ABSTRACT
Cholera continues to be a global threat, with high rates of morbidity and mortality. In 2011, a cholera outbreak occurred in Palawan, Philippines, affecting more than 500 people, and 20 individuals died. Vibrio cholerae O1 was confirmed as the etiological agent. Source attribution is critical in cholera outbreaks for proper management of the disease, as well as to control spread. In this study, three V. cholerae O1 isolates from a Philippines cholera outbreak were sequenced and their genomes analyzed to determine phylogenetic relatedness to V. cholerae O1 isolates from recent outbreaks of cholera elsewhere. The Philippines V. cholerae O1 isolates were determined to be V. cholerae O1 hybrid El Tor belonging to the seventh-pandemic clade. They clustered tightly, forming a monophyletic clade closely related to V. cholerae O1 hybrid El Tor from Asia and Africa. The isolates possess a unique multilocus variable-number tandem repeat analysis (MLVA) genotype (12-7-9-18-25 and 12-7-10-14-21) and lack SXT. In addition, they possess a novel 15-kb genomic island (GI-119) containing a predicted type I restriction-modification system. The CTXΦ-RS1 array of the Philippines isolates was similar to that of V. cholerae O1 MG116926, a hybrid El Tor strain isolated in Bangladesh in 1991. Overall, the data indicate that the Philippines V. cholerae O1 isolates are unique, differing from recent V. cholerae O1 isolates from Asia, Africa, and Haiti. Furthermore, the results of this study support the hypothesis that the Philippines isolates of V. cholerae O1 are indigenous and exist locally in the aquatic ecosystem of the Philippines.
IMPORTANCE
Genetic characterization and phylogenomics analysis of outbreak strains have proven to be critical for probing clonal relatedness to strains isolated in different geographical regions and over time. Recently, extensive genetic analyses of V. cholerae O1 strains isolated in different countries have been done. However, genome sequences of V. cholerae O1 isolates from the Philippines have not been available for epidemiological investigation. In this study, molecular typing and phylogenetic analysis of Vibrio cholerae isolated from both clinical and environmental samples in 2011 confirmed unique genetic features of the Philippines isolates, which are helpful to understand the global epidemiology of cholera.
INTRODUCTION
Cholera is a life-threatening diarrheal disease caused by Vibrio cholerae, a bacterium autochthonous to the aquatic environment. The disease is endemic in many developing countries of Asia, Africa, and South America. Furthermore, cholera poses a serious health risk for those residents of countries where cholera is endemic and also non-endemic countries where the public health infrastructure is compromised (1). The recent cholera epidemic in Haiti has drawn the attention of epidemiologists interested in identifying the origin and transmission of V. cholerae, since cholera had been reported to have been absent in that region previous to the current epidemic (2–4). Asiatic cholera has been reported to be endemic in the Ganges delta of Bangladesh and India, whereas the occurrence of cholera in countries where it is not endemic is usually attributed to imported cases, i.e., travel-associated dissemination of the bacteria. Molecular typing, genomic analysis, and epidemiological data enable identification of a probable source of an organism causing a given outbreak.
Traditionally, V. cholerae isolates have been classified serologically, based on the somatic O antigen, with >200 serogroups identified to date (5). However, only V. cholerae serogroups O1 and O139 have been linked to cholera pandemics. V. cholerae serogroup O1 has two biotypes, El Tor and classical, each showing biotype-specific phenotypic and genetic traits. Allelic variation is evident between classical and El Tor biotypes for genes encoding the major protein subunit of toxin coregulated pilus (tcpA), cholera toxin subunit B (ctxB), regulatory region for phage lysogeny (rstR), and hemolysin (hlyA). Moreover, the classical biotype lacks Vibrio seventh-pandemic islands (VSP-I and -II) and RS1 satellite phage (5, 6).
Historically, V. cholerae O1 is linked to seven distinct pandemics, of which the sixth, and presumably earlier ones, was caused by the classical biotype, while the ongoing seventh pandemic, which started in 1961 in Indonesia, is attributed to the El Tor biotype (1). Recently, genetic analysis of an archival intestinal specimen of a victim who died of cholera in 1849 during an outbreak in Philadelphia confirmed association of the classical biotype with the second cholera pandemic (7). Over the past two decades, V. cholerae O1 El Tor strains have undergone substantial genetic change, and new variants, including altered El Tor, with the potential to cause a more severe cholera have emerged (6, 8, 9). El Tor cholera occurred in the Philippines during late September 1961, with large outbreaks in several different provinces (10). In the following years, until 1969, cholera occurred frequently in the Philippines during the rainy season (11). During the last decade, outbreaks of cholera in the Philippines have occurred immediately after water-related disasters, i.e., floods and typhoons. Although cholera is not considered endemic to the Philippines, sudden increases in cholera cases were reported at times when sanitation and hygienic practices were disturbed due to a natural calamity. Recently, extensive genetic analyses of V. cholerae O1 strains isolated in different geographical locations were done, but molecular analysis of V. cholerae O1 isolates from the Philippines has not yet been accomplished.
In April 2011, a diarrheal outbreak occurred in Palawan, Philippines, and an epidemiological investigation determined V. cholerae to be the causative agent. In the study reported here, V. cholerae O1 clinical and environmental isolates from the region of the Philippines affected by cholera were subjected to serological, bacteriological, and whole-genome sequencing to determine the source of the outbreak. Comparative genomics was done to determine the phylogenetic relationship of these isolates with V. cholerae O1 strains currently circulating in different regions of the world.
RESULTS AND DISCUSSION
MLVA.
Multilocus variable-number tandem repeat analysis (MLVA) was used to discriminate V. cholerae isolates from various geographic locations and distinct populations within a single geographic cluster (12, 13). MLVA of the Philippines isolates revealed two genotypes: 12-7-9-18-25 and 12-7-10-14-21. Genotype 12-7-9-18-25 comprised the environmental isolate (PhRBD_VcEnv) and one of the clinical isolates (PhRBD_Vc311), while the other clinical isolate (PhRBD_Vc326) displayed the 12-7-10-14-21 genotype. The MLVA genotypes of the Philippines isolates did not match previously published MLVA genotypes of V. cholerae from different countries (3, 12, 14–16). However, MLVA is suitable mainly for outbreak investigations and spatiotemporal analysis of V. cholerae strains because of the relatively higher rate of mutation of the small-chromosome (Chr II) loci, which encompass the last two loci in the MLVA nomenclature. If only the three stable MLVA loci of the large chromosome (Chr I) are considered and the two variable loci of the small chromosome are ignored, the genotypes 12-7-9-X-X and 12-7-10-X-X continue to be unique for V. cholerae O1, compared with the profiles of V. cholerae O1 isolates from other countries. Interestingly, genotype 12-7-9-X-X matched that of a V. cholerae O139 strain isolated in 1992 in India. Overall, the MLVA genotypes of the Philippines isolates suggest that a distinct subpopulation of indigenous V. cholerae O1 most likely caused the outbreak of interest in this study.
Toxin gene cluster analysis.
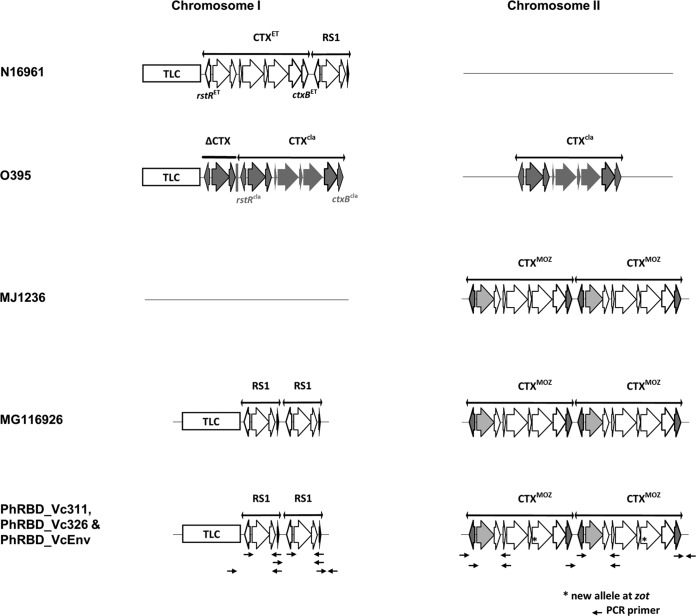
Pathogenicity of V. cholerae has been associated with production of cholera toxin, an enterotoxin encoded by genes carried by lysogenic CTXΦ. CTXΦ consists of two components, a “core” region (with cholera toxin subunit genes ctxA and ctxB and phage morphogenesis proteins) and an RS2 cluster (containing rstA, rstB, and rstR). Satellite phage RS1 is often found in toxigenic V. cholerae carrying an additional rstC gene and the entire RS2. The location of CTX prophage and its orientation can vary among strains. Therefore, it can be used as an indicator of both relatedness and evolution of V. cholerae strains. PCR assays, using a combination of phage-related and chromosome-specific primers, were performed to determine the presence of phage elements and RS1-CTX organization of the Philippines V. cholerae genomes. The three isolates were found to carry the rstC gene, indicating the presence of an RS1 element. In addition, further analysis by PCR confirmed the RS1 element to be present on Chr I (Fig. 1) and CTXΦ on Chr II (Fig. 1). The RS1 element contained V. cholerae El Tor biotype rstRET, but CTXΦ possessed V. cholerae classical-biotype ctxB and rstR, suggesting hybrid El Tor. Moreover, the Philippines isolates carried two copies of the RS1 element and TLC (toxin-linked cryptic plasmid) on Chr I and two copies of CTXΦ on Chr II. A similar RS1-CTXΦ array was identified previously in V. cholerae O1 MG116926 isolated in Bangladesh during 1991 (12, 17). Similar V. cholerae hybrid El Tor isolates have been associated with cholera in Bangladesh, India, Thailand, Vietnam, and Mozambique (17–20).
FIG 1 .
CTXΦ-RS1 array of Philippines V. cholerae isolates (PhRBD_Vc311, PhRBD_Vc326, and PhRBD_VcEnv) with El Tor (N16961), classical (O395), and hybrid El Tor (MJ1236 and MG116926) strains. Philippines isolates carry two copies of the RS1 element and TLC (toxin-linked cryptic plasmid) on the large chromosome (Chr I) and two copies of CTXΦ on the small chromosome (Chr II), similar to MG116926.
Shotgun genome sequence data complemented the RS1-CTX finding for the Philippines isolates, confirming a CTXΦ composition that included rstA, rstB, cep, orfU, ace, zot, ctxA, and ctxB. The deduced amino acid sequences of ctxB showed histidine at position 39 and threonine at position 68, identical to classical ctxB, while rstA contained partial polymorphisms of classical and El Tor hybrid rstA (Table 1). Four copies of heptamer repeats (TTTTGAT) were observed in the promoter-binding region of ctxAB (between zot and ctxA) in the Philippines V. cholerae genomes. These heptamer repeats (TTTTGAT) directly influence the affinity of ToxR binding and activation of the ctxAB promoter (21). The number of heptamer repeats was similar to those in V. cholerae El Tor (four) and altered El Tor (three or four). However, they differed from those in classical (seven) and Haitian (five) V. cholerae O1 strains (3, 22). The three Philippines strains shared a unique point mutation in zot (nonsynonymous; Arg to Cys, C to T at 1057 nucleotides). The genomic data confirmed the CTX prophage of Philippines isolates to be similar to that of V. cholerae hybrid El Tor strains, except in rstA and zot, which showed some unique polymorphisms. Therefore, it is concluded to be a variant of seventh-pandemic V. cholerae El Tor.
TABLE 1 .
Sites of nucleotide polymorphism in CTX prophages
| Strain | Prophagea | rstRa | Nucleotide at position in geneb |
No. of heptamers in zot-ctxA (1197~74)c | ctxB | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
rstA |
rstB |
zot |
||||||||||||||||||||
| 27 | 162 | 183 | 258 | 345 | 516 | 540 | 579 | 609 | 774 | 77-79 | 90 | 96 | 108 | 192 | 288 | 291 | 1057 | |||||
| N16961 | CTXET | ET | C | C | C | G | G | G | A | T | T | C | GTA | A | T | G | A | A | C | C | 4 | ctxB3 |
| O395 | CTXCL | CL | T | T | A | C | T | A | G | C | C | T | – | T | C | . | . | G | T | . | 7 | ctxB1 |
| MJ1236 | CTXHyb | CL | T | T | A | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 4 | ctxB1 |
| HCO1 | CTXHyb | CL | . | . | . | . | . | . | . | . | . | . | – | . | . | . | . | . | . | . | 5 | ctxB7 |
| PhRBD_Vc326 | CTXHyb | CL | T | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | 4 | ctxB1 |
| PhRBD_Vc311 | CTXHyb | CL | T | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | 4 | ctxB1 |
| PhRBD_VcEnv | CTXHyb | CL | T | T | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | 4 | ctxB1 |
ET, El Tor type; CL, classical type; Hyb, hybrid.
Dots indicate sequence identical to that of V. cholerae N16961; dashes indicate deletions.
Number of ToxR-binding site repeats.
Genomic islands.
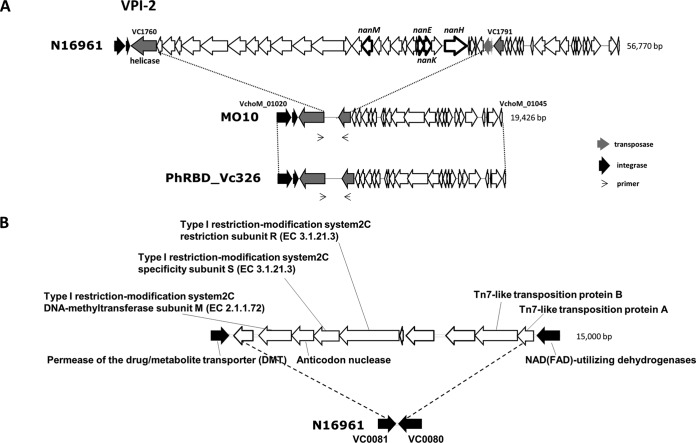
Vibrio pathogenicity island 1 (VPI-1) is a 41.2-kb genetic element encoding one of the major virulence factors, namely, toxin-coregulated pilus (TCP), that serves as a receptor for lysogenic CTXΦ and promotes bacterial colonization of the small intestine (23, 24). The Philippines isolates possess an intact VPI-1; however, they also harbor a truncated VPI-2 of about 16.5 kb, similar to several of the V. cholerae O139 strains isolated in India and Bangladesh (Fig. 2A) (25). Notably, among genes present on VPI-2, the gene for sialidase/neuraminidase (NanH), which has a putative role in pathogenesis, and the sialic acid catabolism gene cluster, which mediates utilization of sialic acid as the sole carbon source, are both missing in Philippines isolates. Moreover, the Philippines isolates were devoid of SXT, which confers resistance to several antibiotics (e.g., streptomycin, sulfamethoxazole/trimethoprim, and chloramphenicol [26]) and is suggested to have evolutionary significance for V. cholerae O139 and altered V. cholerae El Tor strains, reported over the last two decades (4). In a previous study, SXT was considered to be a major genetic element providing a selective advantage for altered V. cholerae El Tor strains in its global dissemination (4). Thus, the lack of SXT in the Philippines isolates may explain a limited distribution of these strains. The three Philippines isolates contained intact Vibrio seventh-pandemic island I (VSP-I; open reading frames [ORFs] VC0175 to VC0185) and Vibrio seventh-pandemic island II (VSP-II; ORFs VC0490 to VC0516), as well as several of the well-documented genomic islands of V. cholerae, including genomic island 1 (GI-1) to GI-10 and a single copy of GI-12 (8). VSP-I and VSP-II are believed to be conserved in the seventh-pandemic El Tor strains, yet the latter was reported to be polymorphic for ORF distribution (27). The presence of an intact VSP-II in the Philippines isolates is an interesting finding and is in contrast to the observation of recently isolated altered El Tor strains from Bangladesh, India, and Haiti carrying a variant VSP-II that lacks ORFs VC0495 to VC0512 (3).
FIG 2 .
(A) Philippines V. cholerae isolates (PhRBD_Vc311, PhRBD_Vc326, and PhRBD_VcEnv) and a V. cholerae O139 strain (MO10) possessing a major truncation in the VPI-2 region compared to seventh-pandemic prototype El Tor (N16961). (B) A new genomic island (GI-119) encoding a type I restriction-modification system is present between VC0081 and VC0080 in Philippines V. cholerae isolates.
Novel type I restriction-modification system.
The Philippines V. cholerae O1 isolates contained a unique mobile element (GI-119) with a predicted type I restriction-modification system (Fig. 2B). As shown in Fig. 2B, the GI is approximately 15 kb and contains 10 unique genes, including three components of a type I restriction-modification system (R, S, and M subunits), interrupted by inclusion of an anticodon nuclease. The type I restriction-modification system-specific genes in the Philippines strains are unique. However, the presence of an anticodon nuclease with a restriction-modification system has been described in Escherichia coli as an anti-T4 phage defense mechanism (28). Although the exact role of a type I restriction-modification system in the Philippines strains is unknown, presumably it could serve as a mechanism protecting against several vibriolytic phages and allowing the bacterium to thrive in an aquatic environment. It should be noted that this novel type I restriction-modification system may have been acquired horizontally by homologous recombination, a possible indication of an evolution of V. cholerae O1 in adapting to the local Philippines niche.
Mutation in housekeeping genes.
The Philippines isolates were found to contain a point mutation (Ser83Ile) in the gyrase gene (gyrA) but revealed no mutations in the topoisomerase gene (parC), as has been shown for V. cholerae O1 CIRS101 (Bangladesh, 2002) and CP1041 (Zambia, 2004). However, the same point mutation in gyrA and another mutation (Ser85Leu) in parC have been reported in V. cholerae isolates from Nigeria, Cameroon, Zimbabwe, Thailand, Bangladesh, and Haiti (3). The presence of the wild-type parC gene in the Philippines isolates is interesting, considering polymorphisms of currently circulating altered V. cholerae El Tor strains from different countries.
Phylogenomics.
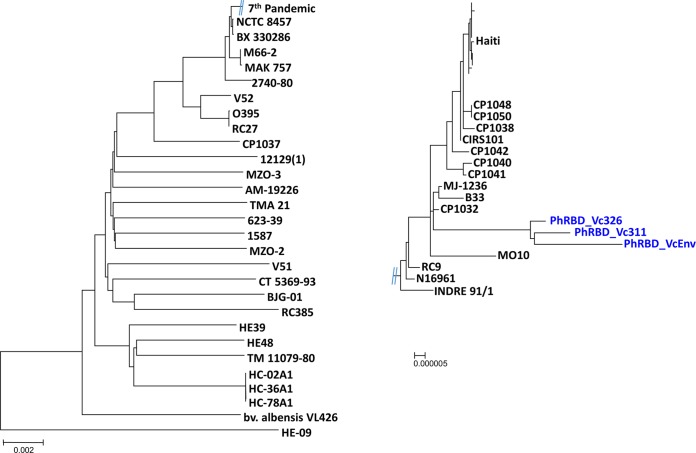
The phylogeny of the Philippines V. cholerae O1 isolates was determined by constructing a genome-relatedness neighbor-joining tree, using homologous alignment of 1,051 orthologs of protein-coding genes (~1,054,653 bp) of 78 V. cholerae genomes. The Philippines isolates clustered with seventh-pandemic V. cholerae El Tor and V. cholerae O139. However, the three isolates (PhRBD_Vc311, PhRBD_Vc326, and PhRBD_VcEnv) formed a distinct monophyletic clade (Fig. 3), distant from chronologically concurrent isolates from Thailand, Bangladesh, Zimbabwe, and Haiti. Moreover, the Philippines V. cholerae O1 strains showed close relatedness with hybrid V. cholerae El Tor strains from Mexico (CP1032, isolated in 1991), Mozambique (B33, isolated in 2004), and Bangladesh (MJ-1236, isolated in 1994) and a V. cholerae O139 strain from India (MO10, isolated in 1992). Interestingly, the time of isolation of these hybrid strains was different from that of the Philippines strains. In the phylogenetic tree, the monophyletic clade of Philippines isolates was positioned between concurrent altered V. cholerae El Tor strains and seventh-pandemic prototype V. cholerae El Tor strains. The Philippines isolates were not identical, and the environmental strain (PhRBD_VcEnv) showed highest homology with clinical strains (PhRBD_Vc311 and PhRBD_Vc326), confirming the environment as the reservoir of V. cholerae. Tight clustering of the Philippines strains in a monophyletic clade suggests that these strains have conserved genetic features and can be assumed to be restricted to the Philippines aquatic ecosystem, the probable source of the outbreak in 2011.
FIG 3 .
Neighbor-joining trees showing phylogenetic relationships of 78 V. cholerae genomes based on 1,051 orthologs of protein-coding genes (~1,054,653 bp). Philippines V. cholerae O1 strains are in blue, showing a tight clustering in a monophyletic clade.
Conclusions.
The Philippines V. cholerae strains isolated during an outbreak of cholera in 2011 belong to the seventh-pandemic clade of V. cholerae O1 and O139 serogroups. The genome sequence and organization of CTXΦ and RS1 indicate that the strains are hybrid V. cholerae El Tor lacking SXT. Although the Philippines strains showed relatedness to previously isolated hybrid V. cholerae El Tor strains from Mexico, Mozambique, and Bangladesh, they possess a novel type I restriction-modification system and truncated VPI-2. Overall, the genomic analyses clearly indicate that the Philippines isolates are novel V. cholerae hybrid El Tor strains, perhaps strains that evolved in the Philippines. Evolution of V. cholerae is very likely occurring not only in regions where cholera is endemic but also in areas where it is not, like the Philippines. In conclusion, the data provided here may be helpful in determining the epidemiology of cholera and the evolution of V. cholerae in regions where cholera is not endemic.
MATERIALS AND METHODS
Bacterial isolation, identification, and classification.
The National Epidemiology Center (NEC), Philippines Department of Health, received a report from the Center for Health Development, Region IV-B, of 90 diarrhea cases, with 15 deaths, from the municipality of Bataraza, Palawan, during the month of March 2011. An NEC team, dispatched to the area in April 2011, identified 562 suspected cholera cases, of which 383 (68%) were among an indigenous tribe of Palawan. Twenty patients suffering from cholera subsequently died (case fatality rate [CFR] = 3.6%). Samples were sent to the Research Institute for Tropical Medicine, Alabang, Philippines, for isolation and identification of the etiological agent. Ten samples from suspected cases were identified as containing V. cholerae O1 Ogawa El Tor, and 27 additional samples were identified as containing Aeromonas spp. In addition, 37 water samples tested positive for Escherichia coli, Vibrio, and Aeromonas by microbiological and serological procedures.
A single environmental and two clinical isolates of presumptive V. cholerae O1 were inoculated into Mueller-Hinton broth and incubated at 37°C overnight (29). Identification was accomplished using the BBL Crystal ID system (Becton, Dickinson) following incubation for 18 to 20 h at 37°C (30). The Crystal ID cartridges were read manually, and the data were entered using Crystal ID software, confirming identification to genus and species levels. DNA was extracted from pure cultures using the Qiagen genomic DNA kit and quantified using a NanoDrop spectrophotometer. The three putative V. cholerae strains, PhRBD_Vc311, PhRBD_Vc326, and PhRBD_VcEnv, were confirmed as V. cholerae based on results obtained using the BBL Crystal enteric/nonfermenter ID system. Each of the three strains metabolized mannose, sucrose, and mannitol as sole carbon sources.
MLVA.
Multilocus variable-number tandem repeat analysis (MLVA) was performed using PCR conditions and primers to amplify five targeted MLVA loci for V. cholerae, namely, VC0147, VC0436-7 (intergenic), VC1650, VCA0171, and VCA0283, as described previously (12). The purified PCR products were sequenced in both directions using a BigDye cycle sequencing kit (Applied Biosystems), and sequencing was performed on an ABI 3770 automatic sequencer according to the manufacturer’s instructions. The number of repeats was determined for each locus, and the MLVA genotypes were assigned by sequentially combining numbers of repeat units in order for five loci.
Genome sequencing.
Isolates were sent to St. Luke’s Medical Center for whole-genome sequencing. Shotgun whole-genome sequencing was performed using Roche GS Junior and Illumina MiSeq, and the output from both technologies was combined for downstream bioinformatic analysis. Roche GS Junior sequencing was accomplished employing protocols developed by Roche (31). Briefly, library DNA was prepared from 1 µg of genomic DNA by physically shearing the DNA by nebulization. The ends of the fragments were repaired employing DNA polymerase (Klenow fragment) and Taq DNA polymerase, and terminal adenosine residues were added to fragments using polynucleotide kinase (PNK). Roche RL adapters containing a terminal thymidine were ligated to the fragments, according to the manufacturer’s protocol. Library quality was determined using Flashgel (Invitrogen) and quantified with fluorometry (Promega QuantiFluor). DNA fragments were annealed to Roche capture beads, emulsified using a Turrax tube drive, distributed onto a 96-tube PCR plate, and amplified using 50 cycles. Beads with amplified DNA were recovered from the emulsion and purified. Beads were packed into a picotiter plate along with the reaction mixture and packing beads, and the plates were loaded into the GS Junior instrument. Sequencing was performed using 200 nucleotide cycles.
Illumina MiSeq sequencing was performed using library DNA prepared from 50 ng of DNA and the Nextera DNA library kit, according to the manufacturer’s protocol. Multiple strains were sequenced simultaneously using the Illumina Multiplex kit. Following amplification and purification of library DNA, libraries were diluted to 6.0 pM, and equal volumes were combined prior to addition to the MiSeq sequencing cartridge. Paired-end, 150-bp reads were generated using the MiSeq and standard protocols (31).
Sequence analysis.
The raw sequencing reads, from both the Roche GS Junior (SFF file) and Illumina MiSeq (FASTA file), were assembled into contigs using GS de novo Assembler software (version 2.7; Roche). Assembled contigs were analyzed using NCBI BLAST to confirm V. cholerae species identification. Annotation of assembled contigs was done using the RAST Annotation Server (8) and the annotation service of the Institute for Genome Sciences (Baltimore, MD). All completed genome projects for Vibrio cholerae in the NCBI database were utilized as reference strains for mapping of reads from the Philippines strains, using GS Reference Mapper software (version 2.7; Roche).
Variant analysis.
Detection of single-nucleotide polymorphisms (SNPs) and structural variants was performed using an in-house pipeline consisting of mapping shotgun sequencing reads from the Philippines isolates to V. cholerae El Tor reference strain N16961 and Roche gsMapper (version 2.7). The BAM file output was sorted and indexed, and reads were aligned using mpileup (SAMTools) to generate variant call files (VCF). VCF were converted to Annovar format files, and the location and type of variant were determined using Annovar and the annotated V. cholerae N16961 genome from NCBI. Comparison of the completed V. cholerae genomes with V. cholerae N16961 was done by in silico read generation using MetaSim, and simulated reads were mapped and processed as described above. In-house scripts were compiled and compared to Annovar output to generate a list of common variants.
Genetic analysis of CTXΦ and flanking regions.
The orientation of CTXΦ and its flanking regions was performed using primers and conditions described previously (3, 12, 32). Sequencing of PCR products was accomplished after purification of the DNA fragments followed by Sanger sequencing (First Base, Singapore).
Comparative genomics and phylogeny.
Genome-to-genome comparison was performed using methods described previously (8). Genomic islands (GIs) were defined as a continuous array of five or more coding sequences (CDSs) discontinuously distributed among genomes of test strains. Identified GIs were annotated using a BLASTP search of member CDSs against the GenBank NR database. Regions orthologous to V. cholerae N16961 were identified by comparisons based on similarity (95%), and the resultant 1,051 orthologs were used to generate a phylogenetic tree. The set of orthologous regions for each CDS of a reference genome was identified according to nucleotide similarity and aligned using CLUSTALW2. The resultant multiple alignments were concatenated to form genome scale alignments, which were then used to generate the neighbor-joining phylogenetic trees (33).
Nucleotide sequence accession number.
This whole-genome shotgun project has been deposited at DDBJ/EMBL/GenBank under accession no. AWWD00000000, AWWE00000000, and AWWF000000 (BioProject PRJNA218554).
ACKNOWLEDGMENTS
We acknowledge the valuable assistance of Lorenz Loyola with sequencing and members of the National Epidemiology Center of the Department of Health in the Philippines: Rowena J. Capistrano, Maria Elaine Joy C. Villareal, Juan M. Lopez, Faith Alberto, and Manuel C. Mapue II.
This work was supported in part by the National Institutes of Health grant no. 1 R01 A139129-01 and by Distinguished Scholar in Oceans and Human Health NOAA grant no. SO660009 for the Advanced Study Institute for Earth System Prediction. Additional funding was provided by Tahoe Research Initiative, Incline Village, NV, and St. Luke’s Medical Center, Quezon City, Philippines.
Footnotes
Citation Klinzing DC, Choi SY, Hasan NA, Matias RR, Tayag E, Geronimo J, Skowronski E, Rashed SM, Kawashima K, Rosenzweig CN, Gibbons HS, Torres BC, Liles V, Alfon AC, Juan ML, Natividad FF, Cebula TA, Colwell RR. 2015. Hybrid V. cholerae El Tor lacking SXT identified as the cause of a cholera outbreak in the Philippines. mBio 6(2):e00047-15. doi:10.1128/mBio.00047-15
REFERENCES
- 1.Sack DA, Sack RB, Nair GB, Siddique AK. 2004. Cholera. Lancet 363:223–233. doi: 10.1016/S0140-6736(03)15328-7. [DOI] [PubMed] [Google Scholar]
- 2.Chin CS, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, Paxinos EE, Yamaichi Y, Calderwood SB, Mekalanos JJ, Schadt EE, Waldor MK. 2011. The origin of the Haitian cholera outbreak strain. N Engl J Med 364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, Hine E, Su Q, Tallon LJ, Prosper JB, Furth K, Hoq MM, Li H, Fraser-Liggett CM, Cravioto A, Huq A, Ravel J, Cebula TA, Colwell RR. 2012. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc Natl Acad Sci U S A 109:E2010–E2017. doi: 10.1073/pnas.1207359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, Niyogi SK, Kim EJ, Ramamurthy T, Chun J, Wood JL, Clemens JD, Czerkinsky C, Nair GB, Holmgren J, Parkhill J, Dougan G. 2011. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaper JB, Morris JG Jr, Levine MM. 1995. Cholera. Clin Microbiol Rev 8:48–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Safa A, Nair GB, Kong RY. 2010. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol 18:46–54. doi: 10.1016/j.tim.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Devault AM, Golding GB, Waglechner N, Enk JM, Kuch M, Tien JH, Shi M, Fisman DN, Dhody AN, Forrest S, Bos KI, Earn DJ, Holmes EC, Poinar HN. 2014. Second-pandemic strain of Vibrio cholerae from the Philadelphia cholera outbreak of 1849. N Engl J Med 370:334–340. doi: 10.1056/NEJMoa1308663. [DOI] [PubMed] [Google Scholar]
- 8.Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, Taviani E, Jeon YS, Kim DW, Brettin TS, Bruce DC, Challacombe JF, Detter JC, Han CS, Munk AC, Chertkov O, Meincke L, Saunders E, Walters RA, Huq A, Nair GB, Colwell RR. 2009. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A 106:15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siddique AK, Nair GB, Alam M, Sack DA, Huq A, Nizam A, Longini IM Jr., Qadri F, Faruque SM, Colwell RR, Ahmed S, Iqbal A, Bhuiyan NA, Sack RB. 2010. El Tor cholera with severe disease: a new threat to Asia and beyond. Epidemiol Infect 138:347–352. doi: 10.1017/S0950268809990550. [DOI] [PubMed] [Google Scholar]
- 10.Dizon JJ, Alvero MG, Joseph PR, Tamayo JF, Mosley WH, Henderson DA. 1965. Studies of cholera El Tor in the Philippines. I. Characteristics of cholera El Tor in Negros Occidental Province, November 1961 to September 1962. Bull World Health Organ 33:627–636. [PMC free article] [PubMed] [Google Scholar]
- 11.Kobari K, Takakura I, Nakatomi M, Sogame S, Uylangco C. 1970. Antibiotic-resistant strains of E1 Tor vibrio in the Philippines and the use of furalazine for chemotherapy. Bull World Health Organ 43:365–371. [PMC free article] [PubMed] [Google Scholar]
- 12.Choi SY, Lee JH, Jeon YS, Lee HR, Kim EJ, Ansaruzzaman M, Bhuiyan NA, Endtz HP, Niyogi SK, Sarkar BL, Nair GB, Nguyen BM, Hien NT, Czerkinsky C, Clemens JD, Chun J, Kim DW. 2010. Multilocus variable-number tandem repeat analysis of Vibrio cholerae O1 El Tor strains harbouring classical toxin B. J Med Microbiol 59:763–769. doi: 10.1099/jmm.0.017939-0. [DOI] [PubMed] [Google Scholar]
- 13.Mohamed AA, Oundo J, Kariuki SM, Boga HI, Sharif SK, Akhwale W, Omolo J, Amwayi AS, Mutonga D, Kareko D, Njeru M, Li S, Breiman RF, Stine OC. 2012. Molecular epidemiology of geographically dispersed Vibrio cholerae, Kenya, January 2009–May 2010. Emerg Infect Dis 18:925–931. doi: 10.3201/eid1806.111774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh R, Nair GB, Tang L, Morris JG, Sharma NC, Ballal M, Garg P, Ramamurthy T, Stine OC. 2008. Epidemiological study of Vibrio cholerae using variable number of tandem repeats. FEMS Microbiol Lett 288:196–201. doi: 10.1111/j.1574-6968.2008.01352.x. [DOI] [PubMed] [Google Scholar]
- 15.Lam C, Octavia S, Reeves PR, Lan R. 2012. Multi-locus variable number tandem repeat analysis of 7th pandemic Vibrio cholerae. BMC Microbiol 12:82. doi: 10.1186/1471-2180-1112-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shakya G, Kim DW, Clemens JD, Malla S, Upadhyaya BP, Dumre SP, Shrestha SD, Adhikari S, Sharma S, Rijal N, Shrestha SK, Mason C, Kansakar P. 2012. Phenotypic and genetic characterization of Vibrio cholerae O1 clinical isolates collected through national antimicrobial resistance surveillance network in Nepal. World J Microbiol Biotechnol 28:2671–2678. doi: 10.1007/s11274-012-1077-3. [DOI] [PubMed] [Google Scholar]
- 17.Lee JH, Choi SY, Jeon YS, Lee HR, Kim EJ, Nguyen BM, Hien NT, Ansaruzzaman M, Islam MS, Bhuiyan NA, Niyogi SK, Sarkar BL, Nair GB, Kim DS, Lopez AL, Czerkinsky C, Clemens JD, Chun J, Kim DW. 2009. Classification of hybrid and altered Vibrio cholerae strains by CTX prophage and RS1 element structure. J Microbiol 47:783–788. doi: 10.1007/s12275-009-0292-6. [DOI] [PubMed] [Google Scholar]
- 18.Chatterjee S, Ghosh K, Raychoudhuri A, Chowdhury G, Bhattacharya MK, Mukhopadhyay AK, Ramamurthy T, Bhattacharya SK, Klose KE, Nandy RK. 2009. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J Clin Microbiol 47:1087–1095. doi: 10.1128/JCM.02026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nair GB, Faruque SM, Bhuiyan NA, Kamruzzaman M, Siddique AK, Sack DA. 2002. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J Clin Microbiol 40:3296–3299. doi: 10.1128/JCM.40.9.3296-3299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Na-Ubol M, Srimanote P, Chongsa-Nguan M, Indrawattana N, Sookrung N, Tapchaisri P, Yamazaki S, Bodhidatta L, Eampokalap B, Kurazono H, Hayashi H, Nair GB, Takeda Y, Chaicumpa W. 2011. Hybrid & El Tor variant biotypes of Vibrio cholerae O1 in Thailand. Indian J Med Res 133:387–394. [PMC free article] [PubMed] [Google Scholar]
- 21.Pfau JD, Taylor RK. 1996. Genetic footprint on the ToxR-binding site in the promoter for cholera toxin. Mol Microbiol 20:213–222. doi: 10.1111/j.1365-2958.1996.tb02502.x. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh P, Naha A, Pazhani GP, Ramamurthy T, Mukhopadhyay AK. 2014. Genetic traits of Vibrio cholerae O1 Haitian isolates that are absent in contemporary strains from Kolkata, India. PLoS One 9:e112973. doi: 10.1371/journal.pone.0112973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyd EF, Moyer KE, Shi L, Waldor MK. 2000. Infectious CTXPhi and the vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect Immun 68:1507–1513. doi: 10.1128/IAI.68.3.1507-1513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karaolis DK, Johnson JA, Bailey CC, Boedeker EC, Kaper JB, Reeves PR. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci U S A 95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jermyn WS, Boyd EF. 2002. Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology 148:3681–3693. [DOI] [PubMed] [Google Scholar]
- 26.Hochhut B, Lotfi Y, Mazel D, Faruque SM, Woodgate R, Waldor MK. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob Agents Chemother 45:2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taviani E, Grim CJ, Choi J, Chun J, Haley B, Hasan NA, Huq A, Colwell RR. 2010. Discovery of novel Vibrio cholerae VSP-II genomic islands using comparative genomic analysis. FEMS Microbiol Lett 308:130–137. doi: 10.1111/j.1574-6968.2010.02008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaufmann G. 2000. Anticodon nucleases. Trends Biochem Sci 25:70–74. doi: 10.1016/S0968-0004(99)01525-X. [DOI] [PubMed] [Google Scholar]
- 29.Gangarosa EJ, Bennett JV, Boring JR III. 1967. Differentiation between Vibrio cholerae and Vibrio cholerae biotype El Tor by the polymyxin B disc test: comparative results with TCBS, Monsur’s, Mueller-Hinton and nutrient agar media. Bull World Health Organ 36:987–990. [PMC free article] [PubMed] [Google Scholar]
- 30.O’Hara CM, Westbrook GL, Miller JM. 1997. Evaluation of Vitek GNI+ and Becton Dickinson Microbiology Systems Crystal E/NF identification systems for identification of members of the family Enterobacteriaceae and other gram-negative, glucose-fermenting and non-glucose-fermenting bacilli. J Clin Microbiol 35:3269–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. 2012. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 32.Nusrin S, Khan GY, Bhuiyan NA, Ansaruzzaman M, Hossain MA, Safa A, Khan R, Faruque SM, Sack DA, Hamabata T, Takeda Y, Nair GB. 2004. Diverse CTX phages among toxigenic Vibrio cholerae O1 and O139 strains isolated between 1994 and 2002 in an area where cholera is endemic in Bangladesh. J Clin Microbiol 42:5854–5856. doi: 10.1128/JCM.42.12.5854-5856.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]