens 13062 residing close to the nucleus of a cortical cell. Nuclear morphology of the plant cells with uniform incorporation of AO (green) over EB (red) and without condensation of chromatin corresponds with cellular viability. Bacterial cells carrying fluorescent GFP and mCherry reporters under the control of constitutive promoters are visualized in white-red to differentiate them from the green nucleus. The white arrow indicates intracellular bacteria congregated around the nucleus of a living cortical cell. Images were collected at intervals of 1 µm (z axis), depicted by the numbering in the top left corner. Co, cortex; E, epiderm; n, nucleus. Images were processed and prepared with Zen lite 2012 software (Carl Zeiss). Download