ABSTRACT
A mechanistic understanding of the purported health benefits conferred by consumption of probiotic bacteria has been limited by our knowledge of the resident gut microbiota and its interaction with the host. Here, we detail the impact of a single-organism probiotic, Lactobacillus rhamnosus GG ATCC 53103 (LGG), on the structure and functional dynamics (gene expression) of the gut microbiota in a study of 12 healthy individuals, 65 to 80 years old. The analysis revealed that while the overall community composition was stable as assessed by 16S rRNA profiling, the transcriptional response of the gut microbiota was modulated by probiotic treatment. Comparison of transcriptional profiles based on taxonomic composition yielded three distinct transcriptome groups that displayed considerable differences in functional dynamics. The transcriptional profile of LGG in vivo was remarkably concordant across study subjects despite the considerable interindividual nature of the gut microbiota. However, we identified genes involved in flagellar motility, chemotaxis, and adhesion from Bifidobacterium and the dominant butyrate producers Roseburia and Eubacterium whose expression was increased during probiotic consumption, suggesting that LGG may promote interactions between key constituents of the microbiota and the host epithelium. These results provide evidence for the discrete functional effects imparted by a specific single-organism probiotic and challenge the prevailing notion that probiotics substantially modify the resident microbiota within nondiseased individuals in an appreciable fashion.
IMPORTANCE
Probiotic bacteria have been used for over a century to promote digestive health. Many individuals report that probiotics alleviate a number of digestive issues, yet little evidence links how probiotic microbes influence human health. Here, we show how the resident microbes that inhabit the healthy human gut respond to a probiotic. The well-studied probiotic Lactobacillus rhamnosus GG ATCC 53103 (LGG) was administered in a clinical trial, and a suite of measurements of the resident microbes were taken to evaluate potential changes over the course of probiotic consumption. We found that LGG transiently enriches for functions to potentially promote anti-inflammatory pathways in the resident microbes.
INTRODUCTION
The consumption of probiotic bacteria has been heralded as a means to promote digestive health, alleviate a range of deleterious conditions, including atopic dermatitis and gastroenterological diseases, and reverse dysbiotic microbiota to restore gut mucosal homeostasis (1–3). In vitro studies to delineate the molecular mechanisms of probiotic species have indicated modulatory capabilities for strain-specific and molecule-specific benefits to the host, which include shifts in anti-inflammatory cytokine profiles and stabilization of epithelial tight junctions (4, 5). Well-characterized effector molecules produced by probiotic organisms include cell surface proteins, lipoteichoic acid, peptidoglycan-derived muropeptides, exopolysaccharides, and pilus-type structures (reviewed in reference 4). However, extrapolation of these mode-of-action studies to in vivo behavior is complicated by a multitude of factors, largely by the complex, reciprocal interaction of the host with the resident microbiota along the gastrointestinal tract. The majority of randomized clinical trials (RCTs) to evaluate the in vivo health benefits of probiotic species have focused on a variety of clinical indicators from the human host; to date, few studies have focused on the impact of probiotic consumption on the resident gastrointestinal microbiota on a community-wide scale (2, 6). There is great need to systematically study the effects of probiotic bacteria on the autochthonous microbial community in the human gut to further elucidate how these organisms confer beneficial outcomes to the host and substantiate commercial health claims.
The most comprehensively studied probiotic strain, Lactobacillus rhamnosus GG ATCC 53103 (LGG), has been shown to have clinical benefits from a variety of cohort studies and is thought to act by (i) competitive colonization advantage through the use of mucus-binding pili, (ii) putative bacteriocin activity identified via bacteriocin-like genomic architecture, and (iii) soluble effector signaling proteins that elicit anti-inflammatory cytokines and activate mitogen-activated protein kinases (MAPKs) (7, 8). Despite the strong biofilm-forming capacity in vitro, LGG colonization in vivo is restricted to discrete ecological niches along the human gastrointestinal tract with limited biofilm formation (9). Recently, the application of a phylogenetic microarray (HITChip) to survey the composition and structure of the fecal community within a healthy adult Finnish cohort revealed no significant difference related to LGG consumption (10).
A major challenge to assess the impact of probiotics on the resident gut consortia is the ability to measure both compositional and functional components on a community-wide scale. Techniques and sequencing technologies to enable sampling of the active microbial fraction have recently become available (11, 12), yet robust bioinformatics and statistical methodologies have yet to be formally implemented for mixed-community RNA sequencing (RNA-seq) data sets. Using a novel pipeline for analysis of metatranscriptomic data, we characterized the structure and functional dynamics (gene expression) of the gut microbiota associated with consumption of the single-organism probiotic Lactobacillus rhamnosus GG ATCC 53103 (LGG), from a study of 12 healthy elderly individuals. The gut microbiota of elderly people provides a unique system to study the fitness landscape of the presumptive healthy, aging microbiota and assess the community stability and dynamics. We found that community composition was not modified due to probiotic intake; however, community-wide transcriptional changes were evident.
RESULTS
Effect of LGG consumption on gut microbial composition and structure in elderly people.
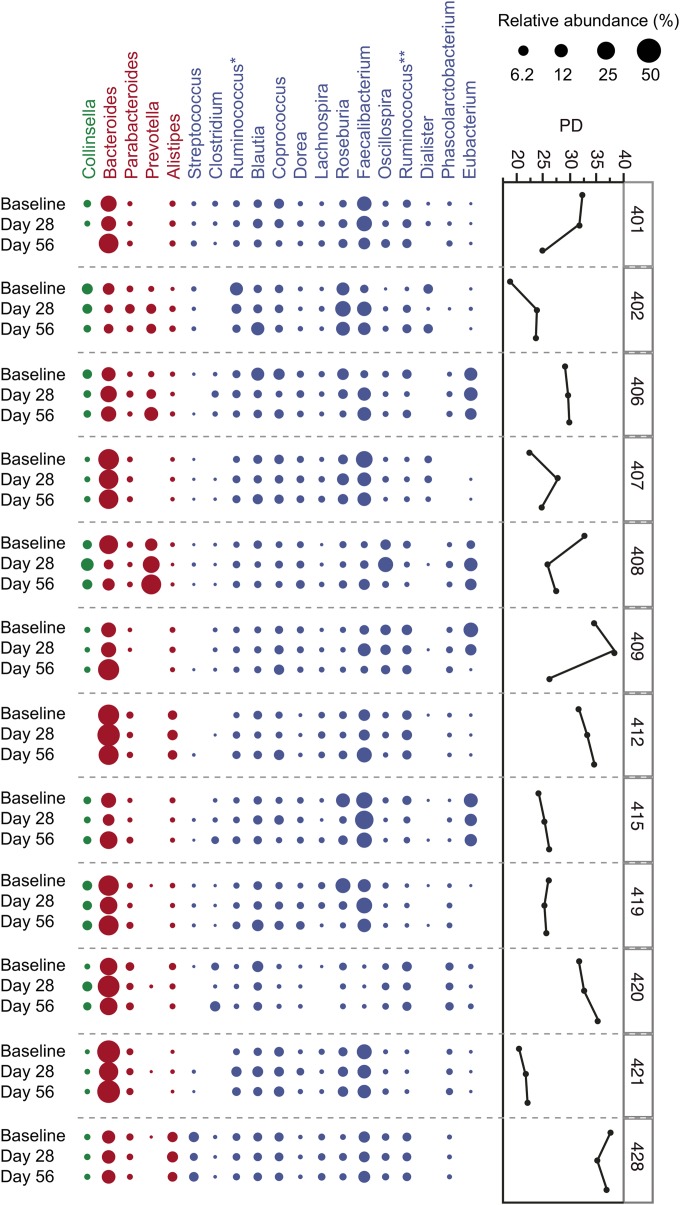
To investigate whether consumption of the probiotic Lactobacillus rhamnosus GG ATCC 53103 (LGG) would exert an influence on the composition and structure of the gut microbiota, we carried out a longitudinal 16S rRNA, metagenomic, and metatranscriptomic analysis of fecal samples obtained from 12 elderly subjects (65 to 80 years old) enrolled in an open-label clinical trial (13). The healthy volunteers included seven females and five males, all of white, non-Hispanic origin, residing in private residences in the greater Boston metropolitan area (further clinical information and demographics can be found in reference 13). Fecal samples were collected prior to probiotic consumption (day 0; baseline), on day 28 after consuming 1010 CFU of LGG twice daily for 28 days, and on day 56, 1 month after stopping LGG consumption. We generated 295,442 high-quality sequence reads from bacterial 16S rRNA gene V1-V2 amplicons, with an average (±standard deviation) of 8,207 (±1,434) reads per fecal sample (see Table S1 in the supplemental material). Consistent with previous studies of the gut microbiota of elderly people (14–16), we observed marked interindividual variability, with longitudinal samples collected from the same individual displaying high bacterial community similarity irrespective of probiotic consumption (Fig. 1; see also Fig. S1 in the supplemental material). No statistically significant differences associated with LGG consumption were found at any phylogenetic level of resolution, as determined by analysis of variance (ANOVA). Moreover, we examined the effect of LGG consumption and medication status on community diversity using a linear mixed-effects model and found that ciprofloxacin was the only measured variable that significantly explained the observed variation (see Table S1).
FIG 1 .
Community composition, temporal stability, and diversity in the gut microbiota in elderly people. Prevalent bacterial genera identified in the 12 elderly individuals at baseline, on day 28, and on day 56, with relative abundance denoted by circle size. Colors represent phylum affiliations as follows, Actinobacteria (green), Bacteroidetes (red), and Firmicutes (blue). Phylogenetic diversity (PD) is shown for each individual over time and was calculated from the rarefied OTUs. Asterisks denote the genus Ruminococcus classified within two separate families, the Lachnospiraceae (*) and the Ruminococcaceae (**).
In addition to 16S rRNA profiling, we also carried out a metagenomic analysis using whole-genome sequencing to assess the gene content of the gut microbiota at all time points for a subset of three subjects. The three subjects were selected based on the 16S rRNA profiles, representing a range of community composition and diversity. Over 1.1 billion sequence reads were generated for three subjects across all time points, with an average of 124.1 million (±13.3 million) reads per fecal sample (see Table S2 in the supplemental material). A robust metagenomic assembly procedure was implemented and yielded an average of 351,662 contigs per sample, with an average N50 of 2,856 (see Table S2; see Materials and Methods). A comparison of the recovered genomes for each of the three individuals revealed strain-resolved patterns of relative abundance (see Table S2). For example, the relative proportion of Bacteroides strains was highly specific across the three sampled individuals and lends support that individuals can be differentiated based on unique strain-resolved gut communities (17, 18).
We reconstructed the probiotic LGG genome at an estimated 62% average genome coverage on day 28, despite an average relative sequence abundance of only 0.4% across the three individuals sampled (see Table S2). The reconstruction of the LGG genome enabled evaluation and verification that many of the functional properties associated with probiotic efficacy were present in LGG. For example, from the annotation of 730, 1,874, and 1,925 LGG-specific genes for subjects 408, 419, and 428, respectively, we were able to positively identify the ABC transporter/permease components required for antimicrobial peptide type IIb bacteriocin synthesis in all three metagenomes and the bacteriocin immunity proteins in subjects 419 and 428 (8). Further, we identified the unique mucus-binding pili gene cluster spaCBA-srtC1 from subjects 419 and 428, indicating that the LGG probiotic administered possessed the capacity to adhere to the host epithelial mucosa (8, 19).
Community-wide gene expression captures community transcriptome groups.
We next used whole-community RNA sequencing (RNA-seq) for all subjects in the study and first sought to evaluate whether the abundance of taxa identified by 16S rRNA profiling correlated with specific patterns of gene expression. In sum, over 4.6 billion sequence reads were generated, with an average of 129.7 million (±26.4 million) reads per fecal sample (see Table S3 in the supplemental material). Importantly, we utilized a robust total RNA extraction method to obtain high-quality intact RNA (RNA integrity number [RIN] averaging 8.5) and an efficient ribosomal depletion protocol that yielded an average 7.3% rRNA out of the total sequenced data set (see Table S3). This represents a high-quality data set to more fully evaluate the functional gene expression within a mixed community from mRNA. Previously reported metatranscriptomics data sets have consisted of greater than 50%, and even in excess of 90%, of ribosomal sequences compared to functional transcripts from fecal and other environments (20, 21). Further, we developed a robust bioinformatics pipeline to exhaustively search reference bacterial, archaeal, viral, and microeukaryotic genomes to classify individual reads as comprehensively as possible and to post-process these data to provide estimates of relative abundance across microbial genera (see Materials and Methods; see also Fig. S2 in the supplemental material). We compared the relative taxonomic abundance predictions between the two data sets and found that the abundance of many prevalent bacterial genera, including Bacteroides, Eubacterium, Faecalibacterium, and Streptococcus, as determined by 16S rRNA profiling, correlated with their respective transcriptional signals (see Table S4 in the supplemental material). However, for other genera, most notably Ruminococcus, there was no significant correlation between data sets. This finding is perhaps not surprising given that 16S rRNA signatures provide a broad view of all bacteria present in the gut microbiota, although with known caveats, such as PCR amplification biases, whereas metatranscriptomics data provide a snapshot of the expressed genes.
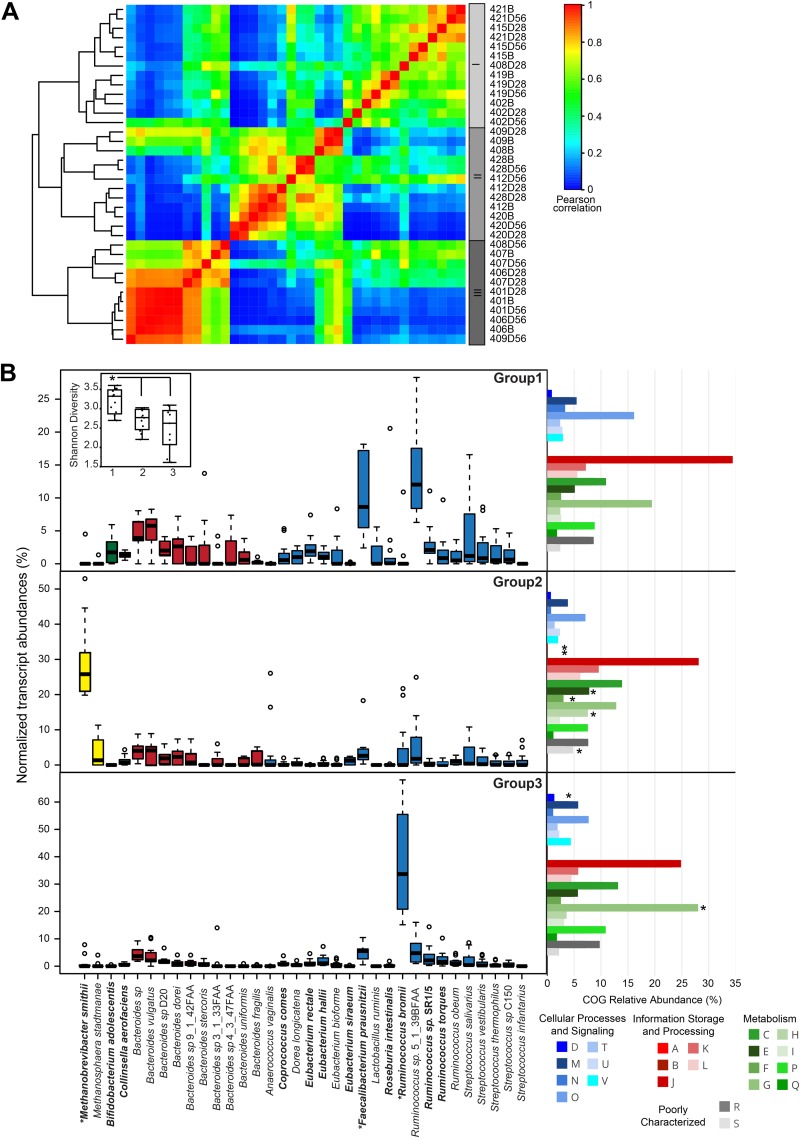
We found that the archaeal transcripts, which were not identified using bacterial-specific 16S rRNA sequencing, contributed to clustering of the samples into three groups, designated transcriptome groups, which displayed distinct patterns of gene expression and transcript diversity (Fig. 2; see also Fig. S3 in the supplemental material). Highly expressed archaeal transcripts from the methanogens, Methanobrevibacter and Methanosphaera, were identified in a subset of individuals (24% average abundance) and, when present, were relatively stable over time (see Fig. S2 in the supplemental material). Each transcriptome group displayed a unique transcript abundance profile from the Firmicutes phylum (Fig. 2B), with the greatest firmicute diversity found within transcriptome group 1. In contrast, within transcriptome group 3, the majority of transcripts were derived from Ruminococcus spp. that significantly differentiated this group from the other two transcriptome groups (see Table S5 in the supplemental material). The transcriptome groups also differed significantly in overall community diversity, as measured by the Shannon diversity index, which is influenced by both the species richness and their relative proportions (Fig. 2B). Transcriptome group 1, which was not dominated by any single taxon, displayed the highest diversity compared with transcriptome groups 2 and 3, which had similar community-wide diversity scores.
FIG 2 .
RNA-seq transcript phylogeny and dynamics. (A) A correlation matrix was constructed from relative transcript abundances mapped to reference species. The scale bar represents the Pearson correlation coefficient (PCC). A histogram for complete linkage clustering of samples is shown, with transcriptome group designations for the bacterial and archaeal fraction (viral and eukaryotic sequences were removed). (B) Distribution of dominant species (>2% relative abundance) across the three identified transcriptome groups. Species listed in bold were found to be discriminant for pairwise transcriptome group comparisons, with an asterisk denoting discriminant species across all three groups. The inset indicates the Shannon diversity for the three transcriptome groups, where the asterisk denotes significantly different diversity measures as determined by the nonparametric Wilcoxon rank sum test (group 1 versus group 2, P = 0.0031; group 1 versus group 3, P = 0.0031). Clusters of orthologous group (COG) category profiles for each transcriptome group are represented, where the asterisks similarly denote discriminant categories across all three transcriptome groups. COG categories are as follows: RNA processing and modification (category A); chromatin structure and dynamics (B); energy production and conversion (C); cell division, chromosome partitioning (D); amino acid transport and metabolism (E); nucleotide transport and metabolism (F); carbohydrate transport and metabolism (G); coenzyme transport and metabolism (H); lipid transport and metabolism (I); translation and biogenesis (J); transcription (K); replication, recombination, and repair (L); cell wall/membrane/envelope (M); cell motility (N); protein turnover, chaperones (O); inorganic ion transport and metabolism (P); secondary metabolism (Q); general function prediction only (R); function unknown (S); signal transduction mechanisms (T); intracellular trafficking and secretion (U); and defense mechanisms (V).
The transcriptome groups were found to be differentiated not only by taxonomic composition but also by functional properties, as defined by clusters of orthologous group (COG) categories (Fig. 2B; see also Table S5). Transcriptome group 2 was differentiated by significantly greater abundances of functions associated with RNA processing and modification (COG category A), chromatin structure and dynamics (B), and amino acid, nucleotide, and coenzyme transport and metabolism (E, F, and H), while transcriptome group 3 was differentiated by carbohydrate transport and metabolism (G) and cell cycle control, cell division, and chromosome partitioning (D).
Global and genus-level differential gene expression associated with community transcriptome groups.
The global gene transcriptional response across the three transcriptome groups was investigated using a robust differential expression analysis applied for mixed-community RNA-seq data. A total of 3,514 unique KEGG orthologs were identified, the majority of which were conserved across the three transcriptome groups. The largest number of unique KEGG orthologs (654) was found within transcriptome group 2, and these largely represented transcripts derived from the methanogenic archaea. Methanobrevibacter-associated KEGG pathways involved in methane (map00680) and nitrogen metabolism (map00910) were found at significantly greater (>10-fold) levels within transcriptome group 2 than within the other transcriptome groups. The methanogens within group 2 were also involved in porphyrin/chlorophyll metabolism (map00860), riboflavin metabolism (map00740), glycine, serine, and threonine metabolism (map00260), pyruvate-ferredoxin oxidoreductase (M00310), lysine biosynthesis (map00300), DNA mismatch repair (map03430), homologous recombination (map03440), and synthesis/degradation of ketone bodies (map00730) (see Table S6 in the supplemental material).
Approximately 55% of the transcripts within transcriptome group 3 were derived from Ruminococcus spp., and differential expression analysis revealed that several Ruminococcus-derived pathways and modules involved in transport and carbohydrate metabolism were highly abundant in this transcriptome group. For example, systems for maltose/maltodextrin transport (M00194), putrescine transport (M00300), and cell division transport (M00256) and pathways for fructose/mannose (etae0051) metabolism from Ruminococcus spp. were highly represented.
As described previously, transcriptome group 1 displayed the greatest level of taxonomic diversity across transcripts, and this was reflected in part in the functional profile. For example, transcripts from several phylogenetic lineages encoding transport systems with specificity for a range of substrates, including maltose/maltodextrin (M00194), putrescine (M00300), and multiple sugars (M00207), as well as enzymes involved in glycolysis (e.g., glucose-6-phosphate isomerase [K01810]) and amino acid metabolism (e.g., aspartate aminotransferase [K00812]), were represented.
While global comparisons enable a top-down approach to identify universal differences across all samples, taxonomically partitioned differential expression analyses provide specific insight into discrete differences among the three transcriptome groups. Therefore, we complemented our global differential expression analysis with genus-level differential expression analyses using a novel pan-genome and orthologous group clustering method (see Materials and Methods; see also Table S6). The equivalent functional differences identified across transcriptome groups globally were largely reflected, as expected, in the genus-level differential expression results. For example, comparison of transcriptome groups 1 and 3 yielded enrichment in functions associated with Firmicutes members Clostridium (122 KEGG orthologs), and Roseburia (34 KEGG orthologs) for group 1, while group 3 was highly enriched in Ruminococcus functions (365 KEGG orthologs) (see Table S6).
Global and genus-level differential gene expression associated with probiotic consumption.
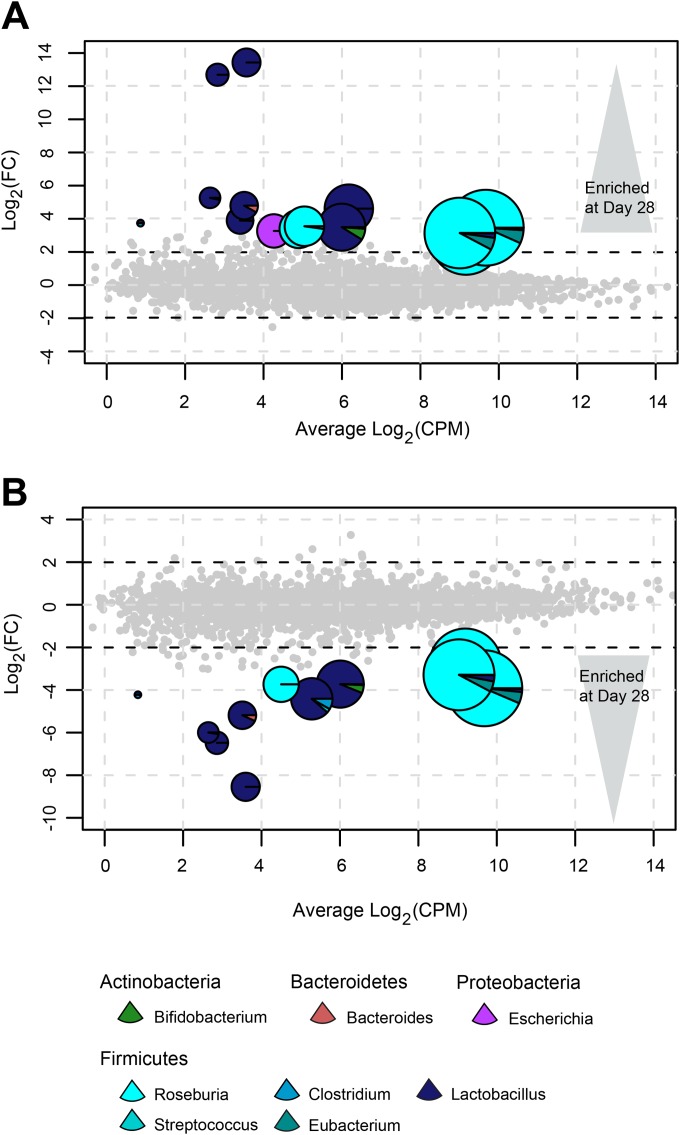
Despite considerable differences in functional dynamics across the three transcriptome groups, global and genus-level alterations associated with the administration of LGG were readily identified. Across all samples, we identified 16 differentially expressed KEGG orthologs (Fig. 3; see also Table S6 in the supplemental material). Half of these globally differentially expressed KEGG orthologs (8/16) were derived from Lactobacillus on day 28 and included functional categories for central glycolytic gene regulation (K05311), 2-oxoisovalerate dehydrogenase components (K00166 and K00167), ribonucleotide reductase (K00524), S1 RNA binding domain protein (K07571), arsenite-transporting ATPase (K01552), UPF0148 protein (K07145), and pyruvate oxidase (K00158). Interestingly, three KEGG orthologs involved in bacterial flagellar motility were differentially expressed on day 28 and were contributed predominantly by Roseburia, with minor contributions by Eubacterium (K02407, flagellar hook associated protein 2; K02397, flagellar hook-associated protein 3 FlgL; and K02396, flagellar hook-associated protein 1 FlgK) (Fig. 3; see also Table S6). Previous studies have suggested that motility of Roseburia and Eubacterium species may allow them to actively penetrate the gastrointestinal mucosa, leading to higher bioavailability of butyrate for the host and eliciting proinflammatory responses in the host with immunostimulatory potential (22).
FIG 3 .
Global differential expression associated with probiotic consumption. Smear plots represent global KEGG orthologs for comparisons between baseline and day 28 (A) and day 28 and day 56 (B). Differentially expressed KEGG orthologs are represented by pie charts compared to non-differentially expressed KEGG orthologs in gray, where colored wedges denote phylogenetic affiliations. Colors represent phylum affiliations as follows: Actinobacteria, green; Proteobacteria, purple; Bacteroidetes, red; and Firmicutes, blue. Circle size denotes average normalized KEGG ortholog counts. Axes represent the log2-fold change (Log2FC) versus the log2 transcript counts per million (Log2CPM).
Next, we complemented our global analysis with genus-level differential expression analyses to identify discrete effects of probiotic consumption on the resident gut microbiota (see Table S6). In sum, 333 KEGG orthologs were differential at baseline compared to on day 28 (238 upregulated on day 28), and an almost equivalent 342 KEGG orthologs were differential on day 28 compared to on day 56 (232 upregulated on day 28). Functions that were differential at baseline were identified for Collinsella (3 KEGG orthologs), Streptococcus (12 orthologs), and Roseburia (31 orthologs), while all Faecalibacterium KEGG orthologs (9) were upregulated on day 28. We identified pilus assembly proteins upregulated on day 28 within Bifidobacterium (K02653) and Ruminococcus (K02283), which might indicate adhesion of certain members of the resident microbiota to the host epithelium induced by the probiotic LGG. Similarly, fibronectin-binding protein 1 (K13734) was upregulated on day 28 for Faecalibacterium and Eubacterium, providing additional evidence for potential host interactions and adherence mediated by LGG. Lastly, a Clostridium-associated lactocepin, a cell envelope protease that has previously been found to have anti-inflammatory effects by selectively degrading proinflammatory chemokines (23), was upregulated on day 28.
Analysis of Lactobacillus-specific expression also revealed more than 200 KEGG orthologs that were differentially expressed on day 28, with many related to carbohydrate uptake (e.g., ABC transporters, PTS system) and utilization (e.g., beta-glucosidase, beta-N-acetyl-hexosaminidase, and secondary metabolism [pyruvate oxidase, 2-oxoisovalerate dehydrogenase E1 component]) (see Table S6). Of the differential Lactobacillus modules and pathways, the probiotic LGG exclusively contributed to the differentially expressed pathway for the biosynthesis of ketone bodies (map00072). Intriguingly, β-hydroxybutyrate and inorganic polyphosphate (polyP) have been linked to cardiovascular health, where probiotic polyP has been shown to protect the intestinal epithelia from oxidant stress and improve epithelial injury as a result of excess inflammation (24, 25).
Transcriptome group-specific differential gene expression reveals metabolic modifications associated with probiotic consumption.
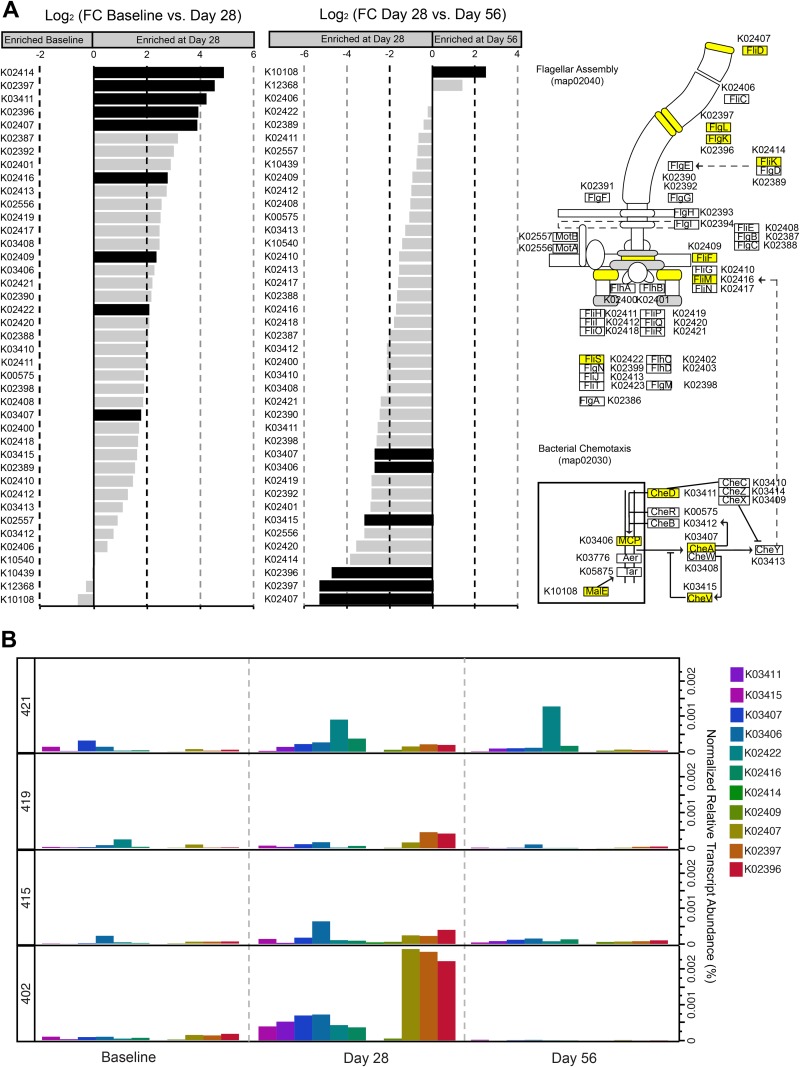
We lastly evaluated differential expression across the transcriptome groups following probiotic administration and found significant differences in differentially expressed KEGG orthologs in each of the three transcriptome groups. A total of 74 global differentially expressed KEGG orthologs and 250 genus-level differentially expressed KEGG orthologs were identified between baseline and day 28 in transcriptome group 1, and the majority of the KEGG orthologs (168/252) were upregulated on day 28 (see Table S6 in the supplemental material). Most of the KEGG orthologs on day 28 were derived from Lactobacillus (144), with the remainder from a limited number of bacterial genera, including Bacteroides, Eubacterium, Faecalibacterium, Roseburia, Ruminococcus, and Streptococcus (see Table S6). Consistent with the global differential expression analysis, we identified an even more pronounced signal for differential expression associated with bacterial motility. Multiple genes for flagellar assembly (fliD, flgL, flgK, fliK, fliF, fliM, and fliS; map02040), bacterial chemotaxis (cheD, cheA, cheV, and the methyl-accepting chemotaxis protein [MCP] gene; map02030), and accessory motility genes (fliW, flgJ, and flbD) were differentially expressed on day 28 and were contributed mainly by the genera Eubacterium and Roseburia (Fig. 4; see also Table S6). Three major motility loci have recently been characterized in Roseburia and Eubacterium isolates from the human gut (22), and mapping our data to these genomes suggests that gene expression from all three of these loci is increased in the presence of LGG. In comparisons between baseline and day 56, there were no significant differences in the levels of relative expression of the motility genes, indicating that the modulation of gene expression associated with administration of LGG was transient.
FIG 4 .
Bacterial motility and chemotaxis differentially expressed during probiotic consumption. (A) Bar charts represent log2-fold change (Log2FC) for flagellar assembly (KEGG map02040) and bacterial chemotaxis (KEGG map02030) KEGG orthologs for comparisons between transcriptome group 1 individuals at baseline and day 28 and on day 28 and day 56. Differentially expressed KEGG orthologs are denoted by black bars. Schematics of the two KEGG pathways are also shown, with differentially expressed KEGG orthologs highlighted in yellow. (B) Distribution of the normalized relative transcript abundance for the 11 KEGG orthologs differentially expressed on day 28 across the four individuals affiliated with transcriptome group 1.
In transcriptome group 2, a total of 20 global differentially expressed KEGG orthologs and 347 genus-level differentially expressed KEGG orthologs were identified between baseline and day 28; of the genus-level comparisons, 29 KEGG orthologs were differentially expressed at baseline and 318 were differentially expressed on day 28 (see Table S6). As observed for transcriptome group 1, the majority of differentially expressed KEGG orthologs on day 28 (337) were derived from Lactobacillus. In Ruminococcus-dominated transcriptome group 3, a total of 62 global differentially expressed KEGG orthologs and 25 genus-level differentially expressed KEGG orthologs were identified between baseline and day 28, with only three KEGG orthologs from Bifidobacterium and a single KEGG ortholog from Ruminococcus differentially expressed on day 28 (see Table S6). This finding was not surprising, since compliance for subject 406 was questionable, with a lack of recruited LGG transcripts, and minor LGG expression levels were detected for subject 407.
In vivo behavior of the probiotic LGG.
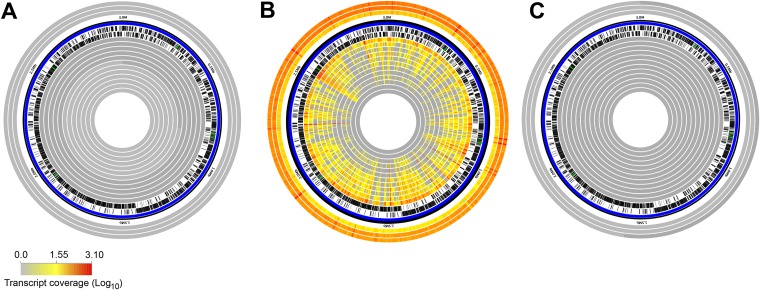
Lastly, to evaluate the in vivo behavior of the probiotic LGG, we examined the transcriptional response specific to LGG (Fig. 5). As expected, transcript coverage of the LGG genome was restricted to the period of probiotic consumption (a total 4,272,346 reads across the entire data set mapped specifically to LGG) (see Table S7 in the supplemental material). We observed a distinct gradient of transcript coverage of the LGG genome and relative abundance of LGG transcripts across the samples, from a high of 71% genome coverage, representing 1.9% of the total transcripts in subject 421 on day 28, to a low of 0.74% coverage in subject 406 (see Table S7). Furthermore, there was a significantly high level of concordance of LGG expression globally (Kendall's coefficient of concordance, W = 0.71), and even greater concordance for transcriptome groups 1 (W = 0.84) and 2 (W = 0.91), despite consistent dosage of LGG administered for each subject (see Table S7). Given the significant concordance across transcriptome groups, we tested whether overall community diversity or the abundance of certain resident microbial taxa contributed to specific patterns of LGG expression. We found no significant correlations for LGG expression related to overall community diversity, transcriptome groups, or specific resident microbial taxa (see Table S7). The reasons for the subject-specific differences in gene expression are not known. It is possible that the differential abundance of LGG transcripts in the day 28 samples reflects a difference in the absolute amount of LGG present in the stool. We found a strong positive correlation between LGG genome recovery and transcript abundance, suggesting that the relative amount of LGG present in the gut might play a strong role in transcriptional activity (r = 0.73, P = 0.0074). Another intriguing hypothesis is that the commensal gut microbiota may differentially direct LGG transcriptional responses across individuals.
FIG 5 .
In vivo expression profile for the probiotic Lactobacillus rhamnosus GG (LGG). Circular genome plots represent metagenomic (outer three tracks) and transcriptomic (inner 12 tracks) coverage at baseline (A), on day 28 during probiotic consumption (B), and on day 56, 1 month after stopping LGG consumption (C). Outer metagenomic tracks from the outermost ring are as follows: subjects 428, 419, and 408. Inner transcriptomic tracks from outermost ring to inner are as follows: subjects 421, 402, 401, 428, 419, 420, 412, 408, 409, 415, 407, and 406. Compliance for subject 406 was questionable, as reflected in the lack of recruited LGG transcripts.
Across the LGG genome, we found the greatest absolute expression levels associated with small noncoding RNAs (Fig. 5). Of the five intergenic regions with greatest transcript coverage, four were previously identified within the Rfam database (bacterial RNase P, RF00010; 6S/SsrS, RF00013; and transfer mRNA, RF00023), although they are not currently annotated within the LGG genome (26). One intergenic region represented a putatively novel sRNA, with transcripts partially overlapping a small 53-amino-acid putative hypothetical protein (LRHM_2096, NCBI gene identifier [ID] 12475226). Downstream of this region is a putative protein (LRHM_2095, NCBI gene ID 12475225) containing a GyrI-like small-molecule binding domain (pfam06445) found in numerous bacterial transcriptional activators that regulate genes involved in resistance to antibiotics, organic solvents, and heavy metals (27). We found little evidence for high transcriptional expression of the many characterized host-associated genes, such as the intestinal adhesins or secreted protein effectors, despite metagenomic evidence for the presence of these genes described previously. A likely explanation could be that transcription of these gene sets is temporally restricted during LGG interaction with the intestinal epithelium and is not captured from sampling of the fecal transcriptional pool.
DISCUSSION
It has been widely proposed that probiotics harbor the potential to reverse dysbiosis in the resident microbial consortia of the human gastrointestinal tract, thereby restoring gut mucosal homeostasis (28). However, direct evidence demonstrating a system-wide restoration of a dysbiotic microbial ecosystem has proven elusive. Few studies have measured the global microbial community composition and structure within an adult human clinical study to evaluate the effects of probiotic administration, with only a single study evaluating the transcriptional response (6, 10). The results of those studies have demonstrated that the composition of the resident bacterial community in adults remains largely unchanged following probiotic administration (10).
Heightened interest in the human microbiome and its relation to human health has led to the hypothesis that administration of a probiotic organism could modulate the resident microbial ecology. There is growing support for taking a system-level approach, or “adaptive management,” to maintain the microbial ecosystems across our body to promote health (29). However, much has yet to be learned about our resident commensal microbes, particularly the mechanisms governing ecological community assembly rules (e.g., deterministic factors, historical contingencies, and stochastic factors) if we are to effectively implement probiotic treatment to elicit a beneficial and long-lasting modification to a given health state. In this study, we provide robust analytical techniques to evaluate, in detail, the effects of a single-organism probiotic on the gut microbiota of elderly people. Through the use of bacterial community profiling using 16S rRNA pyrosequencing, whole-community expression profiling using RNA-seq, and metagenomic sequencing, we investigated the role that probiotics may play on the structure and function of the resident microbiota.
No statistically significant differences associated with LGG consumption were identified from the 16S rRNA analysis, which is consistent with recent studies evaluating the fecal microbiota before and after LGG intervention from an adult Finish cohort (10) and seven adult female monozygotic twin pairs (6). Given the depth of 16S rRNA sequence coverage in our study, there is a possibility that rarer organisms in the community might be impacted by LGG consumption. Our findings indicate that the dominant microbial taxa are not modified by probiotic consumption, yet additional deep sequencing of the community is needed to resolve whether rare members are impacted.
Our inclusion of reference genomes for viruses and human-associated microeukaryotes enabled us to taxonomically identify a subset of transcripts affiliated with these additional microbial components of the community, capturing the community-wide taxonomic profile. While fungal transcripts were not well represented within the community (on average, 0.00018%), highly represented transcripts mapping to the plant-pathogenic single-stranded RNA viruses Pepper mild mottle virus (PMMoV) and Tomato mosaic virus (ToMV) were identified, consistent with previous findings (30, 31), suggesting that a signature for the consumption of virus-infected produce might be readily captured from microbial RNA-seq studies. Further, we found distinct dynamics from the viral and fungal fraction that did not follow the intraspecific clustering patterns observed for the bacterial and archaeal expression profiles. The uncoupled dynamics observed in the viral and fungal fractions could perhaps reflect strong dietary influences compared to host-specific influences.
Comparison of transcriptional profiles relative to taxonomic composition yielded three distinct transcriptome groups that displayed differences in functional dynamics. The transcriptome groups were differentiated both by unique transcript taxonomic composition and distinctive functional properties and also differed significantly in overall community diversity, as measured by the Shannon diversity index. Transcriptome group 2 was characterized by highly abundant transcripts from the dominant archaeon, Methanobrevibacter, which lends further support for the importance of this microbe in a subset of individuals and underscores the need to complement 16S rRNA surveys that use bacterial-specific primer sets to fully capture the extant microbial community.
While differentially expressed functions across the three identified transcriptome groups were more pronounced than differences during probiotic consumption, we nevertheless identified a suite of global and genus-level KEGG orthologs that were differentially expressed during consumption of LGG. We found evidence for differential expression of genes involved in adhesion and a large number of genes involved in bacterial motility during probiotic consumption that were predominantly associated with Roseburia and Eubacterium motile gut species. These commensal gut bacteria are notable as producers of the short-chain fatty acid butyrate, which is the preferred nutrient for human colonocytes, has anti-inflammatory properties, and decreases epithelial permeability. Previous studies have suggested that motility of Roseburia species is an important factor in colonization of the gastrointestinal mucosa, and the presence of flagella may allow them to actively penetrate into the mucus layer, which in turn may lead to higher bioavailability of butyrate for the host (22). It has recently been reported that increased levels of butyrate following administration of the probiotic VSL#3 stimulated the release of glucagon-like peptide 1 (GLP-1) from intestinal L cells in a mouse model of obesity, resulting in reduced food intake and improved glucose tolerance (32).
Furthermore, we were able to directly evaluate the in vivo behavior of LGG and found that the transcript coverage of the LGG genome was restricted to the period of probiotic consumption, a finding that lends support for the transient properties of this probiotic. Despite minimal genomic and absolute transcript abundances of LGG relative to the resident microbiota, we were able to reconstruct, on average, 62% of the genome from the metagenomic data and 34% from the transcriptomic data. We observed noteworthy concordance across study subjects, particularly within transcriptome groups, despite considerable interindividual resident microbiota profiles. These findings are intriguing and might suggest that the commensal gut microbiota may play a role in differentially directing LGG expression patterns across individuals. Further studies to more fully investigate this hypothesis are clearly warranted, as well as further exploration to delineate the functional contribution of the resident microbiota to the metabolic capacity of the host. Taken together, our results provide evidence for discrete, transient functional effects imparted by a specific single-organism probiotic and challenge the prevailing notion that probiotics substantially modify the resident microbiota within nondiseased individuals in an appreciable fashion.
MATERIALS AND METHODS
Volunteer recruitment, clinical evaluation, and sample collection.
Fecal samples were collected from healthy elderly volunteers recruited from the Boston area for an open-label clinical trial to assess the safety and tolerability of LGG (13). Written informed consent was obtained, and all procedures were approved by the Partners Human Research Committee (IRB no. 2010P001695/MGH). The clinical trial was registered at Clinicaltrials.gov (NCT01274598). Approximately 10 g stool was mixed with 20 ml RNAlater, homogenized, aliquoted into cryovials (400 µl), and stored at −80°C until further processing.
DNA extraction, pyrosequencing of barcoded 16S rRNA gene amplicons, and analysis.
Total DNA was extracted from 250 µl homogenized stool using the ZR fecal DNA isolation kit (catalog no. D6010; Zymo Research Corp.) with modifications as described previously (33). Methods for amplification of the bacterial V1-V2 16S rRNA gene region and multiplex 454 pyrosequencing have been described (33) (see Table S1 in the supplemental material). Raw sequences were binned and quality trimmed using the split_libraries.py script from the QIIME software package (version 1.6) (34) with the following criteria: (i) no mismatches in primer sequence and bar code tag, (ii) minimum and maximum read lengths of 200 and 400 bp (base pairs), and (iii) an average read quality score of 25 over a 50-bp sliding window. Sequences were then dereplicated, de novo chimera checked using UCHIME (35), and clustered into operational taxonomic units (OTUs) at a 97% identity level using the usearch quality filtering pipeline implemented within QIIME (34). Representative sequences (most abundant) were selected for each OTU and aligned using PyNAST (36), and a phylogenetic tree was built using RAxML (37). Taxonomic classifications were assigned using the RDP naive Bayesian classifier with the Greengenes 97% OTU database (October 2012) (38). Rarefied OTUs (4,091 sequences per sample) were used to calculate alpha (within-sample) and beta (between-sample) diversity for each sample over time. Dot plots of taxon relative abundances were generated using a custom Perl script (39). Linear mixed-effects modeling was performed using the function “lmer” in the R-package “lme4” (40). Akaike’s information criterion (AIC) values and a chi-square test were used to select the best model. Analysis of variance (ANOVA) was used to test for differences associated with LGG consumption.
Reference microbial and viral databases and expanded KEGG annotations.
A custom-curated database was constructed from draft and finished reference genomes as follows: (i) 739 bacterial and archaeal genomes part of the Human Microbiome Project’s Data Analysis and Coordination Center (HMP DACC; October 2012), (ii) an additional 18 finished Bacteroides and Prevotella genomes from NCBI’s GenBank (August 2012) which were not already represented in the HMP DACC, (iii) nonredundant IMG bacterial and archaeal genomes (IMG v4; November 2012), (iv) 3,332 viral genomes from NCBI’s RefSeq (August 2012), and (v) 64 human-associated fungal and microeukaryotic genomes (see Table S2B in the supplemental material). In sum, our database consisted of 6,809 genomes of 5,720 microbial species representing 925 genera spanning 39 phyla.
All predicted proteins from the reference genome database were annotated with KEGG orthologs using the KEGG database (June 2011) (41). Further, we developed a novel pan-genome and orthologous group clustering method to expand the genus-level annotations for the reference nonviral genome database. Each annotated gene was binned based on genus and aligned against its own genus’ BLAST database with criteria of 85% nucleotide identity and a ≤15% difference in length. Genes and previously unannotated regions with sufficiently close homology were grouped into paralog/ortholog clusters and annotated with KEGG ortholog identifiers. Paralog/ortholog clusters and copy number by strain were retained for downstream count normalization for RNA-seq data processing. This method enabled approximately 50% of all reference genes (both previously annotated and newly identified) to be assigned with KEGG ortholog labels.
Metagenomic sequencing, assembly, and annotation.
The same extracted total DNA used for 16S rRNA gene amplicon sequencing was used for whole-community genomic sequencing. Genomic DNA libraries were constructed using NEBNext DNA sample prep master mix set 1 (New England Biolabs). DNA was fragmented with the Covaris E210 (Covaris), and libraries were prepared using a modified version of the manufacturer’s protocol. The DNA was then purified between enzymatic reactions, and the size selection of the library was performed with AMPure XT beads (Beckman Coulter Genomics). The PCR amplification step was performed with primers containing a 7-nucleotide (nt) index. Sequencing was performed using the Illumina HiSeq2000 platform with three samples multiplexed per lane. Assembly details are provided in Table S2 in the supplemental material.
An in-house quality control (QC) pipeline was used for quality trimming and adaptor removal. The trimmed reads for each sample were filtered via digital normalization (42, 43) and assembled using IDBA-UD (44). The QC-trimmed, non-digitally normalized reads for all samples were then mapped with Bowtie (45) to the assembled contigs, as well as to reference genomes to provide coverage estimates. Contigs for which ≥90% of aligned reads uniquely mapped to the target genome were binned together and summed for coverage estimates and relative abundance profiles. Each contig was run through an in-house automated gene annotation pipeline utilizing MetaGeneMark (46).
RNA extraction, ribosomal depletion, and RNA-seq.
Total RNA was extracted from 250 µl homogenized stool using a modified protocol described by Zoetendal et al. (12). Briefly, a combination of acid phenol, SDS, and aggressive bead beating by using lysing matrix tubes (Qbiogene) and a FastPrep FP120 instrument (Qbiogene) was utilized to lyse cells. Subsequently, three rounds of phenol-chloroform isoamyl alcohol extraction, ethanol precipitation, and DNase treatment using an Ambion Turbo DNA-free kit (Invitrogen catalog no. Am1907) yielded total RNA. RNA was further purified using a Qiagen RNeasy kit (catalog no. 74104) and quality checked using an Agilent 2100 Expert bioanalyzer. This protocol typically yielded an average of 50 µg of high-quality total RNA with an RNA integrity number (RIN) averaging 8.5 (see Table S3 in the supplemental material). Genomic DNA contamination was assessed by 16S rRNA PCR, and a second round of DNase treatment was performed if residual DNA was present. Next, efficient rRNA depletion was achieved using a combined Gram-positive and Gram-negative Ribo-Zero rRNA removal kit (Epicentre Technologies) with a final purification step using a Zymo RNA clean and concentrator kit (catalog no. R1015). Ribosomally depleted mRNA samples were quality checked using an Agilent RNA 6000 Nano kit (catalog no. 5067-1511) and generally ranged from 150 to 300 ng from an initial input of 5 µg total RNA. Illumina RNA-seq libraries containing 6-bp indexes were prepared with the TruSeq RNA sample prep kit (Illumina) by following a variation of the manufacturer’s protocol. cDNA was purified between enzymatic reactions, and library size selection was performed with AMPure XT beads (Beckman Coulter Genomics). Sequencing was performed using the Illumina HiSeq2000 platform with three samples multiplexed per lane.
RNA-seq taxonomic identification and functional annotation.
Raw sequence data were processed using in-house QC pipelines to filter and truncate low-quality reads. A customized Bowtie (45) alignment pipeline was implemented to map transcriptomic read sets. The filtered reads were first searched against the complete Silva bacterial, archaeal, and eukaryotic small subunit (SSU) and large subunit (LSU) ribosomal database containing a total of 2,762,151 sequences (47) and were subsequently culled from the data set. NCBI’s best match tagger (BMTagger) (48) was next used to search and remove human-associated reads. Using the reference genome database described above, reads were then mapped with a criteria of no more than two base pair mismatches over the entire length of each trimmed read and allowing for multiple reference matches. Post-processing of alignments included profiling the results to identify redundant reference strains for each strain X in the reference database as follows: (i) we computed how many reads aligned to X, (ii) how often X was uniquely matched by reads (i.e., reads which aligned to no other genome), (iii) the set of organisms Y (if any existed) that were hit by the same reads aligning to X (and how often), (iv) the coverage of X by matched reads, in terms of average number of reads aligning to each reference base pair, and (v) the overall coverage of X, in terms of the fraction of base pairs in X’s reference genome that were hit by at least one read. After this filtration step, a single representative strain for each species was selected, redundant references were removed, and taxonomic and functional assignments were generated. Sequencing statistics, number of ribosomal and human reads, and reads mapping to reference microbial and viral databases are detailed in Table S3 in the supplemental material. Circular genome plots with mapped transcripts and metagenomic coverage were generated using the program Circleator (49). LDA effect size (LEfSe) analysis was used to identify discriminant species and COGs across comparison transcriptome groups (50).
Differential expression analysis.
Mapped reads were totaled using htseq-count (51) for orthologous gene clusters and KEGG orthologs. Copy number normalization for paralog/ortholog groups was informed by per-sample expression abundance, and pseudolength calculation for the KEGG ortholog group was performed by abundance-weighing the lengths of the most highly expressed genes. Each gene’s length was multiplied by its raw read count; these counts were then summed and divided by the total number of reads aligning to the most-expressed genes. The abundance-weighted, pseudolength-normalized KEGG ortholog counts were then summed to produce aggregated count estimates for KEGG modules and KEGG pathways globally and for each genus.
To examine both global and genus-level differential expression patterns, we utilized the extensive statistical methodologies developed specifically for RNA-seq data within the software package edgeR (52). The trimmed mean of M values (TMM) was utilized to estimate appropriate scaling factors for normalization among samples with the inherent assumption that the majority of counts are not differentially expressed (53). We used generalized linear model (glm) methods in order to account for technical variation characteristic of RNA-seq data, as well as to factor in the complexity of a mixed microbial community data set derived from multiple individuals over three separate sampling times (54). Within this framework, the Cox-Reid profile-adjusted likelihood (CR) method was implemented to estimate dispersions and model the biological coefficient of variation (BCV) across samples. Transcriptome group designations were calculated based on complete-linkage hierarchical clustering of the Pearson correlation coefficients (PCC) for the species-level transcript abundances (LGG transcripts were removed prior to calculation). Transcriptome group membership was validated by three separate algorithms, the average silhouette index (SI), prediction strength, and the Caliński-Harabasz (CH) statistic (see Fig. S3 in the supplemental material).
A suite of independent differential expression calculations were performed using the negative binomial model with an adjusted P value of <0.05 (Benjamini and Hochberg multiple-testing correction) between all pairs of transcriptome groups, the study time points, and across the study time points for each transcriptome group for the global KEGG orthologs, global KEGG modules and pathways, and genus-level KEGG orthologs, modules, and pathways.
Nucleotide sequence accession numbers.
All data have been deposited in NCBI’s database of Genotypes and Phenotypes (dbGaP) under study accession no. phs000896.v1.p1 and are accessible to authorized users following the NIH dbGAP system policies and procedures.
SUPPLEMENTAL MATERIAL
Unweighted (A) and weighted (B) UniFrac measures of beta diversity. UniFrac is a phylogenetically aware measure of beta diversity, where the weighted measure additionally takes into account phylogenetic tree branch length. Download
Genome and transcript coverage for abundant bacterial and archaeal genera across all individuals. Circular genome plots represent metagenomic (outer three tracks) and transcriptomic (inner 12 tracks) coverage at baseline, on day 28 during probiotic consumption, and on day 56, 1 month after stopping LGG consumption. (A) Alistipes; (B) Anaerococcus; (C) Bacteroides; (D) Bifidobacterium; (E) Collinsella; (F) Coprococcus; (G) Dorea; (H) Eubacterium; (I) Faecalibacterium; (J) Lactobacillus; (K) Methanobrevibacter; (L) Methanosphaera; (M) Parabacteroides; (N) Roseburia; (O) Ruminococcus; (P) Streptococcus. The outer metagenomic tracks from the outermost ring are as follows: subjects 428, 419, and 408. The inner transcriptomic tracks from the outermost ring to inner are as follows: subjects 421, 402, 401, 428, 419, 420, 412, 408, 409, 415, 407, and 406. Download
RNA-seq transcriptome group assignments and validation. (A) Pearson correlation coefficient (PCC) values were used to assign transcriptome groups, with group number selection based on maximizing the prediction strength (PS), silhouette index (SI), and the Caliński-Harabasz (CH) statistic; (B) principle coordinate plot for grouping, where the two-component axis explains 84.77% of the total variance. Download
Bacterial 16S rRNA sequencing statistics and linear mixed-effects analysis. (A) 16S rRNA bar code and primer sequences, along with total number of sequences for each sample; (B) summary of a linear mixed-effects analysis on the response of bacterial community diversity. The factors tested are medication status (aspirin, statins, proton pump inhibitors [PPI], nonsteroidal anti-inflammatory drug [NSAIDs], ciprofloxacin), blood pressure, suspected norovirus infection, and probiotic consumption (LGG). The base model takes into account variability within subjects only, with fixed variables added to the model to test for fit. The full model includes all fixed variables, where the change in model fit is a result of exclusion of each fixed effect based on Akaike’s information criterion (AIC). Statistically significant variables are in bold.
Metagenomic information and reference mapping information. (A) Metagenomic assembly statistics for nine samples. (B) Reference microbial, viral, and microeukaryote genomes used in the analysis. For draft genomes, accession numbers represent the first contig/scaffold only. Taxonomic lineage reflects NCBI taxonomy. (C) Strain-level genome abundances and coverage from metagenomic assemblies.
RNA-seq statistics.
Correlations between 16S rRNA and RNA-seq relative taxonomic distributions. PCC, Pearson correlation coefficient. Highlighted in green are genera displaying a significant correlation between 16S rRNA and RNAseq data sets.
Discriminant species and COG categories for transcriptome group designations. (A) Discriminant species for transcriptome group 1 versus transcriptome group 2; (B) discriminant species for transcriptome group 1 versus transcriptome group 3; (C) discriminant species for transcriptome group 2 versus transcriptome group 3 (no discriminant species were identified across the time points sampled); (D) discriminant COG categories for transcriptome group 1 versus transcriptome group 2; (E) discriminant COG categories for transcriptome group 1 versus transcriptome group 3; (F) discriminant COG categories for transcriptome group 2 versus transcriptome group 3. Highlighted rows are as follows: blue, discriminative species/COGs for transcriptome group 1 in all comparisons; green, discriminative species/COGs for transcriptome group 2 in all comparisons; red, discriminative species/COGs for transcriptome group 3 in all comparisons.
Global and genus-level KEGG ortholog differential expression for all pairwise comparisons. (A) Global comparison of transcriptome group 1 versus transcriptome group 2; (B) global comparison of transcriptome group 1 versus transcriptome group 3; (C) global comparison of transcriptome group 2 versus transcriptome group 3; (D) genus-level comparison of transcriptome group 1 versus transcriptome group 2; (E) genus-level comparison of transcriptome group 1 versus transcriptome group 3; (F) genus-level comparison of transcriptome group 2 versus transcriptome group 3; (G) global comparison of baseline and day 28; (H) global comparison of day 28 and day 56; (I) genus-level comparison of baseline and day 28; (J) genus-level comparison of day 28 and day 56; (K) global comparison of baseline and day 28 for transcriptome group 1; (L) global comparison of day 28 and day 56 for transcriptome group 1; (M) genus-level comparison of baseline and day 28 for transcriptome group 1; (N) genus-level comparison of day 28 and day 56 for transcriptome group 1; (O) global comparison of baseline and day 28 for transcriptome group 2; (P) global comparison of day 28 and day 56 for transcriptome group 2; (Q) genus-level comparison of baseline and day 28 for transcriptome group 2; (R) genus-level comparison of day 28 and day 56 for transcriptome group 2; (S) global comparison of baseline and day 28 for transcriptome group 3; (T) global comparison of day 28 and day 56 for transcriptome group 3; (U) genus-level comparison of baseline and day 28 for transcriptome group 3; (V) genus-level comparison of day 28 and day 56 for transcriptome group 3.
Lactobacillus rhamnosus GG (LGG) transcript statistics and comparison to resident genera on day 28. (A) LGG genome coverage, abundance, and concordance across all subjects on day 28. Concordance was calculated using Kendall coefficient of concordance. (B) Correlations among LGG relative expression and resident genera on day 28. LGG relative expression is calculated for all genome-mapped transcripts. PCC, Pearson correlation coefficient.
ACKNOWLEDGMENTS
We acknowledge the tremendous effort of NIH Program Officer Linda Duffy, for contributions in structuring the U01 study design and implementation of protocols under phased regulatory approved Investigational New Drug (IND).
This research was conducted under National Science Foundation-funded MRI-R2 project no. DBI-0959894. Funding support was provided by the National Institutes of Health, National Center for Complementary and Alternative Medicine (NIH/NCCAM) (3U01AT002952 and 3K24AT003683).
Footnotes
Citation Eloe-Fadrosh EA, Brady A, Crabtree J, Drabek EF, Ma B, Mahurkar A, Ravel J, Haverkamp M, Fiorino A-M, Botelho C, Andreyeva I, Hibberd PL, Fraser CM. 2015. Functional dynamics of the gut microbiome in elderly people during probiotic consumption. mBio 6(2):e00231-15. doi:10.1128/mBio.00231-15.
REFERENCES
- 1.Van Baarlen P, Wells JM, Kleerebezem M. 2013. Regulation of intestinal homeostasis and immunity with probiotic lactobacilli. Trends Immunol 34:208–215. doi: 10.1016/j.it.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Sanders ME, Guarner F, Guerrant R, Holt PR, Quigley EM, Sartor RB, Sherman PM, Mayer EA. 2013. An update on the use and investigation of probiotics in health and disease. Gut 62:787–796. doi: 10.1136/gutjnl-2012-302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonnenburg JL, Fischbach MA. 2011. Community health care: therapeutic opportunities in the human microbiome. Sci Transl Med 3:78ps12. doi: 10.1126/scitranslmed.3001626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bron PA, van Baarlen P, Kleerebezem M. 2012. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat Rev Microbiol 10:66–78. doi: 10.1038/nrmicro2690. [DOI] [PubMed] [Google Scholar]
- 5.Lebeer S, Vanderleyden J, De Keersmaecker SC. 2010. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol 8:171–184. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- 6.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, Chervaux C, Knights D, Lozupone CA, Knight R, Duncan AE, Bain JR, Muehlbauer MJ, Newgard CB, Heath AC, Gordon JI. 2011. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med 3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerritsen J, Smidt H, Rijkers GT, de Vos WM. 2011. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr 6:209–240. doi: 10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx AP, Lebeer S, De Keersmaecker SC, Vanderleyden J, Hämäläinen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjärvi T, Auvinen P, de Vos WM. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc Natl Acad Sci U S A 106:17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lebeer S, Verhoeven TL, Claes IJ, De Hertogh G, Vermeire S, Buyse J, Van Immerseel F, Vanderleyden J, De Keersmaecker SC. 2011. FISH analysis of Lactobacillus biofilms in the gastrointestinal tract of different hosts. Lett Appl Microbiol 52:220–226. doi: 10.1111/j.1472-765X.2010.02994.x. [DOI] [PubMed] [Google Scholar]
- 10.Lahti L, Salonen A, Kekkonen RA, Salojärvi J, Jalanka-Tuovinen J, Palva A, Orešič M, de Vos WM. 2013. Associations between the human intestinal microbiota, Lactobacillus rhamnosus GG and serum lipids indicated by integrated analysis of high-throughput profiling data. PEER J 1:e32. doi: 10.7717/peerj.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giannoukos G, Ciulla DM, Huang K, Haas BJ, Izard J, Levin JZ, Livny J, Earl AM, Gevers D, Ward DV, Nusbaum C, Birren BW, Gnirke A. 2012. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol 13:R23. doi: 10.1186/gb-2012-13-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zoetendal EG, Booijink CC, Klaassens ES, Heilig HG, Kleerebezem M, Smidt H, de Vos WM. 2006. Isolation of RNA from bacterial samples of the human gastrointestinal tract. Nat Protoc 1:954–959. doi: 10.1038/nprot.2006.143. [DOI] [PubMed] [Google Scholar]
- 13.Hibberd PL, Kleimola L, Fiorino AM, Botelho C, Haverkamp M, Andreyeva A, Poutsiaka D, Fraser C, Solano-Aguilar G, Snydman DR. 2014. No evidence of harms of probiotic Lactobacillus rhamnosus GG ATCC 53103 in healthy elderly—a phase I open label study to assess safety, tolerability and cytokine responses. PLoS One 9:e113456. doi: 10.1371/journal.pone.0113456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, Stanton C, van Sinderen D, O’Connor M, Harnedy N, O’Connor K, Henry C, O’Mahony D, Fitzgerald AP, Shanahan F, Twomey C, Hill C, Ross RP, O’Toole PW. 2011. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A 108(Suppl 1):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O’Sullivan O, Fitzgerald GF, Deane J, O’Connor M, Harnedy N, O’Connor K, O’Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O’Toole PW. 2012. Gut microbiota composition correlates with diet and health in the elderly. Nature 488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 16.Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, Nikkïla J, Monti D, Satokari R, Franceschi C, Brigidi P, De Vos W. 2010. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One 5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, Waller A, Mende DR, Kultima JR, Martin J, Kota K, Sunyaev SR, Weinstock GM, Bork P. 2013. Genomic variation landscape of the human gut microbiome. Nature 493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eloe-Fadrosh EA, Rasko DA. 2013. The human microbiome: from symbiosis to pathogenesis. Annu Rev Med 64:145–163. doi: 10.1146/annurev-med-010312-133513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Douillard FP, Ribbera A, Järvinen HM, Kant R, Pietilä TE, Randazzo C, Paulin L, Laine PK, Caggia C, von Ossowski I, Reunanen J, Satokari R, Salminen S, Palva A, de Vos WM. 2013. Comparative genomic and functional analysis of Lactobacillus casei and Lactobacillus rhamnosus strains marketed as probiotics. Appl Environ Microbiol 79:1923–1933. doi: 10.1128/AEM.03467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosalbes MJ, Durbán A, Pignatelli M, Abellan JJ, Jiménez-Hernández N, Pérez-Cobas AE, Latorre A, Moya A. 2011. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One 6:e17447. doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gifford SM, Sharma S, Booth M, Moran MA. 2013. Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J 7:281–298. doi: 10.1038/ismej.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neville BA, Sheridan PO, Harris HM, Coughlan S, Flint HJ, Duncan SH, Jeffery IB, Claesson MJ, Ross RP, Scott KP, O’Toole PW. 2013. Pro-inflammatory flagellin proteins of prevalent motile commensal bacteria are variably abundant in the intestinal microbiome of elderly humans. PLoS One 8:e68919. doi: 10.1371/journal.pone.0068919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Von Schillde M.-A, Hörmannsperger G, Weiher M, Alpert C-A, Hahne H, Bäuerl C, van Huynegem K, Steidler L, Hrncir T, Pérez-Martínez G, Kuster B, Haller D. 2012. Lactocepin secreted by Lactobacillus exerts anti-inflammatory effects by selectively degrading proinflammatory chemokines. Cell Host Microbe 11:387–396. doi: 10.1016/j.chom.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Segawa S, Fujiya M, Konishi H, Ueno N, Kobayashi N, Shigyo T, Kohgo Y. 2011. Probiotic-derived polyphosphate enhances the epithelial barrier function and maintains intestinal homeostasis through integrin–p38 MAPK pathway. PLoS One 6:e23278. doi: 10.1371/journal.pone.0023278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dedkova EN, Blatter LA. 2014. Role of β-hydroxybutyrate, its polymer poly-β-hydroxybutyrate and inorganic polyphosphate in mammalian health and disease. Front Physiol 5:1–22. doi: 10.3389/fphys.2014.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A. 2013. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res 41:D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon HJ, Bennik MH, Demple B, Ellenberger T. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat Struct Biol 7:424–430. doi: 10.1038/75213. [DOI] [PubMed] [Google Scholar]
- 28.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang T, Breitbart M, Lee WH, Run J-Q, Wei CL, Soh SW, Hibberd ML, Liu ET, Rohwer F, Ruan Y. 2006. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol 4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yadav H, Lee J.-H, Lloyd J, Walter P, Rane SG. 2013. Beneficial metabolic effects of a probiotic via butyrate induced GLP-1 secretion. J Biol Chem 288:25088–25097. doi: 10.1074/jbc.M113.452516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eloe-Fadrosh EA, McArthur MA, Seekatz AM, Drabek EF, Rasko DA, Sztein MB, Fraser CM. 2013. Impact of oral typhoid vaccination on the human gut microbiota and correlations with S. Typhi-specific immunological responses. PLoS One 8:e62026. doi: 10.1371/journal.pone.0062026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 38.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaikova E, Walsh DA, Stilwell CP, Mohn WW, Tortell PD, Hallam SJ. 2010. Microbial community dynamics in a seasonally anoxic fjord: Saanich Inlet, British Columbia. Environ Microbiol 12:172–191. doi: 10.1111/j.1462-2920.2009.02058.x. [DOI] [PubMed] [Google Scholar]
- 40.Bates D, Mächler M, Bolker B, Walker S. 2014. Fitting linear mixed-effects models using lme4. Cornell University Library, Ithaca, NY: http://arxiv.org/abs/1406.5823. [Google Scholar]
- 41.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res 32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howe AC, Jansson JK, Malfatti SA, Tringe SG, Tiedje JM, Brown CT. 2014. Tackling soil diversity with the assembly of large, complex metagenomes. Proc Natl Acad Sci U S A 111:4904–4909. doi: 10.1073/pnas.1402564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown CT, Howe AC, Zhang Q, Pyrkosz A, Brom T. 2012. A reference of free algorithm for computational normalization of shotgun sequencing data. Cornell University Library, Ithaca, NY: http://arxiv.org/abs/1203.4802. [Google Scholar]
- 44.Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 45.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu W, Lomsadze A, Borodovsky M. 2010. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res 38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. Silva: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agarwala R, Rotmistrovsky K. 2011. BMTagger: best match tagger for removing human reads from metagenomics datasets. ftp://ftp.ncbi.nlm.nih.gov/pub/agarwala/bmtagger/.
- 49.Crabtree J, Agrawal S, Mahurkar A, Myers GS, Rasko DA, White O. 2014. Circleator: flexible circular visualization of genome-associated data with BioPerl and SVG. Bioinformatics 30:3125–3127. doi: 10.1093/bioinformatics/btu505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anders S, Pyl PT, Huber W. 2015. HTSeq—a python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Unweighted (A) and weighted (B) UniFrac measures of beta diversity. UniFrac is a phylogenetically aware measure of beta diversity, where the weighted measure additionally takes into account phylogenetic tree branch length. Download
Genome and transcript coverage for abundant bacterial and archaeal genera across all individuals. Circular genome plots represent metagenomic (outer three tracks) and transcriptomic (inner 12 tracks) coverage at baseline, on day 28 during probiotic consumption, and on day 56, 1 month after stopping LGG consumption. (A) Alistipes; (B) Anaerococcus; (C) Bacteroides; (D) Bifidobacterium; (E) Collinsella; (F) Coprococcus; (G) Dorea; (H) Eubacterium; (I) Faecalibacterium; (J) Lactobacillus; (K) Methanobrevibacter; (L) Methanosphaera; (M) Parabacteroides; (N) Roseburia; (O) Ruminococcus; (P) Streptococcus. The outer metagenomic tracks from the outermost ring are as follows: subjects 428, 419, and 408. The inner transcriptomic tracks from the outermost ring to inner are as follows: subjects 421, 402, 401, 428, 419, 420, 412, 408, 409, 415, 407, and 406. Download
RNA-seq transcriptome group assignments and validation. (A) Pearson correlation coefficient (PCC) values were used to assign transcriptome groups, with group number selection based on maximizing the prediction strength (PS), silhouette index (SI), and the Caliński-Harabasz (CH) statistic; (B) principle coordinate plot for grouping, where the two-component axis explains 84.77% of the total variance. Download
Bacterial 16S rRNA sequencing statistics and linear mixed-effects analysis. (A) 16S rRNA bar code and primer sequences, along with total number of sequences for each sample; (B) summary of a linear mixed-effects analysis on the response of bacterial community diversity. The factors tested are medication status (aspirin, statins, proton pump inhibitors [PPI], nonsteroidal anti-inflammatory drug [NSAIDs], ciprofloxacin), blood pressure, suspected norovirus infection, and probiotic consumption (LGG). The base model takes into account variability within subjects only, with fixed variables added to the model to test for fit. The full model includes all fixed variables, where the change in model fit is a result of exclusion of each fixed effect based on Akaike’s information criterion (AIC). Statistically significant variables are in bold.
Metagenomic information and reference mapping information. (A) Metagenomic assembly statistics for nine samples. (B) Reference microbial, viral, and microeukaryote genomes used in the analysis. For draft genomes, accession numbers represent the first contig/scaffold only. Taxonomic lineage reflects NCBI taxonomy. (C) Strain-level genome abundances and coverage from metagenomic assemblies.
RNA-seq statistics.
Correlations between 16S rRNA and RNA-seq relative taxonomic distributions. PCC, Pearson correlation coefficient. Highlighted in green are genera displaying a significant correlation between 16S rRNA and RNAseq data sets.
Discriminant species and COG categories for transcriptome group designations. (A) Discriminant species for transcriptome group 1 versus transcriptome group 2; (B) discriminant species for transcriptome group 1 versus transcriptome group 3; (C) discriminant species for transcriptome group 2 versus transcriptome group 3 (no discriminant species were identified across the time points sampled); (D) discriminant COG categories for transcriptome group 1 versus transcriptome group 2; (E) discriminant COG categories for transcriptome group 1 versus transcriptome group 3; (F) discriminant COG categories for transcriptome group 2 versus transcriptome group 3. Highlighted rows are as follows: blue, discriminative species/COGs for transcriptome group 1 in all comparisons; green, discriminative species/COGs for transcriptome group 2 in all comparisons; red, discriminative species/COGs for transcriptome group 3 in all comparisons.
Global and genus-level KEGG ortholog differential expression for all pairwise comparisons. (A) Global comparison of transcriptome group 1 versus transcriptome group 2; (B) global comparison of transcriptome group 1 versus transcriptome group 3; (C) global comparison of transcriptome group 2 versus transcriptome group 3; (D) genus-level comparison of transcriptome group 1 versus transcriptome group 2; (E) genus-level comparison of transcriptome group 1 versus transcriptome group 3; (F) genus-level comparison of transcriptome group 2 versus transcriptome group 3; (G) global comparison of baseline and day 28; (H) global comparison of day 28 and day 56; (I) genus-level comparison of baseline and day 28; (J) genus-level comparison of day 28 and day 56; (K) global comparison of baseline and day 28 for transcriptome group 1; (L) global comparison of day 28 and day 56 for transcriptome group 1; (M) genus-level comparison of baseline and day 28 for transcriptome group 1; (N) genus-level comparison of day 28 and day 56 for transcriptome group 1; (O) global comparison of baseline and day 28 for transcriptome group 2; (P) global comparison of day 28 and day 56 for transcriptome group 2; (Q) genus-level comparison of baseline and day 28 for transcriptome group 2; (R) genus-level comparison of day 28 and day 56 for transcriptome group 2; (S) global comparison of baseline and day 28 for transcriptome group 3; (T) global comparison of day 28 and day 56 for transcriptome group 3; (U) genus-level comparison of baseline and day 28 for transcriptome group 3; (V) genus-level comparison of day 28 and day 56 for transcriptome group 3.
Lactobacillus rhamnosus GG (LGG) transcript statistics and comparison to resident genera on day 28. (A) LGG genome coverage, abundance, and concordance across all subjects on day 28. Concordance was calculated using Kendall coefficient of concordance. (B) Correlations among LGG relative expression and resident genera on day 28. LGG relative expression is calculated for all genome-mapped transcripts. PCC, Pearson correlation coefficient.