ABSTRACT
Resistance following antiviral therapy is commonly observed in human influenza viruses. Although this evolutionary process is initiated within individual hosts, little is known about the pattern, dynamics, and drivers of antiviral resistance at this scale, including the role played by reassortment. In addition, the short duration of human influenza virus infections limits the available time window in which to examine intrahost evolution. Using single-molecule sequencing, we mapped, in detail, the mutational spectrum of an H3N2 influenza A virus population sampled from an immunocompromised patient who shed virus over a 21-month period. In this unique natural experiment, we were able to document the complex dynamics underlying the evolution of antiviral resistance. Individual resistance mutations appeared weeks before they became dominant, evolved independently on cocirculating lineages, led to a genome-wide reduction in genetic diversity through a selective sweep, and were placed into new combinations by reassortment. Notably, despite frequent reassortment, phylogenetic analysis also provided evidence for specific patterns of segment linkage, with a strong association between the hemagglutinin (HA)- and matrix (M)-encoding segments that matches that previously observed at the epidemiological scale. In sum, we were able to reveal, for the first time, the complex interaction between multiple evolutionary processes as they occur within an individual host.
IMPORTANCE
Understanding the evolutionary forces that shape the genetic diversity of influenza virus is crucial for predicting the emergence of drug-resistant strains but remains challenging because multiple processes occur concurrently. We characterized the evolution of antiviral resistance in a single persistent influenza virus infection, representing the first case in which reassortment and the complex patterns of drug resistance emergence and evolution have been determined within an individual host. Deep-sequence data from multiple time points revealed that the evolution of antiviral resistance reflects a combination of frequent mutation, natural selection, and a complex pattern of segment linkage and reassortment. In sum, these data show how immunocompromised hosts may help reveal the drivers of strain emergence.
INTRODUCTION
Influenza A viruses are characterized by rapidly accumulating genetic diversity caused by a combination of error-prone replication (1), frequent reassortment during coinfections (2), and strong natural selection (3). These processes are also central to the host adaptation of newly emerging influenza viruses following cross-species transmission and to the evolution of drug resistance in human populations. However, despite their obvious importance, the frequencies, patterns, and consequences of mutation, reassortment, and natural selection as they occur within individual hosts are poorly understood. Indeed, most of what is known about the evolution of influenza A virus comes from population-level epidemiological studies based on the analysis of viral consensus sequences (reviewed in reference 4). In contrast, studying the drivers of human influenza virus genetic diversity within an infected host is challenging because the infection period is usually short: the typical incubation period for influenza A virus averages 2 days, with shedding of virus occurring from a day before the appearance of symptoms to approximately 7 to 10 days after onset of illness (5, 6). Hence, longitudinal sampling from influenza A virus-infected patients usually represents a limited set of time points for study. However, in severely immunocompromised patients virus shedding can continue for months (7, 8). Similar long-term shedding has been observed in a number of other normally acute human viral infections, including those by norovirus (9) and respiratory syncytial virus (10), in which immunosuppressed individuals appear to serve as a reservoir for the generation and spread of novel viral variants. In such immunocompromised patients, antiviral therapy also constitutes a major selection pressure, with drug resistance a common occurrence (11).
Millions of people worldwide are immunocompromised due to uncontrolled HIV infections or because of immunosuppressive chemotherapy to prevent organ transplant rejection, to control autoimmune diseases, or to treat inflammatory diseases. Patients with chronic virus shedding may effectively act as superspreaders, and because the viruses that they harbor can accumulate mutations under minimal immune pressure, in theory they may also initiate the emergence of novel strains.
Human influenza A viruses have a propensity for the rapid emergence of resistance following antiviral therapy (7, 8, 12–15). There are two classes of antiviral drugs approved for the treatment of influenza virus infections. The adamantanes (amantadine and rimantadine) block the viral ion channel protein. Resistance to these agents appears rapidly under treatment, and essentially all circulating strains of human influenza A viruses are now intrinsically resistant to these compounds, primarily due to the S31N substitution in the M2 protein (8, 13). Most therapeutic interventions are based on the use of the neuraminidase (NA) inhibitors (NAIs) oseltamivir and zanamivir, which are active against all influenza A virus subtypes and B strains. Neuraminidase mutations conferring resistance to these agents are subtype and drug specific (16). Importantly, immunocompromised subjects receiving prolonged therapy may develop multidrug-resistant influenza virus infections (7, 11, 17). However, the pathway of drug resistance evolution in such patients, as well as the compatibility of viral mutations, using sensitive detection methods such as deep sequencing, has been poorly studied. More generally, understanding the evolutionary forces that shape RNA virus genetic diversity and promote the emergence of drug resistance within infected hosts may ultimately assist in the design and deployment of antiviral therapies.
To determine the evolutionary processes that lead to the emergence of resistant viruses, we analyzed in detail, and over an extended 21-month period, influenza A virus genetic diversity by performing deep sequencing of H3N2 virus populations from a patient with severe combined immunodeficiency disease (SCID) who received prolonged antiviral therapy (7). This unique natural experiment provides a rare view into the patterns, dynamics, and mechanisms of drug resistance of influenza virus.
RESULTS
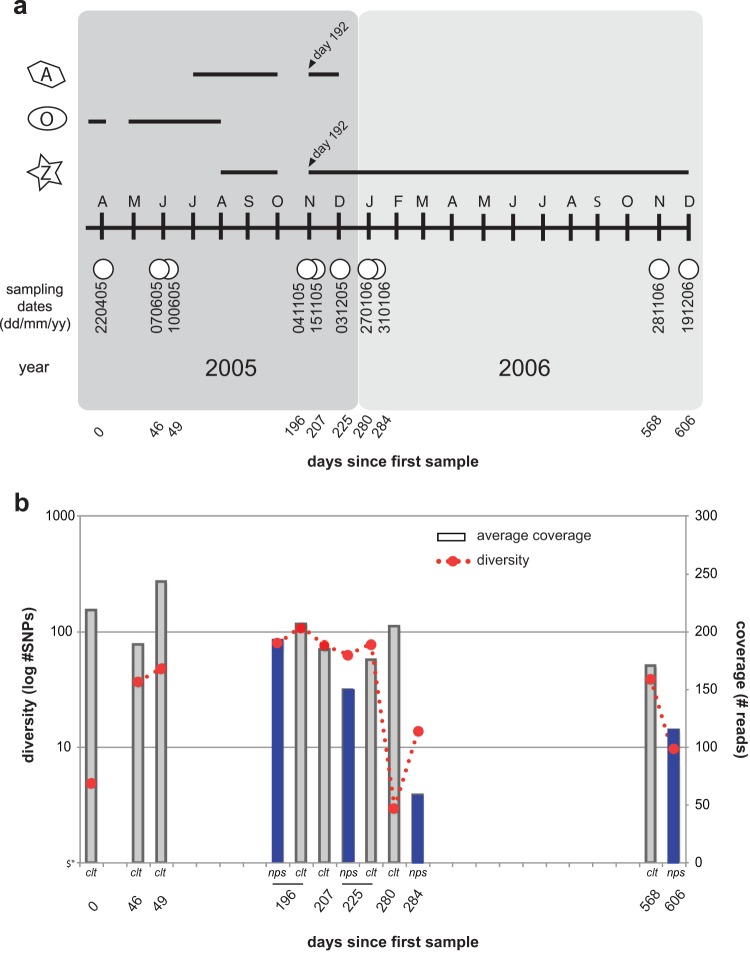
We analyzed 10 samples collected over a 21-month period in a rare case of extreme prolonged virus shedding from a 3-year-old child with SCID (7) (Fig. 1a). The infection was treated sequentially with two neuraminidase inhibitors—oral oseltamivir (Hoffman-La Roche) and nebulized zanamivir (Glaxo Wellcome)—as well as amantadine (Fig. 1a). While a number of original primary nasopharyngeal specimens were available for sequencing, the majority of the specimens had to be cultured in MDCK cells (maximum of 3 passages). Only a small number of amino acid differences were observed at the consensus level between the primary specimens and the isolates (see Table S1 in the supplemental material) for the two time points (days 196 and 225) for which we had both types of samples. However, in both instances 15 to 20% of the minor variants present in the primary specimens appear to be lost in cell culture (see Table S2 in the supplemental material and also see Materials and Methods).
FIG 1 .
Timeline of sampling, drug treatment, and genetic diversity. (a) Sampling and drug treatment. White circles correspond to dates at which samples were collected from the patient. Letters correspond to sampling month, starting with April. A, amantadine treatment; O, oseltamivir treatment; Z, zanamivir treatment. Black lines track the length of time for which the patient was treated with each drug. Days are numbered based on time since first sample. (b) Total genetic diversity based on SNPs present within each sample (identified by LoFreq) and average sequence coverage. nps, nasopharyngeal swabs; clt, culture isolates.
Genetic diversity is reduced by selective sweeps.
To quantify genetic diversity within the patient at each time point, we identified alternate nucleotides present in the underlying sequence reads that had been mapped back to the consensus assemblies. The number of gene positions containing variant nucleotides was summed across all segments to capture allele richness (Fig. 1b; see also Table S2 in the supplemental material). Notably, such allele richness appeared to increase over time until a drastic loss occurred within an 8-week period between days 225 and 280 (Fig. 1b). This decrease in diversity was not associated with the type of specimen analyzed (i.e., cell culture or primary swab). It could be attributed to the establishment of a new infection or could be the result of a selection event that would have reduced diversity while favoring a small number of strains, i.e., a selective sweep. The frequency of individual variants present in the virus populations captured for each sample shows that there is a mix of synonymous and nonsynonymous mutations (see Fig. S2 in the supplemental material).
To determine whether the patient was persistently infected with the original strain or had acquired a new, secondary infection, we conducted phylogenetic analyses of the consensus hemagglutinin (HA) and NA assemblies using global sequences sampled over the three influenza seasons that bracket the time scale of infection (January 2004 to December 2007). This revealed that all patient samples cluster together regardless of the sampling date (see Fig. S1 in the supplemental material, blue clusters), indicative of a single-source infection. Thus, the patient appears to have been persistently infected with the same original virus.
Drug resistance appears at multiple sites and across the virus population.
Four types of drug resistance mutations appear in the consensus sequences over the course of the infection: (i) that converting serine to asparagine at position 31 (S31N) of the M2 protein (8, 13); (ii) that converting glutamine to valine at position 119 (E119V) (7, 15) or (iii) isoleucine to valine at position 222 (I222V) of the NA (7, 15); and (iv) deletion of residues 245 to 248 of the NA, also associated with resistance to the neuraminidase inhibitors (12, 14). To investigate whether these mutations were present as minor variants at earlier time points, we determined the frequency of the alternate consensus base call found in the underlying sequence reads. In M2, a minor variant of another drug resistance mutation (V27A) emerges along with the S31N mutation in response to amantadine treatment (Fig. 2a; see also Table S3 in the supplemental material). The same scenario is observed for the NA, where the E119V mutation is seen by day 46 (Fig. 2b), which corresponds to day 36 of oseltamivir treatment, and becomes dominant 3 days later (day 49). Because of drug resistance, therapy was switched to zanamivir. While the frequencies of the 119V mutation fluctuate over the course of the infection, it is maintained in the viral population until the latter part of 2006, when it disappears completely. I222V appears by day 49 (Fig. 2b) and subsequently varies in frequency and disappears. Also of note was the deletion of amino acids 245 to 248 in the NA protein, which briefly became dominant at day 196 and then disappeared (Fig. 2b) as oseltamivir treatment was stopped.
FIG 2 .
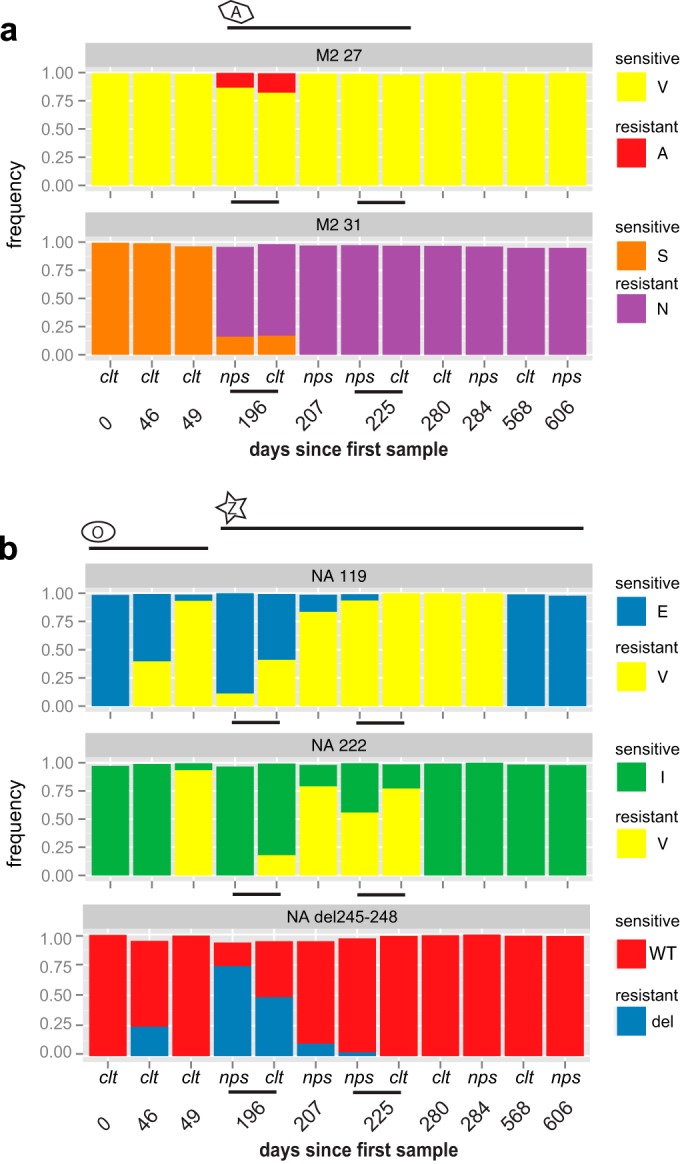
Plots of drug resistance frequency. Histograms representing the frequency of ion channel blocker mutations (a) and neuraminidase inhibitor mutations (b) on Ion Torrent sequence reads. A, amantadine treatment; O, oseltamivir treatment; Z, zanamivir treatment. Black lines track the length of time for which the patient was treated with each drug. nps, nasopharyngeal swabs; clt, culture isolates.
Based on the mutation frequencies observed here, we suspected that multiple drug resistance mutations could cooccur in the same virus genomes. To determine their compatibility, we examined the presence of drug resistance mutations in single Ion Torrent sequence reads and by PacBio single-molecule sequencing (see Table S3 in the supplemental material). Notably, the amantadine drug resistance mutations 27A and 31N were never found on the same M segment (i.e., same reads). For the NA, 222V is present in many cases as a double mutation with 119V. The cooccurrence of these mutations is thought to have a synergistic effect on resistance to oseltamivir (7) and on conserving sensitivity to zanamivir. Strikingly, at day 225 the majority of the viruses present carry a triple mutation comprised of 31N in the M2 and 119V/222V in the NA. However, the 119V mutation was rarely found paired with del245–248 in the same NA segment in the nps samples but was found at a 5% frequency in 2 of the culture samples (see Table S3 in the supplemental material). Although we observe this double mutation at low frequency in our analysis, there does not appear to be incompatibility between 119V and del245–248 mutations, as a recent study reports the double mutant as dominant in primary samples collected from an immunocompromised patient (18). The mutations 222V and del245–248, on the other hand, appeared exclusive of each other, as they were never found on the same NA segment, and the double mutation has not been reported in other studies.
Natural selection on reassortants breaks patterns of segment linkage.
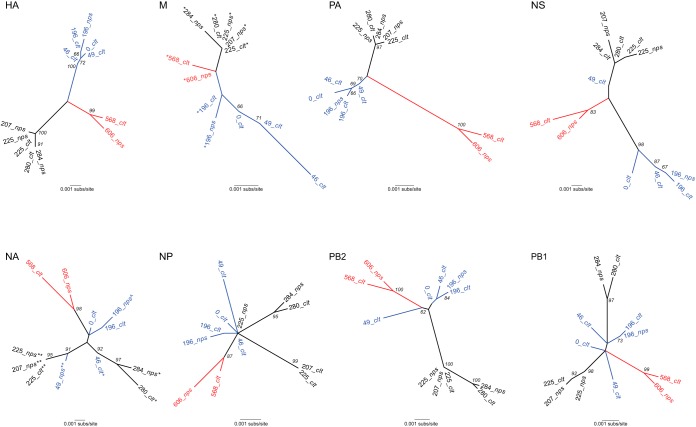
To determine whether reassortment had occurred in these sequences and was linked to antiviral resistance, we inferred phylogenetic trees with the consensus assemblies for each sample (Fig. 3). Strikingly, there were major topological differences across the 8 segment trees, in which samples for some of the segments clearly clustered into three separate groups (represented as black, blue, and red clades), while others did not, strongly indicative of reassortment. Critically, however, reassortment did not occur freely among all segments, with the HA, PA, and M segments possessing very similar tree topologies, and closely related phylogenetic patterns also observed in PB2 and NS. Such phylogenetic congruence is compatible with physical linkage among segments such that they are generally inherited together in the absence of reassortment. In contrast, more complex tree topologies were observed in the other segments, suggestive of multiple reassortment events.
FIG 3 .
Phylogenetic trees of consensus assemblies for all eight segments. All trees are shown as unrooted. Branch lengths correspond to the number of nucleotide substitutions per site (same scale for each tree), and bootstrap values of >60% are shown. The three clades, defined on the basis of the HA tree topology, are colored red, blue, and black, respectively. M tree symbols: *, S31N. NA tree symbols: *, E119V; **, E119V/I222V; ^, del245–248. Segments and the proteins that they encode: HA, hemagglutinin; M, matrix (M1) and ion channel (M2); NA, neuraminidase; NP, nucleoprotein; NS, nonstructural proteins 1 and 2 (NEP and NS2, respectively); PB2, PB1, and PA, polymerases (PB2 = 2,280 nt, PB1 = 2,274 nt, PA = 2,151 nt, HA = 1,701 nt, NP = 1,497 nt, NA = 1,410 nt, M = 982 nt, NS1 = 863 nt). nps, nasopharyngeal swabs; clt, culture isolates.
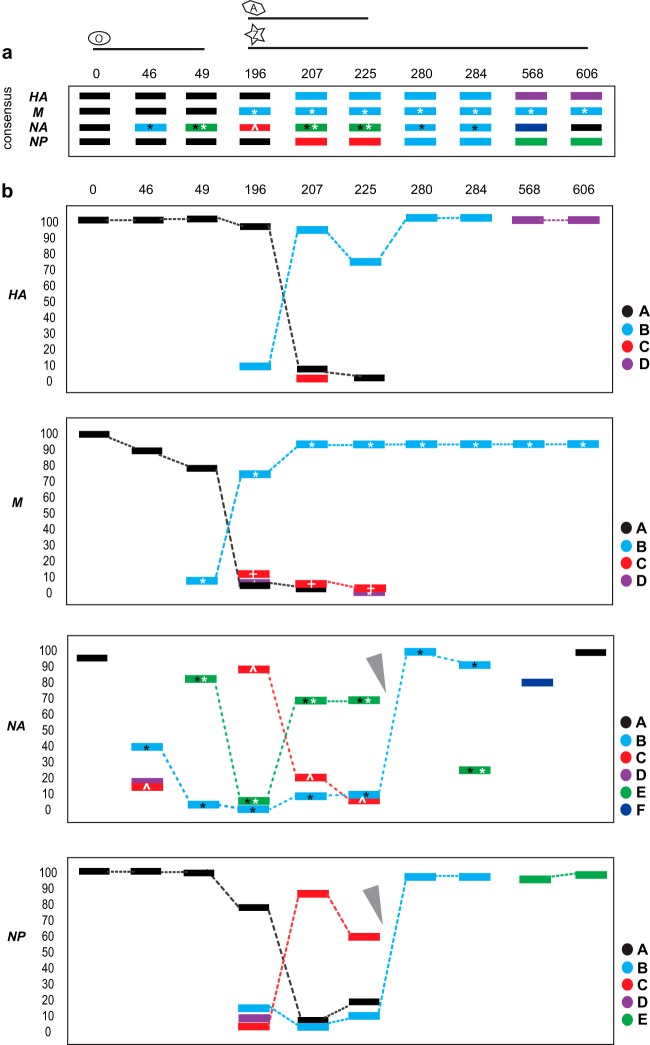
To characterize reassortment in more detail, we reconstructed haplotypes for HA, NA, NP, and M using the single-molecule data (see Table S4 in the supplemental material). We calculated the relationship between all haplotypes by clustering using their pairwise distances and determining cocirculating lineages at each time point (see Fig. S3 in the supplemental material). For HA, a minor lineage initially present at day 196 (Fig. 4; see also Fig. S3 in the supplemental material) became dominant at day 207 and remained dominant at days 225, 280, and 284. The emergence of this new HA appears to be largely driven by linkage to a drug-resistant M segment that carries the S31N mutation (Fig. 4), although the slower fixation of the HA variant may be indicative of more complex epistatic interactions. For NA and NP, a minor lineage at day 196 was dominant at days 207 and 225; this variant carried a double NA mutation (E119V and I222V). However, at day 280 a new lineage took over, which carried only the NA E119V mutation; it was already present as a minor variant at earlier time points (Fig. 4). The parallel emergence of a minor lineage in both NA and NP with a dynamic and phylogenetic pattern different from that seen in HA/M is indicative of a reassortment event between days 225 and 280 (Fig. 4, gray arrowheads). Based on the drug resistance mutations identified in the NA, reassortment seems to have led to emergence of the single E119V mutation lineage, replacing the lineage carrying both 119V and 222V. As this new reassortant emerged, haplotype diversity was greatly reduced, reflected in both the total genetic diversity measured in the single nucleotide polymorphism (SNP) data and the Shannon entropy (19) for all haplotypes of a given segment (Fig. 1b; see also Fig. S3), and hence indicative of a major selective sweep. While there were no changes to patient therapy between days 225 and 280, amantadine and zanamivir were reinitiated on 1 November of that year (Fig. 1a), corresponding to day 192. It is unclear whether this was the event that led to the emergence of the new minor variants.
FIG 4 .
Schematic representation of lineage dynamics for segments HA, M, NA, and NP. Consensus (a) and haplotype (b) frequencies across time points (see Table S4 and Fig. S2 in the supplemental material). M segment symbols: white asterisk, S31N; white cross, V27A. NA segment symbols: black asterisk, E119V; white asterisk, I222V; white caret, del245–248. Gray arrowheads point to the suspected reassortment event. Note that time points 196 and 225 show the data from the primary swabs and not from the culture isolates. Numbers above each panel are days since first sample was collected; numbers to the left of each graph in panel b are percents.
DISCUSSION
The evolution of antiviral resistance in this persistent influenza virus infection reveals a complex interplay between resistance mutations at differing frequencies, multiple cocirculating lineages that have acquired the same resistance mutation in parallel (i.e., the NA lineage B [E119V] and lineage E [E119V/I222V] and M lineages B and D [S31N]) (Fig. 4), selective sweeps that result in genome-wide reductions in genetic diversity, possible epistasis, and patterns of intrinsic segment linkage that are broken down by reassortment. Notably, the patterns of phylogenetic congruence seen here, particularly that between HA and M, correspond to those seen in comparisons of H3N2 influenza viruses at the broader phylogenetic scale (20), suggesting that they reflect a direct physical association between these segments. Another important observation in this context was that although the greatest genetic diversity and most rapid evolution are usually observed in the viral HA, in this case the selection associated with antiviral (adamantane) resistance in M is clearly the dominant evolutionary process, with their congruent phylogenetic patterns (Fig. 3) suggesting that the HA is tracking M segment evolution through linkage. Hence, the selective pressure associated with antiviral resistance is stronger than that due to antibody escape. Whether the lack of HA-driven evolution noted here is a common phenomenon in the case of selection for antiviral resistance at the epidemiological scale or reflects immunosuppression in this particular host is uncertain and evidently merits further investigation.
One limitation of the study is that not all primary specimens were available, such that cultured isolates had to be used in the analysis, leading to some culture-associated variability; this, however, does not affect the overall observations that (i) there is an abrupt decrease in genetic diversity between days 225 and 280 and (ii) there are reassortment events that appear to be linked to the emergence of drug-resistant strains. These data clearly show that mutations appeared de novo in minor variants many weeks before they became dominant and, in response to drug pressure, that mutations became fixed following reassortment of drug-resistant segments with other segments that likely conferred better fitness (21). Reassortment leading to the rise of drug-resistant viral lineages has been reported previously in the context of epidemiological surveillance studies to explain the global emergence of adamantane-resistant H3N2 (22, 23) and oseltamivir-resistant seasonal H1N1 (24) viruses. However, the data presented here are the first in which reassortment and the complex patterns of drug resistance emergence have been mapped within an individual host and which paradoxically reveal the strength of some segment linkages. Hence, although reassortment is evidently a major process of influenza virus evolution, it may not always be frequent enough to break all patterns of segment linkage.
While the results from this study and those of others (17, 18) may help in the design of better antiviral drug therapy regimens that limit the emergence of drug resistance, they hint at important evolutionary forces that should be characterized further. Indeed, exploring the patterns and drivers of segment linkage, reassortment, and epistasis within infected hosts, as well their impact on viral fitness, could help inform targeted surveillance programs for the early identification of new emerging strains and hence assist genomic risk assessment. In particular, a better understanding of segment linkage and reassortment, and whether they differ between mammalian and avian influenza viruses, may enable more accurate predictions of the rapidity with which particular genomic combinations can be obtained, including those mediating drug resistance, antigenic escape, and host adaptation.
MATERIALS AND METHODS
Sequence analyses.
Total RNA was extracted from primary nasopharyngeal swabs (nps) and cell culture isolates (clt). Multisegment reverse transcription-PCR (M-RTPCR) (25) was used to amplify influenza virus-specific segments. The M-RTPCR products were sheared according to the Ion Torrent library preparation manual recommendations for long amplicons and using a 200-base insert size. Equimolar amounts of each sample were mixed to create a final bar-coded sample pool. To prepare the final pool for sequencing, it was diluted in low-Tris-EDTA (low-TE) buffer and clonally amplified using the automated One Touch system (Invitrogen), followed by sequencing on a 314 chip with 200-base chemistry.
The raw Ion Torrent reads were mapped against A/California/7/2004(H3N2) using bowtie 2 (26). These were then demultiplexed into their respective identities based on their bar code signature. Each resulting BAM file was filtered based on an average Phred quality of 25 and analyzed using SAMtools (27). The SAMtools rmdup function was used to remove PCR duplicates, generated during the amplification steps. Consensus sequences for each sample were generated by reporting the dominant nucleotide at each position of the mapped sequence reads. Consensus assemblies were then inspected manually and curated by visualizing BAM file alignments using Tablet (28). For the identification of SNPs, SMALT (http://www.sanger.ac.uk/resources/software/smalt) was used to align reads to the coding sequences (CDS) of each assembly, with an identity threshold of 0.9. Using the default parameters, LoFreq (29) was then used to identify low-frequency variants from these alignments and to further correct assembly errors due to platform errors. LoFreq flags strand bias and treats each base as a Bernoulli trial with an associated sequencing error probability derived from a Phred-scaled quality value. A Poisson-binomial distribution is used to identify variant bases in a given column of n bases, so that each Bernoulli trial has a distinct success probability.
To determine how much variability is introduced by cell culture, we compared the two time points (day 196 and day 225) for which we had both primary specimen and cell culture isolates (see Table S2 in the supplemental material). In both instances, 15 to 20% of the variants present in the primary specimens appear to be lost in cell culture, and conversely, multiple variants appear in cell culture that were not observed in the primary specimens. However, the latter is likely to be a function of the low coverage obtained from the Ion Torrent data such that LoFreq excludes these variant sites. When we phased the SNPs identified by LoFreq using single-molecule sequencing (see text below on PacBio), the majority of the variants seen in culture were also present in the original primary specimens but at a very low frequency that was not captured by the Ion Torrent sequence data.
We performed single-molecule sequencing using PacBioSMRT cell sequencing (30) for haplotype reconstruction. The M-RTPCR protocol was repeated from the extracted RNA using Uni12/13 primer sets containing 10-base bar codes but without the MBT (MluI-Bcl1-TOPO) stabilizing bases. DNA library preparation and sequencing were performed according to the manufacturer’s instructions and reflect the P4-C2 sequencing enzyme and chemistry, respectively. Eight of the tagged samples (day0_220405_clt, day46_070605_clt, day49_100605_clt, day196_041105_nps, day196_041105_clt, day207_151105_nps, day225_031205_clt, and day280_270106_clt) were pooled prior to library preparation, while 4 of the samples were processed individually without tagging (day225_031205_nps, day284_310106_nps, day568_281106_clt, and day606_191206_nps). Primer was annealed to the SMRTbell libraries for sequencing. The polymerase-template complex was then bound to the P4 enzyme using a ratio of 10:1 (polymerase to SMRTbell) at 0.5 nM for 4 h at 30°C and then held at 4°C until ready for magbead loading, prior to sequencing. The magbead-loaded, polymerase-bound, SMRTbell libraries were placed onto the RSII machine at a sequencing concentration of 50 pM and configured for a 180-min continuous sequencing run to allow for the maximum number of passes for consensus error correction. Sequencing was conducted across 2 SMRTcells for the pooled samples, totaling ~59,000 reads, and ~22,000 to 32,000 postfiltered reads were generated from a single SMRTcell for each of the individual samples. Continuous long-read data with 12 to 28 single-molecule passes on average were then generated and passed through the RS_ReadsOfInsert.1 pipeline using a 10-pass minimum and 95% accuracy cutoff to achieve higher consensus QV FASTA and FASTQ files for variant calling purposes. FASTQ format PacBio reads were demultiplexed using FastX_barcode_splitter. Reads were aligned against a composite version of the genome, from the A/California/NHRC0006/2005(H3N2) strain. The alignment was performed with BWA-SW (31), using the following parameters: -b5 -q2 -r1 -z1. Reads that mapped against each segment were retrieved using SAMtools and converted to FASTA format. Resulting reads were BLAST aligned against their reference segment, and reads producing alignments of <1,000 bp, or with more than 40 gap openings, were removed from the analysis. We examined LoFreq predicted sites in each read of the multiple alignment and generated matrices of haplotype predictions with counts for each sample. We applied a conservative approach of using only PacBio reads that spanned the entirety of the predicted sites and discarded reads where there were apparent alignment issues surrounding the LoFreq predicted sites.
Phylogenetic analyses.
Phylogenetic trees of global H3N2 influenza virus sequences were inferred for the HA and NA segments downloaded from the FluDB database available at NCBI (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) for samples dated January 2004 to December 2007 from any geographical region. Only complete, or near-complete, segments were queried, and identical sequences were collapsed into a single sequence. This resulted in a total of 1,148 HA and 1,223 NA sequences that were aligned using MAFFT (32) and visually inspected using SeaView (33). Character sets were chosen to exclude the missing data from the segment ends. These data were then used to infer “panoramic” phylogenetic trees using the maximum likelihood (ML) method available in the RAxML package (34). This analysis was based on 1,701 nucleotides (nt) for the HA and 1,411 nt for the NA, in both cases employing the general time-reversible (GTR) model of sequence evolution, with rate variation among sites modeled using a gamma (Γ) distribution. Phylogenies for the consensus assemblies of all segments at each time point were inferred using the ML method available in the PhyML (v2.4.5) (35) program. Because of the high level of sequence similarity in this case, this analysis utilized the simpler HKY85 model of nucleotide substitution and a combination of nearest neighbor interchange (NNI) and subtree pruning and regrafting (SPR) branch-swapping. Bootstrap resampling (1,000 replications) was used to assess the statistical support for each grouping.
Haplotype reconstruction.
For a given segment, each sequence was reconstructed by substituting the haplotype SNPs into the reference founding sequence (day 0, 220405). Cluster analysis was performed on all major haplotypes as listed in Table S4 in the supplemental material. From these sequences, clustergrams were created using the Bioinformatics Software Toolbox for Matlab, version R2014a. The Euclidean distance between segments was calculated, and a clustergram was created by calculating the average segment linkage. At each time point, haplotype diversity and frequency of that haplotype within the sample were calculated for all segments and represented as a measure of entropy (19). This value was calculated at a time point, t, and for a given segment, s, where s may be the HA, NP, NA, or M segments. For a given segment and time, there are a set of major haplotypes. The proportion of these haplotypes was normalized to a given haplotype frequency for the ith haplotype, fi(t,s). The entropy is then given by
where n is the total number of haplotypes at time t for segment s. The entropy gives a measure of the diversity of haplotypes and typically conservatively underestimates the true diversity as a consequence of Jensen’s inequality.
Nucleotide sequence accession numbers.
Ion Torrent and PacBio sequences were submitted to the sequence read archives at NCBI under accession number PRJNA253584. Consensus sequences were submitted to NCBI under accession numbers KM438082 to KM438177.
SUPPLEMENTAL MATERIAL
Panoramic phylogenies of HA and NA genes. The samples in this study form a monophyletic group and are surrounded by a blue box; the remaining taxa comprise human H3N2 strains collected between January 2004 and December 2007; sequences were downloaded from the NCBI FluDB. Trees were midpoint rooted for purposes of clarity only, and branch lengths are drawn according to the number of nucleotide substitutions per site. Download
Frequencies of minor variants across proteins for each sample analyzed. Nonsynonymous mutations are highlighted in red, while synonymous mutations are in black. The dashed boxes highlight the time points for which primary specimens and culture isolates are available. nps, nasopharyngeal swabs; clt, culture isolates. Download
Clustergrams of distance-based pairwise comparisons of haplotypes for HA, M, NA, and NP. Each sequence was reconstructed by substituting the haplotype SNPs into the reference founding sequence (220405). Colored boxes represent each haplotype; boxes with black outlines represent the major variant. Haplotypes were clustered into lineages (A, B, C, D, E, and F). For the M segment, the following symbols represent drug resistance mutations: +, V27A; *, S31N. For the NA segment, the following symbols represent specific drug resistance mutations: *, E119V; **, E119V/I222V. At each time point, haplotype diversity and frequency of that haplotype within the sample were calculated for all segments and represented as a measure of entropy (inset graphs within each clustergram). Download
Point mutation differences between primary swabs and culture isolates observed in the consensus assembly of the Ion Torrent sequences.
SNP data from LoFreq analysis.
Cooccurrence of drug resistance mutations on single reads (PacBio and/or Ion Torrent).
PacBio haplotypes from the phasing of LoFreq variant sites (accompanies Fig. S3).
ACKNOWLEDGMENTS
T.S. is a predoctoral trainee supported by NIH T32 training grant T32 EB009403 as part of the HHMI-NIBIB Interfaces Initiative. L.C. is a visiting scholar from Tsinghua University and supported in part by the China Scholarship Council. This research was supported in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S. Department of Health and Human Services, under grant no. U01AI111598 (E.G.); by the National Institute of General Medical Sciences, NIH, under award no. U54GM088491 (E.G.); and by a Canadian Institutes of Health Research grant (G.B.). E.C.H. is supported by an NHMRC Australia Fellowship (AF30).
Footnotes
Citation Rogers MB, Song T, Sebra R, Greenbaum BD, Hamelin M-E, Fitch A, Twaddle A, Cui L, Holmes EC, Boivin G, Ghedin E. 2015. Intrahost dynamics of antiviral resistance in influenza A virus reflect complex patterns of segment linkage, reassortment, and natural selection. mBio 6(2):e02464-14. doi:10.1128/mBio.02464-14.
REFERENCES
- 1.Drake JW. 1993. Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci U S A 90:4171–4175. doi: 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desselberger U, Nakajima K, Alfino P, Pedersen FS, Haseltine WA, Hannoun C, Palese P. 1978. Biochemical evidence that “new” influenza virus strains in nature may arise by recombination (reassortment). Proc Natl Acad Sci U S A 75:3341–3345. doi: 10.1073/pnas.75.7.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush RM, Fitch WM, Bender CA, Cox NJ. 1999. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol Biol Evol 16:1457–1465. doi: 10.1093/oxfordjournals.molbev.a026057. [DOI] [PubMed] [Google Scholar]
- 4.Viboud C, Nelson MI, Tan Y, Holmes EC. 2013. Contrasting the epidemiological and evolutionary dynamics of influenza spatial transmission. Philos Trans R Soc Lond B Biol Sci 368:20120199. doi: 10.1098/rstb.2012.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leekha S, Zitterkopf NL, Espy MJ, Smith TF, Thompson RL, Sampathkumar P. 2007. Duration of influenza A virus shedding in hospitalized patients and implications for infection control. Infect Control Hosp Epidemiol 28:1071–1076. doi: 10.1086/520101. [DOI] [PubMed] [Google Scholar]
- 6.Suess T, Buchholz U, Dupke S, Grunow R, an der Heiden M, Heider A, Biere B, Schweiger B, Haas W, Krause G, Robert Koch Institute Shedding Investigation Group . 2010. Shedding and transmission of novel influenza virus A/H1N1 infection in households—Germany, 2009. Am J Epidemiol 171:1157–1164. doi: 10.1093/aje/kwq071. [DOI] [PubMed] [Google Scholar]
- 7.Baz M, Abed Y, McDonald J, Boivin G. 2006. Characterization of multidrug-resistant influenza A/H3N2 viruses shed during 1 year by an immunocompromised child. Clin Infect Dis 43:1555–1561. doi: 10.1086/508777. [DOI] [PubMed] [Google Scholar]
- 8.Boivin G, Goyette N, Bernatchez H. 2002. Prolonged excretion of amantadine-resistant influenza A virus quasi species after cessation of antiviral therapy in an immunocompromised patient. Clin Infect Dis 34:e23–e25. doi: 10.1086/338870. [DOI] [PubMed] [Google Scholar]
- 9.Vega E, Donaldson E, Huynh J, Barclay L, Lopman B, Baric R, Chen LF, Vinjé J. 2014. RNA populations in immunocompromised patients as reservoirs for novel norovirus variants. J Virol 88:14184–14196. doi: 10.1128/JVI.02494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geis S, Prifert C, Weissbrich B, Lehners N, Egerer G, Eisenbach C, Buchholz U, Aichinger E, Dreger P, Neben K, Burkhardt U, Ho AD, Kräusslich HG, Heeg K, Schnitzler P. 2013. Molecular characterization of a respiratory syncytial virus outbreak in a hematology unit in Heidelberg, Germany. J Clin Microbiol 51:155–162. doi: 10.1128/JCM.02151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Vries E, Stittelaar KJ, van Amerongen G, Veldhuis Kroeze EJ, de Waal L, Fraaij PL, Meesters RJ, Luider TM, van der Nagel B, Koch B, Vulto AG, Schutten M, Osterhaus AD. 2013. Prolonged influenza virus shedding and emergence of antiviral resistance in immunocompromised patients and ferrets. PLoS Pathog 9:e1003343. doi: 10.1371/journal.ppat.1003343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abed Y, Baz M, Boivin G. 2009. A novel neuraminidase deletion mutation conferring resistance to oseltamivir in clinical influenza A/H3N2 virus. J Infect Dis 199:180–183. doi: 10.1086/595736. [DOI] [PubMed] [Google Scholar]
- 13.Hayden FG, Belshe RB, Clover RD, Hay AJ, Oakes MG, Soo W. 1989. Emergence and apparent transmission of rimantadine-resistant influenza A virus in families. N Engl J Med 321:1696–1702. doi: 10.1056/NEJM198912213212502. [DOI] [PubMed] [Google Scholar]
- 14.Memoli MJ, Hrabal RJ, Hassantoufighi A, Eichelberger MC, Taubenberger JK. 2010. Rapid selection of oseltamivir- and peramivir-resistant pandemic H1N1 virus during therapy in 2 immunocompromised hosts. Clin Infect Dis 50:1252–1255. doi: 10.1086/651605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samson M, Pizzorno A, Abed Y, Boivin G. 2013. Influenza virus resistance to neuraminidase inhibitors. Antiviral Res 98:174–185. doi: 10.1016/j.antiviral.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Abed Y, Baz M, Boivin G. 2006. Impact of neuraminidase mutations conferring influenza resistance to neuraminidase inhibitors in the N1 and N2 genetic backgrounds. Antivir Ther 11:971–976. [PubMed] [Google Scholar]
- 17.Ruiz-Carrascoso G, Casas I, Pozo F, González-Vincent M, Pérez-Breña P. 2010. Prolonged shedding of amantadine- and oseltamivir-resistant influenza A(H3N2) virus with dual mutations in an immunocompromised infant. Antivir Ther 15:1059–1063. doi: 10.3851/IMP1657. [DOI] [PubMed] [Google Scholar]
- 18.Eshaghi A, Shalhoub S, Rosenfeld P, Li A, Higgins RR, Stogios PJ, Savchenko A, Bastien N, Li Y, Rotstein C, Gubbay JB. 2014. Multiple influenza A (H3N2) mutations conferring resistance to neuraminidase inhibitors in a bone marrow transplant recipient. Antimicrob Agents Chemother 58:7188–7197. doi: 10.1128/AAC.03667-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenbaum BD, Li OT, Poon LL, Levine AJ, Rabadan R. 2012. Viral reassortment as an information exchange between viral segments. Proc Natl Acad Sci U S A 109:3341–3346. doi: 10.1073/pnas.1113300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ince WL, Gueye-Mbaye A, Bennink JR, Yewdell JW. 2013. Reassortment complements spontaneous mutation in influenza A virus NP and M1 genes to accelerate adaptation to a new host. J Virol 87:4330–4338. doi: 10.1128/JVI.02749-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson MI, Simonsen L, Viboud C, Miller MA, Holmes EC. 2009. The origin and global emergence of adamantane resistant A/H3N2 influenza viruses. Virology 388:270–278. doi: 10.1016/j.virol.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simonsen L, Viboud C, Grenfell BT, Dushoff J, Jennings L, Smit M, Macken C, Hata M, Gog J, Miller MA, Holmes EC. 2007. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol 24:1811–1820. doi: 10.1093/molbev/msm103. [DOI] [PubMed] [Google Scholar]
- 24.Yang JR, Lin YC, Huang YP, Su CH, Lo J, Ho YL, Yao CY, Hsu LC, Wu HS, Liu MT. 2011. Reassortment and mutations associated with emergence and spread of oseltamivir-resistant seasonal influenza A/H1N1 viruses in 2005–2009. PLoS One 6:e18177. doi: 10.1371/journal.pone.0018177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Wentworth DE. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza A viruses. J Virol 83:10309–10313. doi: 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milne I, Stephen G, Bayer M, Cock PJ, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 29.Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 34.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Panoramic phylogenies of HA and NA genes. The samples in this study form a monophyletic group and are surrounded by a blue box; the remaining taxa comprise human H3N2 strains collected between January 2004 and December 2007; sequences were downloaded from the NCBI FluDB. Trees were midpoint rooted for purposes of clarity only, and branch lengths are drawn according to the number of nucleotide substitutions per site. Download
Frequencies of minor variants across proteins for each sample analyzed. Nonsynonymous mutations are highlighted in red, while synonymous mutations are in black. The dashed boxes highlight the time points for which primary specimens and culture isolates are available. nps, nasopharyngeal swabs; clt, culture isolates. Download
Clustergrams of distance-based pairwise comparisons of haplotypes for HA, M, NA, and NP. Each sequence was reconstructed by substituting the haplotype SNPs into the reference founding sequence (220405). Colored boxes represent each haplotype; boxes with black outlines represent the major variant. Haplotypes were clustered into lineages (A, B, C, D, E, and F). For the M segment, the following symbols represent drug resistance mutations: +, V27A; *, S31N. For the NA segment, the following symbols represent specific drug resistance mutations: *, E119V; **, E119V/I222V. At each time point, haplotype diversity and frequency of that haplotype within the sample were calculated for all segments and represented as a measure of entropy (inset graphs within each clustergram). Download
Point mutation differences between primary swabs and culture isolates observed in the consensus assembly of the Ion Torrent sequences.
SNP data from LoFreq analysis.
Cooccurrence of drug resistance mutations on single reads (PacBio and/or Ion Torrent).
PacBio haplotypes from the phasing of LoFreq variant sites (accompanies Fig. S3).