ABSTRACT
Spiroplasmas are helical and motile members of a cell wall-less eubacterial group called Mollicutes. Although all spiroplasmas are associated with arthropods, they exhibit great diversity with respect to both their modes of transmission and their effects on their hosts; ranging from horizontally transmitted pathogens and commensals to endosymbionts that are transmitted transovarially (i.e., from mother to offspring). Here we provide the first genome sequence, along with proteomic validation, of an endosymbiotic inherited Spiroplasma bacterium, the Spiroplasma poulsonii MSRO strain harbored by Drosophila melanogaster. Comparison of the genome content of S. poulsonii with that of horizontally transmitted spiroplasmas indicates that S. poulsonii has lost many metabolic pathways and transporters, demonstrating a high level of interdependence with its insect host. Consistent with genome analysis, experimental studies showed that S. poulsonii metabolizes glucose but not trehalose. Notably, trehalose is more abundant than glucose in Drosophila hemolymph, and the inability to metabolize trehalose may prevent S. poulsonii from overproliferating. Our study identifies putative virulence genes, notably, those for a chitinase, the H2O2-producing glycerol-3-phosphate oxidase, and enzymes involved in the synthesis of the eukaryote-toxic lipid cardiolipin. S. poulsonii also expresses on the cell membrane one functional adhesion-related protein and two divergent spiralin proteins that have been implicated in insect cell invasion in other spiroplasmas. These lipoproteins may be involved in the colonization of the Drosophila germ line, ensuring S. poulsonii vertical transmission. The S. poulsonii genome is a valuable resource to explore the mechanisms of male killing and symbiont-mediated protection, two cardinal features of many facultative endosymbionts.
IMPORTANCE
Most insect species, including important disease vectors and crop pests, harbor vertically transmitted endosymbiotic bacteria. These endosymbionts play key roles in their hosts’ fitness, including protecting them against natural enemies and manipulating their reproduction in ways that increase the frequency of symbiont infection. Little is known about the molecular mechanisms that underlie these processes. Here, we provide the first genome draft of a vertically transmitted male-killing Spiroplasma bacterium, the S. poulsonii MSRO strain harbored by D. melanogaster. Analysis of the S. poulsonii genome was complemented by proteomics and ex vivo metabolic experiments. Our results indicate that S. poulsonii has reduced metabolic capabilities and expresses divergent membrane lipoproteins and potential virulence factors that likely participate in Spiroplasma-host interactions. This work fills a gap in our knowledge of insect endosymbionts and provides tools with which to decipher the interaction between Spiroplasma bacteria and their well-characterized host D. melanogaster, which is emerging as a model of endosymbiosis.
INTRODUCTION
Spiroplasma bacteria belong to the class Mollicutes, which includes cell wall-less bacteria related to Firmicutes (1, 2). Initially discovered as the causative agents of important plant and insect diseases (3–5), Spiroplasma bacteria are widely associated with arthropods, and an estimated 5 to 10% of all insect species harbor these bacteria (6, 7). Spiroplasmas are especially diverse with respect to their modes of transmission: Spiroplasma lineages are either endosymbiotic, exhibiting transovarial vertical transmission from mother to offspring, or “infectious” in the classical sense, being transmitted horizontally between hosts through plants. Endosymbiotic spiroplasmas are found in the insect hemolymph, and the only intracellular stage in their cycle reported to date is during vertical transmission, when they are found in the cytoplasm of the nascent oocyte and in the early syncytial embryo. Nonendosymbiotic spiroplasmas are either insect commensals that are restricted to the gut lumen or “infectious” in that they invade hemolymph and organs, including the salivary glands, muscles, nerves, and connective tissue (5, 8, 9). The “infectious” category includes pathogens of shrimp and crabs that pose serious problems in the aquaculture industry, as well as pathogens of bees, such as Spiroplasma melliferum and S. apis (8–12). In addition, three Spiroplasma species (S. citri, S. kunkelii, and S. phoeniceum) are known plant pathogens that proliferate in the plant phloem and are vectored by phloem-feeding insects (3–5). Some, notably S. citri, can be grown in vitro and are amenable to genetic studies, unlike maternally transmitted strains, which are highly fastidious. To date, the genomes of 11 Spiroplasma species have been totally or partially sequenced, including two plant pathogens, S. citri and S. kunkelii (13, 14); two bee pathogens, S. melliferum and S. apis (10–12); commensals of deerflies and syrphid flies, S. chrysopicola and S. syrphidicola, respectively (15); and four commensals of mosquitoes S. taiwanense, S. culicicola, S. diminutum, and S. sabaudiense (16, 17). Spiroplasma genomes are small, ranging from 1 to 2 Mb, and AT rich. To date, no vertically transmitted Spiroplasma species have been sequenced. This represents the last major lineage of vertically transmitted insect endosymbiont to have its genome sequenced. Sequencing of the S. poulsonii genome required overcoming challenges associated with the sequencing and alignment of genomes that are highly AT rich and overrun with viruses and other mobile elements. Here we report the draft genome sequence of the Drosophila melanogaster endosymbiont S. poulsonii MSRO (here referred to as S. poulsonii).
Together with Wolbachia bacteria, Spiroplasma bacteria are the only known natural inherited symbionts of Drosophila flies (18). At least 16 Drosophila species have been found to be infected with inherited Spiroplasma spp. (19). S. poulsonii MSRO was isolated from an infected D. melanogaster female collected in Uganda (20). S. poulsonii resides primarily in the hemolymph of D. melanogaster and has only been observed inside cells as it is being transmitted vertically in the oocyte cytoplasm. Vertical transmission occurs by subversion of the yolk transport and uptake pathway (21). Although horizontal transmission of S. poulsonii does occur over evolutionary time scales, it is rare (22). Within natural populations of D. melanogaster, S. poulsonii prevalence is variable but generally less than 5% (20, 23). In other species of Drosophila, related S. poulsonii strains are found over a broad prevalence range, in some cases as high as 80% (19, 24). S. poulsonii MSRO affects its D. melanogaster host in two striking ways. Like a number of other inherited endosymbionts, it manipulates host reproduction in order to increase the frequency of infected female hosts. It does this by killing males at the embryonic stage (23). Male killing has evolved independently in a number of inherited symbionts, including some strains of Wolbachia and Rickettsia (25, 26). A number of inherited Spiroplasma strains kill male flies, beetles, butterflies, and aphids (19, 27–29). The mechanistic basis of male killing induced by Spiroplasma bacteria, or by any inherited symbionts, remains largely enigmatic. This phenotype has captured the imagination of many biologists, as it is quite incredible that the symbiont is able to specifically recognize and kill one sex of its host at such an early stage. Notably, recent sequencing of the genome of butterfly male-killing Wolbachia sp. strain wBol1-b has revealed some fast-evolving sequences and horizontal gene transfer associated with viral sequences not present in other, closely related, genomes of non-male-killing Wolbachia strains (30).
Recently, it has also been established that several Spiroplasma strains, including a number of S. poulsonii strains, can protect their hosts against eukaryotic parasites. For example, the S. poulsonii strain that infects Drosophila neotestacea protects its host against nematode infection (24) and the strains infecting D. hydei (SPHY) and D. melanogaster (MSRO) protect their hosts against parasitoid wasps (31, 32). There is much interest in determining how symbionts confer protection, including understanding how symbionts target natural enemies without causing collateral damage in their hosts. In the case of S. poulsonii MSRO, there appear to be only minor effects on infected female D. melanogaster longevity and rates of egg laying. Intriguingly, old symbiont-infected females show an apparent lack of coordination and tremors (33).
In this study, we purified S. poulsonii strain MSRO from D. melanogaster hemolymph and sequenced its genome. We provide the first sequenced genome of an inherited Spiroplasma bacterium. By comparing the S. poulsonii genome content with the genome contents of closely related horizontally transmitted (i.e., nonendosymbiotic) Spiroplasma bacteria, our study provides insights into the molecular mechanisms involved in the endosymbiotic mode of life and provides an important resource for deciphering the mechanism of male killing and symbiont-mediated protection.
RESULTS AND DISCUSSION
Sequencing and assembly of the genome of S. poulsonii strain MSRO.
Male-killing S. poulsonii strain MSRO was initially isolated from an infected D. melanogaster female collected in Uganda (20) and subsequently transferred to and maintained in an Oregon-R wild-type strain (34). Although a strain of S. poulsonii (DW-1) infecting Drosophila willistoni was once cultivated in cell-free medium (35), it was subsequently lost. Currently, there is no cell-free culture of endosymbiotic Spiroplasma bacteria. Instead, bacterial cells were obtained by manual collection of hemolymph from infected female flies with a microinjector, followed by filtration through a 0.45-µm-pore-size filter, which is expected to block host cells and other possible bacterial contaminants but not Spiroplasma bacteria (36). Three micrograms of pure DNA was extracted by the classical phenol-chloroform DNA extraction technique.
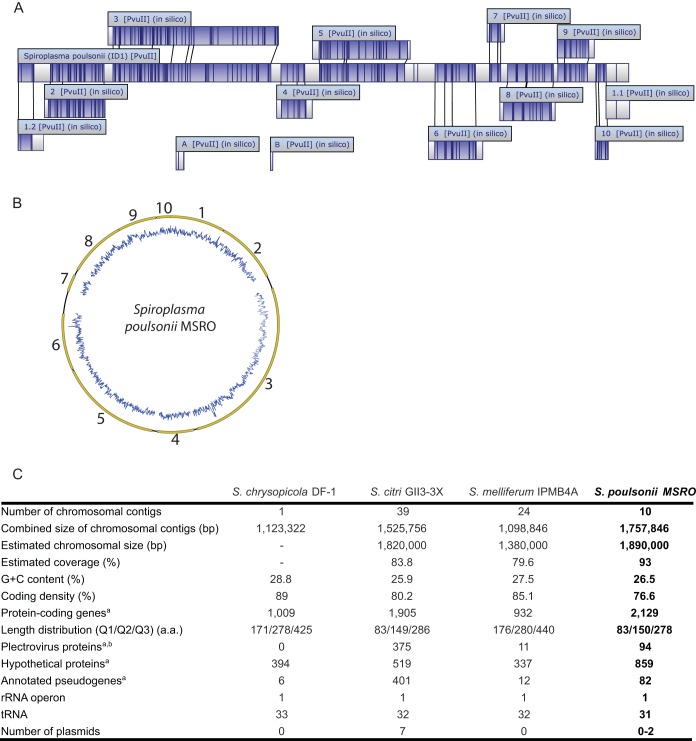
The sequence generated by a PacBio RSII smart cell sequencing run produced an assembly of 12 contigs. To determine the total genome length and to position the 12 contigs, we performed an in silico alignment with optical mapping (OpGen) technology. This allowed us to obtain a schematic view of the genome and place contigs 1 to 10 along a single circular chromosome. The presence of numerous viral sequences was the main obstacle preventing a complete genome assembly. Contigs 1 to 10 covered ~93% (1.76 Mb) of the ~1.9-Mb S. poulsonii chromosome length predicted by optical mapping (Fig. 1). The estimated size of the S. poulsonii chromosome is similar to the estimated genome size of S. citri strain gii3, 1.82 Mb (14), and larger than those of other spiroplasmas of the same clade, 1.38 and 1.55 Mb for S. melliferum IPMB4A (11) and S. kunkelii (37), respectively. Nevertheless, it is significantly smaller than the 2-Mb size estimated by pulsed-field gel electrophoresis for S. poulsonii strain DW-1 (38). Variation in the number of prophage sequences is thought to be partially responsible for the difference in chromosome size between these two S. poulsonii strains. It is notable that S. citri and related genomes contain numerous viral sequences and range from 1.6 to 1.9 Mb (39–41).
FIG 1 .
Draft assembly of the S. poulsonii MSRO genome. (A) Locations of the 12 contigs on the S. poulsonii chromosome as determined by optical mapping with MapSolver software (OpGen). Contigs A and B could not be placed on the chromosome. (B) Schematic representation of the S. poulsonii chromosome. Contigs (in yellow) were placed in order on the basis of the in silico alignment by optical mapping technology. The estimated length of the chromosome is ~1.89 Mb. GC content in depicted in blue. (C) Genome assembly statistics. The table shown was modified from reference 15. The superscript letter a indicates that for S. chrysopicola, S. melliferum, and S. poulsonii, putative pseudogenes were annotated with the “/pseudo” tag in gene feature as suggested by the NCBI GenBank guidelines and were not included in the total number of protein-coding genes. For S. citri, putative pseudogenes were annotated by adding the term “truncated” in the CDS product description field and were included in the total number of protein-coding genes. The superscript letter b indicates that most of the plectrovirus-related regions were excluded from the S. melliferum IPMB4A assembly because of unresolvable polymorphism, resulting in a lower number of plectroviral genes. The genome of S. chrysopicola does not contain any identifiable plectroviral fragments; this lineage was likely to have diverged prior to the plectroviral invasion of the common ancestor of S. citri, S. melliferum, and S. poulsonii. a.a., amino acids.
The draft S. poulsonii genome has a G+C content of 26.6%, which is in agreement with a previous determination done with S. poulsonii DW-1 (38). The G+C content in S. poulsonii is similar to that observed in the other spiroplasmas belonging to the same clade (25.9 to 27.5%). G+C content variation in spiroplasmas from the Citri clade is usually attributed to the proportion of prophage sequences, which are AT biased. In general, insect bacterial symbiont genomes tend to be highly AT biased.
In addition to the 10 chromosomal contigs, two small contigs, A and B, could not be placed on the chromosome by optical mapping (Fig. 1A). These contigs correspond either to extrachromosomal DNA (plasmid) or to chromosomal sequences that are too small to be placed on the chromosome by the MapSolver software. Spiroplasma species that are members of the Citri clade, S. melliferum KC3 and S. citri GII3-3X (but not S. melliferum IPMB4A), contain plasmids that often carry potential virulence factors (10, 11, 14). The fact that contigs A and B contain several genes that are found in plasmids in other Spiroplasma species (42), like Soj and those for several ARPs (adhesion-related proteins), suggests that they could also be extrachromosomal DNA sequences. However, the facts that plasmids have not previously been found in Spiroplasma group IV (to which S. poulsonii belongs) and that we were not able to circularize contigs A and B suggest that these contigs may be part of the S. poulsonii chromosome.
Genome rearrangement and virus sequence.
Spiroplasma genomes from the Citri clade contain a vast amount of plectroviral sequences; with roughly 20% of their chromosomes being of viral origin (10, 13, 14). Comparative genomic evidence suggests that viral invasion took place in the common ancestor of the Citri clade, since genomes from the Chrysopicola clade do not contain any viral sequences (15). Repetitive viral sequences are thought to increase rates of horizontal gene transfer and genome rearrangement. In line with this, Spiroplasma species that have genomes with few or no viral sequences show almost no genome rearrangements compared with each other (15, 16). In the S. poulsonii genome, we found 94 predicted coding gene sequences corresponding to viral sequences from the Plectrovirus SpV1 family (SpV1-R8A2, SpV1-C74, and SVTS2). Previous studies have revealed the presence of SpV3-like virus sequences in various Spiroplasma species infecting Drosophila flies. As the genome of an SpV3-like virus has not been sequenced, it could not be annotated in the S. poulsonii genome (43). The lengths of the viral sequences found in the S. poulsonii chromosome range from a few hundred base pairs to more than 3,000 bp. In addition, the S. poulsonii genome contains about 235 related genes encoding four different families of transposases, which are generally presumed to be involved in catalyzing integration and excision of DNA sequences. As previously reported for S. melliferum IPMB4A (11), a high level of gene decay and chromosomal rearrangements was observed in the S. poulsonii genome compared to genomes of S. citri and S. melliferum (see Fig. S1 in the supplemental material). This is likely a consequence of abundant viral sequences. Proliferation of viruses and other mobile elements is a common feature of the early evolution of insect symbiont genomes (44), and it should be noted that Wolbachia, the other endosymbiont of Drosophila, also harbors multiple virus and mobile-element sequences that are also thought to facilitate genome rearrangements (30, 45).
Horizontal transfer of genes can occur as a result of viral integration. It has been demonstrated that species-specific regions of the genomes of S. citri and S. melliferum were flanked by viral sequences, suggesting that they originated by horizontal transfer (15). For instance, several genes encoding transmembrane proteins in S. melliferum strain KC3 are flanked by viral sequences, forming a functional mobile genetic unit thought to be acquired from other bacteria (10). We did not uncover any functional gene units of this type in the genome of S. poulsonii. All of the sequences found in S. poulsonii were related to other spiroplasmas. This indicates that horizontal transfer between S. poulsonii and other bacterial endosymbionts associated with Drosophila (e.g., Wolbachia) is absent or rare.
Gene annotations.
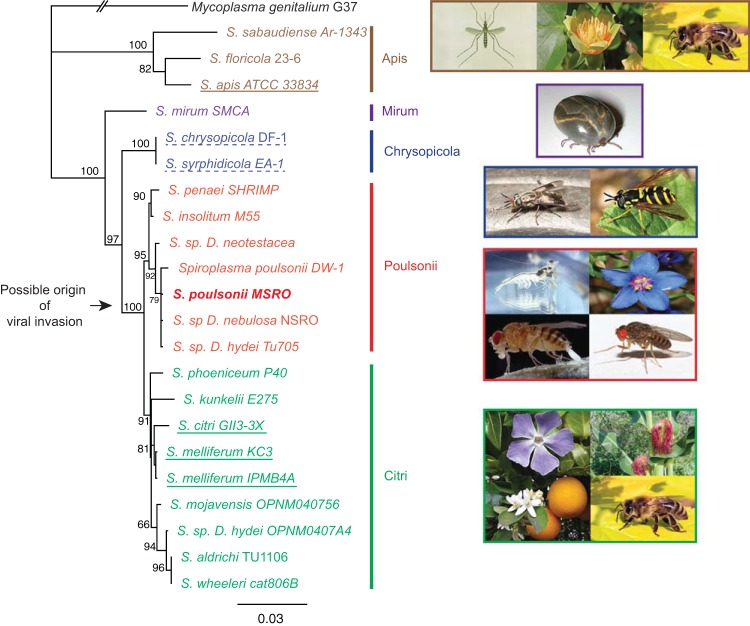
The genome of S. poulsonii encodes 31 tRNAs that are organized in five clusters and cover all 20 amino acids. It contains a single 16S-23S-50S ribosomal operon (Fig. 1C; see Table S1 in the supplemental material). The 16S rRNA gene sequence of S. poulsonii MSRO confirms its position in the Poulsonii clade and is identical to that of DW1 isolated from D. willistoni, NRSO isolated from D. nebulosa, and SPHY isolated from D. hydei (Fig. 2) (22, 46).
FIG 2 .
Phylogenetic tree of Spiroplasma species. This maximum-likelihood phylogenetic tree was inferred by using the 16S rRNA gene. GenBank accession numbers are listed in Materials and Methods. The values on internal branches are percentages of bootstrap support based on 1,000 replicates. Color codes represent the different clades, and the black arrow denotes the probable point of viral invasion of the lineage (15). Representative images of Spiroplasma hosts are shown to the right of each clade. Species previously sequenced are underlined. Solid underlining denotes an infectious pathogen, and dashed underlining denotes a mutualistic relationship with the insect host. Of note, all of the genomes sequenced are from horizontally transmitted spiroplasmas. The following pictures were obtained from Wikipedia: mosquito, http://zh.wikipedia.org/wiki/%E8%9A%8A; tulip, http://en.wikipedia.org/wiki/Liriodendron_tulipifera; honey bee, http://en.wikipedia.org/wiki/Western_honey_bee; tick, http://en.wikipedia.org/wiki/Haemaphysalis_longicornis; Syrphidae, http://en.wikipedia.org/wiki/File:Syrphidae_poster.jpg; deer fly, http://commons.wikimedia.org/wiki/File:Chrysops_callidus.jpg; giant tiger prawn, http://commons.wikimedia.org/wiki/File:CSIRO_ScienceImage_2992_The_Giant_Tiger_Prawn.jpg; scarlet pimpernel, http://commons.wikimedia.org/wiki/File:Flower_poster_2.jpg. The remaining images are from our personal lab collection.
Among the 2,263 predicted genes found in the S. poulsonii draft genome, only 401 encode well-characterized proteins, 859 encode hypothetical proteins, 82 are pseudogenes, 94 are viral coding sequences, 3 encode rRNA, and 31 encode tRNAs. The length distribution of predicted amino acid sequences in S. poulsonii is lower than that in other spiroplasmas, indicating that the number of pseudogenes is probably underestimated. However, since these short hypothetical proteins have no functional domain/database hit, it is extremely difficult to determine their function (Fig. 1C). According to the clusters of homologous genes (COG) classification, ~81% of the S. poulsonii genes encode unknown proteins. Among the 401 well-characterized proteins identified in the genome, the three most highly represented classes of proteins were involved in translation/ribosome (class J) (~25%), replication/recombination/repair (class L) (~23%), and carbohydrate transport and metabolism (class G) (~8%), (Fig. 3). Of note, the large size of class L is due mainly to the presence of putative transposases that fall in this category (see Table S1 in the supplemental material).
FIG 3 .
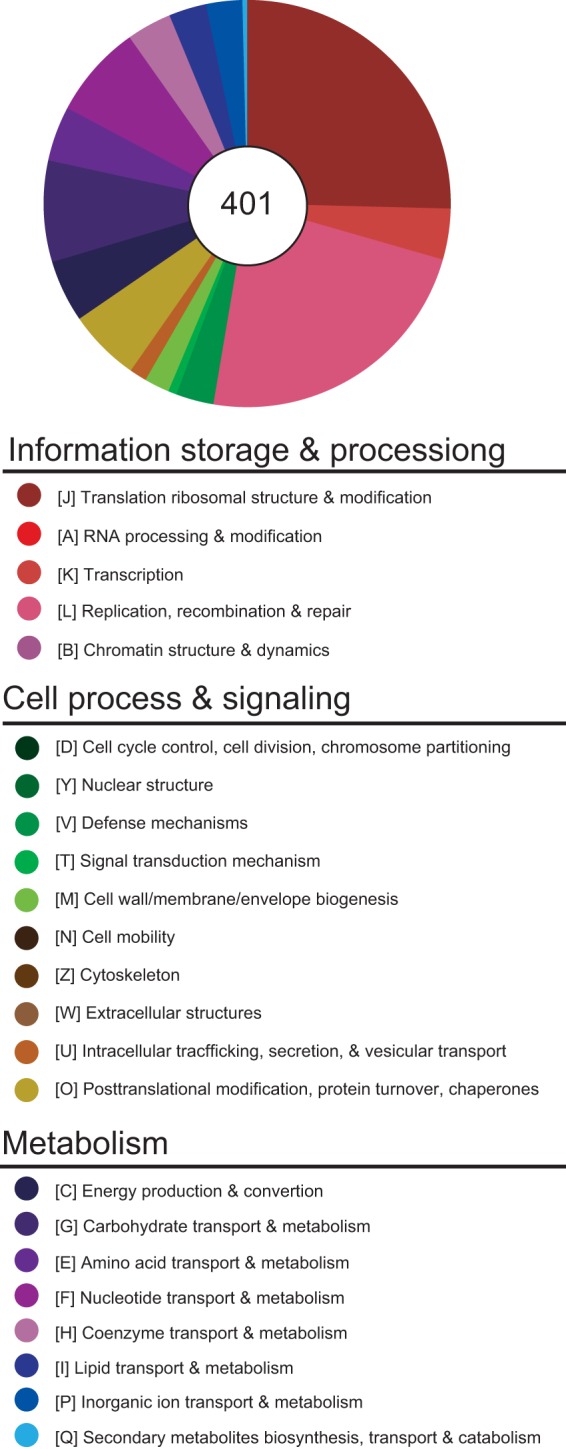
Only 25% of protein-coding genes have a functional classification. The functional categorization of each protein-coding gene was done according to the COG assignment; genes that did not have any inferred COG annotation were assigned to a custom category (X). Among the total of 2,263 genes, only the 401 protein-coding genes that have specific functional category assignments are represented in the pie chart.
The genome contains numerous genes involved in DNA replication, recombination, and translation, including dnaA, dnaB, dnaC, dnaE, dnaG, dnaI, dnaJ, dnaK, dnaN, dnaX, gyrA, gyrB, ruvB, ruvA, uvrA, uvrB, uvrC, uvrD, ligA, rhnB, and rnhC. Additionally, it includes the complete Sec secretory system (secA, secD/F, secG, secY, ffh, ftsY, and yidC). Most of the genes encoding mismatch repair proteins, such as mutL, mutH, exoI, exoX, and exoJ, are missing; only mutM is still present. The majority of the genes encoding proteins involved in homologous recombination are missing as well; only recA, recE, recD, recO, recU, recR, and recT were found; however, the recE and recA genes were truncated. As previously reported for S. citri and S. melliferum (11, 14), S. poulsonii has a premature stop codon in recA. It was hypothesized that a functional recA gene could facilitate homologous recombination and therefore genome rearrangements (47). The fact that recA is truncated seems to contradict this notion since extensive genomic rearrangements were found in S. poulsonii compared to other, closely related, spiroplasmas (see Fig. S1 in the supplemental material). However, there is some evidence that the pseudogenization of recA is recent (codon shifts have different locations on the recA gene in the different strains) and may be caused by independent events in each Spiroplasma lineage. Thus, the possibility cannot be excluded that the genome rearrangements we observed occurred before the loss of recA (48).
To gain further insights into the genomic adaptation associated with the endosymbiotic life, we compared the gene contents of S. poulsonii with those of the three horizontally transmitted spiroplasmas S. citri GII3-X, S. melliferum, and S. chrysopicola. We found that they all have 618 genes in common, among which more than a third correspond to putative virus coding sequences. In addition, S. poulsonii has 92, 21, and 18 genes in common specifically with S. citri, S. melliferum, and S. chrysopicola, respectively (Fig. 4). Besides genes coding for viral sequences, the most-represented classes common to all three spiroplasmas are translation/ribosome (J), replication/DNA recombination/repair (L), and nucleotide transport/metabolism (F). Among the genes absent from S. chrysopicola and S. syrphidicola but also present in the other closely related Spiroplasma species, we found genes involved in sugar metabolism (treA, fruR, fruF, deoD, and kgdA) (see Table S3 in the supplemental material) (15). Thus, the evolutionary divergence of the Citri-Poulsonii and Chrysopicola clades was accompanied by divergence in genes involved in carbohydrate metabolism. Interestingly, among spiroplasmas belonging to the Citri clade, the COG class of which S. poulsonii lacks the most genes compared to S. citri and S. melliferum is also carbohydrate transport and metabolism (G) (see Table S3). This reduction in the number of genes involved in carbohydrate transport and metabolism could be related to the fact that S. poulsonii is an endosymbiotic Spiroplasma bacterium that is confined in its insect host while S. citri and S. melliferum can also be found outside their insect host. Similar to other members of the Citri-Chrysopicola-Mirum clade, S. poulsonii contains five copies of mreB (49), which encodes a functional analogue of actin in bacteria and has been shown to play an important role in the helical shape of the Spiroplasma cytoskeletal structure (50).
FIG 4 .
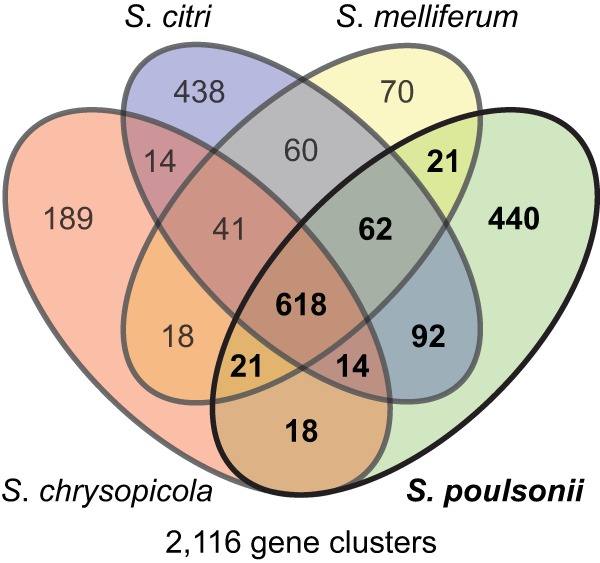
Numbers of shared and genome-specific homologous gene clusters. The Venn diagram shows the numbers of shared and genome-specific homologous gene clusters among the S. poulsonii, S. citri, S. melliferum, and S. chrysopicola genomes.
Carbon and energy metabolism.
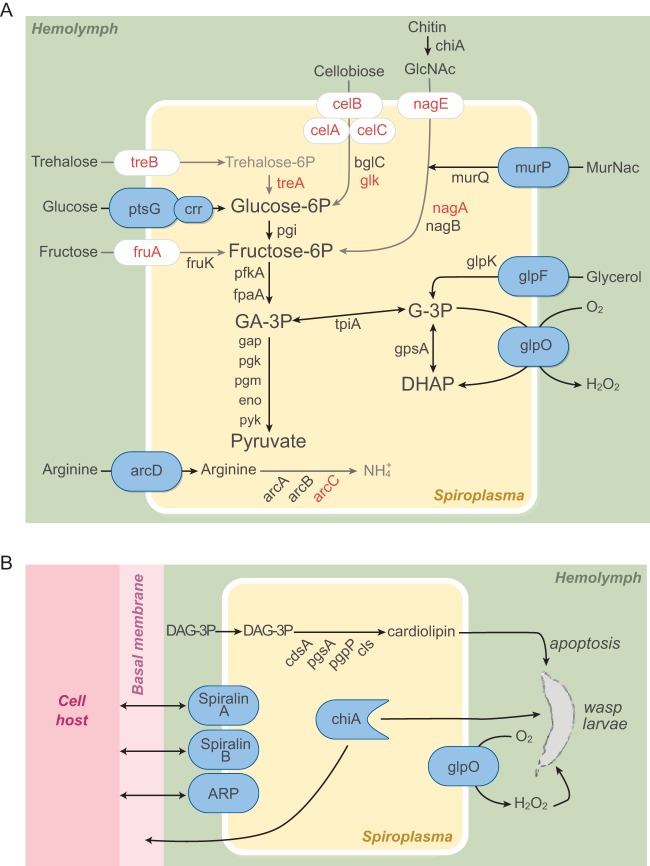
The S. poulsonii genome encodes the well-conserved F0F1 ATP synthase operon (atpB, atpE, atpF, atpH, atpA, atpD, and atpC), the entire glycolysis pathway, and four enzymes of the nonoxidative phase of the pentose cycle (rpe, 2 tkt, prs, and rpiB). Its genome also encodes the phosphoenolpyruvate phosphotransferase transport systems (PTSs) of glucose (ptsG), 2-O-a-mannosyl-d-glycerate (mngA), maltose, and ascorbate (ulaA) (Fig. 5A). It is notable that the genes encoding N-acetylglucosamine PTSs and FruA transporters were pseudogenes. We were not able to identify a trehalose transporter gene (treB), and the trehalase treA gene is truncated. This is an interesting finding because trehalose is the most abundant sugar of the Drosophila hemolymph, followed by glucose and mannose (51). The absence of trehalose transporters from S. poulsonii may be important in the regulation of symbiont densities, as it may limit endosymbiont overproliferation, which could be detrimental to its host. treB is found in S. citri, S. apis, and S. melliferum (11, 12, 14) but is absent from the S. chrysopicola and S. syrphidicola genomes (15). S. apis and S. melliferum, but not S. chrysopicola and S. syrphidicola, are pathogenic to their host (52). Another observation that is in line with this hypothesis is that S. citri is much more pathogenic than S. poulsonii when injected into D. melanogaster (34). The pseudogenization of two fructose transporter genes (fruA) is not surprising since fructose is found at very low levels in the fly hemolymph (51). We speculate that the evolutionary pressure to maintain these genes in S. poulsonii might be relaxed relative to spiroplasmas that infect plants (e.g., S. citri) because fructose is an important source of carbon in plant phloem (53).
FIG 5 .
S. poulsonii sugar metabolism and virulence genes. (A) Metabolic scheme representing enzymes and transporters encoded by the S. poulsonii genome that are involved in sugar metabolism. Enzymes or transporters that are absent from or nonfunctional in S. poulsonii are red. (B) Potential entomopathogenic factors and surface proteins encoded by the S. poulsonii genome. Three potential virulence factors (chitinase A, GlpO, and cardiolipin) may play a role in S. poulsonii-mediated protection against parasitoid wasps. Two spiralin-like proteins and one ARP might play important roles in endosymbiosis.
A polymer found in fly hemolymph that could potentially be used as a source of nitrogen and carbon by S. poulsonii is chitin. The presence of a chitinase gene belonging to the GH18 superfamily suggests that S. poulsonii might be able to cleave different forms of chitin, chitosan, and peptidoglycan to produce N-acetylglucosamine (54). However, it is unlikely that chitin cleavage plays a role in metabolite acquisition, since a gene essential for N-acetylglucosamine metabolism, nagA (GlcNAc-6P deacetylase), is truncated because of a viral sequence insertion. Moreover, genes encoding the PTS involved in the import of N-acetylglucosamine are also absent from this draft genome (Fig. 5A; see Table S1 in the supplemental material).
S. poulsonii harbors a gene cluster involved in arginine metabolism (arcA, arcB, arc, and arcD) that is present in all sequenced spiroplasmas. However, in contrast to the arcC gene of other spiroplasmas, that of S. poulsonii is truncated. This suggests that arginine degradation via the arginine deiminase pathway is not required for S. poulsonii persistence in Drosophila (Fig. 5A; see Table S3). Production of ATP by arginine degradation is not essential in S. citri either (55). The lack of a functional arcC gene in the S. poulsonii genome would prevent the metabolism of carbamoyl-phosphate and thus the synthesis of NH3 for ATP production. While a complete arginine deiminase pathway is not essential for S. poulsonii, we cannot rule out the possibility that carbamoyl-phosphate could be the substrate of another enzyme and that arginine could indeed be implicated in energy production.
The main classes of lipids that are present in Spiroplasma bacteria are phosphatidylglycerols, cardiolipins, sterols, and sphingolipids (56). Cardiolipins are a class of lipids found exclusively in eubacteria and in the mitochondria of eukaryotic cells and play a role in the curvature of the bacterial membrane (57). In contrast to other major lipidic components of Spiroplasma membranes, cardiolipins are not present at detectable levels in Drosophila hemolymph and therefore would need to be synthesized by S. poulsonii (58). A more detailed investigation of the lipid metabolic capacity of S. poulsonii revealed a lack of enzymes involved in the citric acid cycle and fatty acid elongation. S. poulsonii does, however, appear to be able to modify some phospholipids. The presence of a phospholipase gene (pldB) and genes involved in the synthesis of phosphatidylglycerol and diphosphatidylglycerol (cardiolipin) indicates that S. poulsonii has the capacity to generate phospholipids from fatty acids or diacylglyceride (DAG) precursors imported from the external environment. The cardiolipin synthetase enzyme gene cls is present in S. poulsonii as in all sequenced genomes of spiroplasmas of the Citri-Chrysopicola-Mirum clade. Genomic analysis confirms that S. poulsonii can synthetize cardiolipin from DAG extracted from the hemolymph, consistent with a previous study (33) (Fig. 5B). S. poulsonii also harbors the complete set of enzymes implicated in terpenoid C55 synthesis (dxs, dxr, ispD, ispE, ispF, ispG, and ispH), which are involved in the nonmevalonate pathway resulting in the synthesis of farnesyl pyrophosphate. This complete pathway is also found in all Spiroplasma genomes of the Apis and Citri-Chrysopicola-Mirum clades (with the exception of S. citri, where ispE could not be identified, [14]). As was found to be the case for other spiroplasmas, these genes are dispersed throughout the S. poulsonii chromosome.
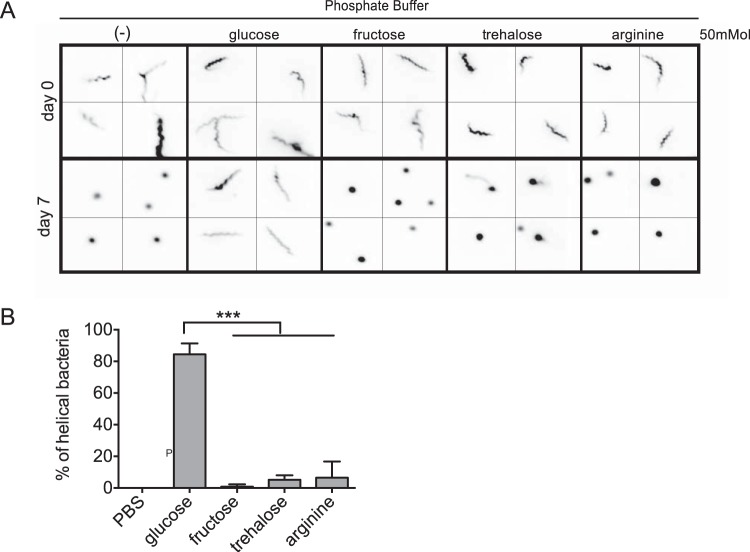
We utilized an in vitro assay to validate our analysis of the metabolic capacities of S. poulsonii. A previous study has shown that the helicoidal shape and motility of spiroplasmas require the maintenance of an electrochemical transmembrane gradient, which consumes ATP (59). We therefore used S. poulsonii shape deformation as an assay to monitor the ability of S. poulsonii to maintain a helicoidal shape and hence its capacity to efficiently generate ATP from several metabolites, including glucose, fructose, trehalose, or arginine. S. poulsonii cells were extracted from D. melanogaster hemolymph and incubated in phosphate buffer alone or complemented with different sugars or arginine. Bacterial shape was then monitored over time by fluorescence microscopy (Fig. 6A). S. poulsonii maintained its membrane integrity and motility in phosphate buffer for more than 24 h, compared to only a few hours for S. citri (data not shown). Nevertheless, S. poulsonii cells lose their helical shape and motility after a few days of incubation in phosphate buffer, with most bacteria appearing as rounded vesicles. The loss of helicity and of motility was also observed when the phosphate buffer was complemented with fructose, trehalose, or arginine. In contrast, most S. poulsonii cells conserved their characteristic helical shape after 7 days in the presence of glucose (Fig. 6). We obtained the same results when we added both glucose and arginine (data not shown); thus, the addition of arginine does not seem to affect shape stability. We therefore conclude that S. poulsonii is able to gain sufficient energy to maintain helicity and motility by importing and metabolizing glucose but not fructose, trehalose, or arginine, as predicted by genome analysis.
FIG 6 .
Deformation test confirming the limited metabolic capacity of S. poulsonii. Drosophila hemolymph containing S. poulsonii was extracted and incubated in phosphate buffer alone or complemented with glucose, fructose, trehalose, or arginine. Bacterial shape was then monitored by fluorescence microscopy. Representative images (A) and percentages of helical cells compared to the total cell counts after 7 days (B) are shown. S. poulsonii maintains its helical shape for 1 week in the presence of glucose but not fructose, trehalose, or arginine. Shown is the mean ± the standard error of the mean of data pooled from three independent experiments (n = >100; ***, P < 0.0001).
Potential virulence factors.
Because S. poulsonii is known to protect flies against parasitoid attacks, we searched the S. poulsonii genome for factors that might play a role in the killing of parasitic wasps that develop inside fly larvae. The S. poulsonii genome contains one gene coding for a chitinase A that could play a role in Spiroplasma-Drosophila interactions (Fig. 5B). One potential target of this chitinase is the basal membranes surrounding the ovaries that S. poulsonii must cross in order to reach the germ line and be vertically transmitted (21). This chitinase could be a virulence factor but could also be beneficial to the host by playing a role in Spiroplasma-mediated protection against parasitoid wasps by targeting the chitin-based cuticle of this hymenopteran.
As previously reported for S. melliferum KC3, S. taiwanense, and S. culicicola, the S. poulsonii genome contains the gene that encodes glycerol-3-phosphate oxidase (glpO) (10, 16, 17). This gene is found in a cluster between glpK and glpF, which encode a kinase and a membrane channel involved in glycerol metabolism, respectively. These three genes (glpF, glpO, and glpK) are absent from S. citri and S. melliferum IPMB4A (11, 14). GlpO has been characterized as a virulence factor in Mycoplasma mycoides subsp. mycoides, inducing tissue inflammation (60). The two substrates of GlpO are glycerol-3-phosphate and oxygen, while the two products of the catalyzed reaction are glycerone phosphate and hydrogen peroxide (H2O2). H2O2 has been reported to be involved in the cellular immune response of insect against parasitoid wasp eggs (61). We therefore hypothesize that GlpO could be involved in protection against parasitoid wasps (Fig. 5B) and possibly in the pathological effects observed in old flies harboring S. poulsonii (33).
As described above, our genomic analysis confirms that S. poulsonii has the set of enzymes to generate cardiolipin from diacylglycerol in fly hemolymph (see Table S1 in the supplemental material; Fig. 5B). A number of studies have demonstrated links between cardiolipin and cell death (62), which raises the possibility that cardiolipin produced by Spiroplasma bacteria could also be involved in the pathological effects observed in old flies.
Adhesins and surface membrane proteins.
Like other members of the class Mollicutes, Spiroplasma bacteria lack a cell wall and their plasma membrane is directly exposed to the external environment. In Mollicutes, many membrane-associated components, including transmembrane proteins and lipoproteins (i.e., acylated proteins at their N-terminal cysteinyl residue), have been shown to be involved in pathogenesis or virulence or to interact with the host immune system (63). In order to identify S. poulsonii surface components that may be putatively involved in interactions with its Drosophila host, we performed a proteomic analysis of the membrane and soluble fraction of S. poulsonii isolated from Drosophila hemolymph. Protein expression profiles were established following TX-114 phase partition and SDS-PAGE separation and further characterized for protein identities by liquid chromatography-tandem mass spectrometry. This analysis led us to identify more than 300 proteins that were validated by our genome annotation. The protein content of the detergent phase was enriched in amphiphilic proteins and contained 24 proteins for which a prokaryotic membrane lipoprotein lipid attachment site (PROSITE PS51527) was predicted. The detected lipoproteins represented 45% of the predicted lipoproteome (53 putative lipoproteins encoded by genes present in the genome; see Tables S1 and S2 in the supplemental material). The large number of lipoproteins exposed in S. poulsonii membrane suggests that S. poulsonii contains a large repertoire of lipoproteins that are likely to shape its surface and to contribute to its capacity to be an endosymbiont. Spiralin is a lipoprotein known only from Spiroplasma bacteria. It is the most abundant lipoprotein found at the surface of S. citri and S. melliferum (64, 65) and has been implicated in the abilities of S. citri to adhere to insect organs and to invade cells (66). While spiralin is highly conserved among spiroplasmas from the Citri clade (about 60% identity among the S. citri, S. melliferum, and S. kunkelii spiralins), the spiralin A-encoding gene (referred to as Spiralin A) found in S. poulsonii (v1c12390) is only 40% identical to those for other, closely related, Spiroplasma spiralins (see Fig. S2A in the supplemental material). Interestingly, the genome of S. poulsonii encodes another spiralin-related protein (v1c09300) that we named spiralin B. This lipoprotein is only ~25% similar to the S. melliferum and S. citri spiralins and S. poulsonii spiralin A (see Fig. S2A). To our surprise, proteomic analysis indicates that spiralin B, not spiralin A, was the most abundant protein detected in the S. poulsonii membrane (relative abundance, >70%). These divergent lipoproteins (spiralins A and B) may play a role in S. poulsonii vertical transmission, including colonization of the female germ line.
We also found a homolog of an ARP that was previously implicated in the ability of S. citri to invade insect cells (67). Genes encoding ARPs have been found in all spiroplasmas in the Citri clade and are found on plasmids (10, 42). As stated earlier, the putative ARP gene in S. poulsonii (v1c22630) was found on contig B, which may be extrachromosomal. Proteomic analysis suggests that this protein is abundant in S. poulsonii membrane (see Table S2 in the supplemental material). The S. poulsonii protein is 60 to 66% similar to S. citri ARPs (ScARPs) (see Fig. S2B) and, as is known to be the case for all characterized ScARPs, contains a signal peptide that is predicted to be cleaved after alanine 23. In addition, the presence of a hydrophobic transmembrane alpha helix located close to the C terminus is predicted in the S. poulsonii protein. Seven repetitions of a 36-amino-acid domain in the N-terminal hydrophilic region correspond to an NCL-1–HT2A–Lin-41 repeat domain that is found in numerous pathogenic bacteria and is known to be involved in eukaryotic cell invasion. This structural motif is found in several ScARPs but also in proteins mediating the invasion of host brain epithelia by Mycobacterium tuberculosis (68). In light of this, it is probable that the S. poulsonii ARP may also play a role in tissue invasion.
Conclusions.
We sequenced the first genome of a vertically transmitted Spiroplasma species, S. poulsonii. Using PacBio technology and optical mapping, we obtained a draft assembly of the S. poulsonii MSRO genome composed of 12 contigs and covering 93% of the total chromosome length. The high number of repeated viral sequences integrated in the chromosome was the main obstacle to obtaining the complete chromosomal sequence. The size of the genome, the presence of many viral sequences, and the evidence of rearrangement indicate that S. poulsonii has retained many genomic features already described for spiroplasmas of the Citri clade. The absence of any sequence deriving from other groups of bacteria indicates that the mechanisms of male killing and vertical transmission have evolved independently in this lineage. The presence of many virus sequences and eroding genes indicates that S. poulsonii, like other facultative endosymbionts (Wolbachia, Sodalis, Serratia) has specific genome features distinct from those of both free-living bacteria and the highly compact obligate endosymbiont genomes (30, 45, 69–71).
Analysis of metabolic pathways reveals striking differences between S. poulsonii and horizontally transmitted spiroplasmas. S. poulsonii has lost many metabolic pathways and transporters, indicating a high level of metabolic integration with its insect host, similar to many insect endosymbiont genomes. Consistent with genome-based predictions, we observed that S. poulsonii metabolizes glucose but not trehalose, fructose, or arginine. The absence of a trehalose transporter is striking given that this sugar is the most abundant one in Drosophila hemolymph. We speculate that this adaptation may prevent S. poulsonii from overproliferating and damaging its host. It was recently demonstrated that the amount of hemolymphatic DAG limits S. poulsonii titers in the hemolymph (33). It was speculated that this greater dependence on lipids and lesser dependence on sugar prevent endosymbiont overproliferation under conditions of host nutrient limitation, since lipid availability depends heavily on the fly’s nutritional state. Our results confirmed that S. poulsonii lacks most of the lipid metabolic enzymes and therefore must acquire lipids intact from its host. Notably, S. poulsonii has the genomic capacity to produce the bacterium-specific lipid cardiolipin. It also possesses the complete nonmevalonate terpenoid synthesis pathway. S. poulsonii also has an incomplete set of enzymes required for arginine metabolism. Although S. poulsonii has a low impact on the general fitness of its host, very high titers of Spiroplasma bacteria in old flies are associated with defects suggestive of neuronal dysfunction (33). This indicates that the S. poulsonii genome codes for factors that have the capacity to be harmful to insects.
S. poulsonii is also a protective endosymbiont in that it can mediate the protection of its host against infestation by parasitoid wasps (31, 32). One possible explanation for this protection is that “immune priming” by S. poulsonii induces a stronger basal immune response in the fly and hence more efficient elimination of wasp eggs/larvae. However, S. poulsonii does not activate a humoral or cellular immune response in the fly (34). The S. poulsonii genome reveals three putative candidate virulence genes as possible virulence factors for the wasp: the gene for the chitinase ChiA, the gene for the H2O2-producing enzyme GlpO, and the gene for the toxic lipid cardiolipin. It is noteworthy that virulence factors have been described as playing a possible role in Spiroplasma-mediated protection against nematodes (72). Another possible hypothesis that cannot be excluded is that Spiroplasma-mediated protection is mediated by nutrient competition. Endosymbiont proliferation depletes the lipid content of the fly hemolymph (33), and both spiroplasmas and wasp larvae colonize this fly compartment. One could speculate that Spiroplasma growth reduces the concentration of nutrients that are essential for wasp larval development.
We have identified several plasma membrane proteins that mediate the interaction with the host, notably, two spiralin-related proteins quite divergent from other Spiroplasma spiralins and an ARP. These adhesion proteins might be implicated in the colonization of the host germ line. It has been established that the ability of spiroplasmas to enter the Drosophila oocyte involves interactions with yolk protein and the yolkless receptor on the exterior of the oocyte membrane, which result in Spiroplasma endocytosis (21). The S. poulsonii membrane proteins involved in mediating these interactions are likely to be spiralins and ARPs; however, the details of these interactions have not yet been elucidated. The current genome could be used as a tool to facilitate the genome annotation of other endosymbiotic spiroplasmas. Further comparisons of non-male-killing and male-killing Spiroplasma genomes could help us to understand the molecular mechanism underlying this phenotype. In conclusion, the completion of the S. poulsonii genome fills a major gap in our knowledge of insect endosymbionts and provides an important resource to decipher the interaction between Spiroplasma bacteria and their well-characterized insect host, D. melanogaster.
MATERIALS AND METHODS
Genomic DNA preparation.
Cell-free cultivation of spiroplasmas is not currently possible. To obtain the required amount of genomic DNA to generate a genomic library (~3 µg), we extracted ~50 µl of pure fly hemolymph with a Nanoject microinjector (Drummond Scientific) from 1-month-old infected flies. The hemolymph extracts were immediately mixed with 500 µl of phosphate-buffered saline (PBS) on ice to avoid melanization. Bacteria were pelleted, washed twice with 500 µl of PBS, and then filtered through a 0.45-µm filter to eliminate other potential bacterial contaminants or Drosophila hemolymph cells (36). Bacteria were subsequently pelleted. For optical mapping, the bacterial pellet was sent to OpGen. For sequencing, DNA was extracted by phenol-chloroform extraction. The DNA library was prepared with the Pacific Biosciences DNA Template prep kit 2.0 (3 to 10 kb) with 3 µg of S. poulsonii genomic DNA with an expected insert length of 25 kb.
Genome sequencing.
Sequencing was performed with a movie of 180 min with the P4-C2 chemistry kit with size selection with the single-molecule real-time (SMRT) MagBead kit. The loading concentration was 0.09 nM in an SMRT cell v3 with the PacBio RS II sequencing machine. Assembly was performed with HGAP (hierarchical genome assembly process) version 3 from the PacBio SMRT analysis pipeline smrtanalysis-2.1.1. The mean coverage is 186.39 with the following parameters: 1 smrtcell 1 movie + 1 smrtcell (V3 + P4-C2) 2 movies, HGAP-3 smrtanalysis_2.2.0, mean coverage of 186.39, length cutoff of 9,450, 47 polished contigs, a maximum contig length of 504,367, an N50 contig length of 163,695, and a contig length sum of 2,354,599. Errors in the PacBio reference assembly were detected and corrected by aligning 454 reads (GATC Biotech AG Titanium Sequencing kit XLR70, genomic library for GS FLX sequencing). At every position of the genome, we computed the proportions of mapped reads supporting the reference base and a potential alternative base. If the alternative base was supported by at least 70% of the reads, the reference was updated to the new more abundant alternative base. Only uniquely mapping reads ≥200 bp long were considered in the analysis, and a coverage of at least 10 reads at a given position was required to proceed with the correction. Alignments were obtained with Bowtie 2. Important metabolic truncated genes were confirmed by PCR and Sanger sequencing.
Optical mapping comparative with other genomes.
Whole-genome mapping was performed by OpGen. Contigs obtained by HGAP alignment were placed into optical mapping by in silico alignment with OpGen MapSolver Software 3.2.0. Figure 1B was obtained with Geneious 7.0.6 software. To examine the extent of genome rearrangement in Spiroplasma and other, related, bacteria, we used MAUVE 2.3.1 (73), S. melliferum IPMB4A (GenBank accession no. AMGI01000000), and S. citri GII3-3X (GenBank accession no. AM285301 to AM285339).
Spiroplasma phylogenetic relationships were reconstructed by the maximum-likelihood method with a Tamura-Nei model in Geneious. In addition to S. poulsonii MSRO, 16S rRNA sequences from the following Spiroplasma bacteria were included in the analysis: S. sabaudiense (Ar-1343) (NR_025710), S. floricola (23-6) (NR_025703), S. apis (ATCC 33834) (GU993267), S. mirum (NR_104955), S. chrysopicola (DF-1) (NR_025699), S. syrphidicola (EA-1) (NR_025711), S. penaei (SHRIMP) (NR_043177), S. insolitum (M55) (NR_025705.1), Spiroplasma strain NSRO from D. nebulosa (AB434486), a Spiroplasma species from D. hydei (TU705) (FJ657200), S. poulsonii MSRO (UGA) (FJ657180), S. phoeniceum (P40) (NR_043178), S. kunkelii (DQ319068), S. citri (GII3-3X) (AM285316), S. melliferum (IPMB4A) (JQ347516), S. melliferum BC-3 (NR_025756), S. mojavensis (OPNM040756) (FJ657216), a Spiroplasma species from D. hydei (OPNM0407A4) (FJ657237), a Spiroplasma species from D. aldrichi (TU1106) (FJ657236), and S. wheeleri (cat806B) (FJ657228). The 16S rRNA of a Spiroplasma species from D. neotestacea was kindly provided by Steve Perlman. Mycoplasma genitalium (G37) (NR_026155) was used as the outgroup. We resampled the tree by bootstrapping (1,000 replicates). Alignment of the spiralin sequences of S. melliferum IPMB4A (ELL44999), S. citri (AAB06629), and S. kunkelii (AAB05468) was performed with the GONNET algorithm.
Genome analysis and annotation.
The procedures used for genome analysis and annotation were based on those described in our previous studies (11, 15) (see Text S1 in the supplemental material for more information).
Electrochemical transmembrane gradient maintenance test.
Five hundred nanoliters of pure S. poulsonii was extracted and diluted in 200 µl of sodium phosphate buffer (50 mM) alone or complemented with 50 mM glucose, fructose, trehalose, or arginine. Images were taken every day for 7 days. To observe S. poulsonii in fly hemolymph, 5 µl of the sample was mixed with 5 µl of PBS containing 0.02 mM SYTO9 (Invitrogen). Slides were then mounted and observed on a Zeiss Axio Imager Z1. Images were captured with an AxioCam MRn camera and AxioVision software.
Nucleotide sequence accession number.
This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under accession no. JTLV00000000. The version described in this paper is version JTLV01000000 (http://www.ncbi.nlm.nih.gov/nuccore/JTLV00000000).
SUPPLEMENTAL MATERIAL
Supplemental methods. Download
Extensive genome rearrangements among closely related Spiroplasma species. The color blocks represent regions of homologous backbone sequences without rearrangement among the genomes compared (top, S. citri GII-3X; middle, S. poulsonii MRSO; bottom, S. melliferum IPMB4A). The vertical red bars indicate the boundaries of individual contigs. The average nucleotide sequence identities were calculated on the basis of single-copy genes that were conserved among the three genomes compared in each group. Download
Phylogenetic tree of spiralin and ARP amino acid sequences. Phylogenetic analysis of whole protein sequences with the MUSCLE algorithm (multiple-sequence comparison by log expectation). Shown are the spiralin (A) and ARP (B) sequences of closely related Spiroplasma species. The number in brackets indicates the contig position. The only functional ARP found in the S. poulsonii genome is ARP S. p. (B); ARP S. p (6) is truncated by a viral insertion; the N-terminal and C-terminal portions were put together for the sequence alignment. GenBank accession numbers are listed in Materials and Methods. Download
List of annotated features.
List of S. poulsonii peptides and proteins identified by proteomics.
List of shared and genome-specific homologous gene clusters. Taps legends: all, all genes; Sch, S. chrysopicola; Sci, S. citri GII-3X; Sme, S. melliferum IPMB4A; Spo, S. poulsonii MSRO; +, presence.
ACKNOWLEDGMENTS
We thank Steve Perlman and Elodie Ramond for stimulating discussions. We thank Keith Hartshman, Maroun Bou Sleiman, and Petr Dostál for biostatistics help and the Lausanne Genomic Technologies Facility (UNIL Switzerland) for DNA sequencing.
The Lemaitre lab work was funded by the ERC advanced grant (339970). Chih-Horng Kuo was supported by research grants from Academia Sinica and the National Science Council of Taiwan (NSC 101-2621-B-001-004-MY3). Laure Béven was supported by grants from Structure Fédérative de Recherche Biologie Intégrative et Ecologie.
Footnotes
Citation Paredes JC, Herren JK, Schüpfer F, Marin R, Claverol S, Kuo C-H, Lemaitre B, Béven L. 2015. Genome sequence of the Drosophila melanogaster male-killing Spiroplasma strain MSRO endosymbiont. mBio 6(2):e02437-14. doi:10.1128/mBio.02437-14.
REFERENCES
- 1.Whitcomb RF. 1980. The genus Spiroplasma. Annu Rev Microbiol 34:677–709. doi: 10.1146/annurev.mi.34.100180.003333. [DOI] [PubMed] [Google Scholar]
- 2.Gasparich GE, Whitcomb RF, Dodge D, French FE, Glass J, Williamson DL. 2004. The genus Spiroplasma and its non-helical descendants: phylogenetic classification, correlation with phenotype and roots of the Mycoplasma mycoides clade. Int J Syst Evol Microbiol 54:893–918. doi: 10.1099/ijs.0.02688-0. [DOI] [PubMed] [Google Scholar]
- 3.Whitcomb RF, Chen TA, Williamson DL, Liao C, Tully JG, Bové JM, Mouches C, Rose DL, Coan ME, Clark TB. 1986. Spiroplasma kunkelii sp. nov.: characterization of the etiological agent of corn stunt disease. Int J Syst Bacteriol 36:170–178. doi: 10.1099/00207713-36-2-170. [DOI] [Google Scholar]
- 4.Saillard C, Vignault JC, Bové JM, Raie A, Tully JG, Williamson DL, Fos A, Garnier M, Gadeau A, Carle P, Whitcomb RF. 1987. Spiroplasma phoeniceum sp. nov., a new plant-pathogenic species from Syria. Int J Syst Evol Microbiol 37:106–115. doi: 10.1099/00207713-37-2-106. [DOI] [Google Scholar]
- 5.Bové JM, Renaudin J, Saillard C, Foissac X, Garnier M. 2003. Spiroplasma citri, a plant pathogenic mollicute: relationships with its two hosts, the plant and the leafhopper vector. Annu Rev Phytopathol 41:483–500. doi: 10.1146/annurev.phyto.41.052102.104034. [DOI] [PubMed] [Google Scholar]
- 6.Hackett KJ, Clark TB. 1979. Ecology of spiroplasmas, p 113–200. In Whitcomb RF, Tully JG (ed), Molecular biology and pathogenicity of mycoplasmas. Academic Press, New York, NY. [Google Scholar]
- 7.Duron O, Bouchon D, Boutin S, Bellamy L, Zhou L, Engelstädter J, Hurst GD. 2008. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol 6:27. doi: 10.1186/1741-7007-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W, Wen B, Gasparich GE, Zhu N, Rong L, Chen J, Xu Z. 2004. A Spiroplasma associated with tremor disease in the Chinese mitten crab (Eriocheir sinensis). Microbiology 150 (Pt 9):3035–3040. doi: 10.1099/mic.0.26664-0. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Gu W, Ding Z, Ren Y, Chen J, Hou Y. 2005. A novel Spiroplasma pathogen causing systemic infection in the crayfish Procambarus clarkii (Crustacea: decapod), in China. FEMS Microbiol Lett 249:131–137. doi: 10.1016/j.femsle.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Alexeev D, Kostrjukova E, Aliper A, Popenko A, Bazaleev N, Tyakht A, Selezneva O, Akopian T, Prichodko E, Kondratov I, Chukin M, Demina I, Galyamina M, Kamashev D, Vanyushkina A, Ladygina V, Levitskii S, Lazarev V, Govorun V. 2012. Application of Spiroplasma melliferum proteogenomic profiling for the discovery of virulence factors and pathogenicity mechanisms in host-associated spiroplasmas. J Proteome Res 11:224–236. doi: 10.1021/pr2008626. [DOI] [PubMed] [Google Scholar]
- 11.Lo W-S, Chen L-L, Chung W-C, Gasparich GE, Kuo C-H. 2013. Comparative genome analysis of Spiroplasma melliferum IPMB4A, a honeybee-associated bacterium. BMC Genomics 14:22. doi: 10.1186/1471-2164-14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ku C, Lo WS, Chen LL, Kuo CH. 2014. Complete genome sequence of Spiroplasma apis B31T (ATCC 33834), a bacterium associated with May disease of honeybees (Apis mellifera). Genome Announc 2. doi: 10.1128/genomeA.01151-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai X, Hogenhout SA. 2002. A genome sequence survey of the mollicute corn stunt Spiroplasma Spiroplasma kunkelii. FEMS Microbiol Lett 210:7–17. doi: 10.1111/j.1574-6968.2002.tb11153.x. [DOI] [PubMed] [Google Scholar]
- 14.Carle P, Saillard C, Carrère N, Carrère S, Duret S, Eveillard S, Gaurivaud P, Gourgues G, Gouzy J, Salar P, Verdin E, Breton M, Blanchard A, Laigret F, Bové J-M, Renaudin J, Foissac X. 2010. Partial chromosome sequence of Spiroplasma citri reveals extensive viral invasion and important gene decay. Appl Environ Microbiol 76:3420–3426. doi: 10.1128/AEM.02954-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ku C, Lo W-S, Chen L-L, Kuo C-H. 2013. Complete genomes of two dipteran-associated spiroplasmas provided insights into the origin, dynamics, and impacts of viral invasion in Spiroplasma. Genome Biol Evol 5:1151–1164. doi: 10.1093/gbe/evt084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo W-S, Ku C, Chen L-L, Chang T-H, Kuo C-H. 2013. Comparison of metabolic capacities and inference of gene content evolution in mosquito-associated Spiroplasma diminutum and S. taiwanense. Genome Biol Evol 5:1512–1523. doi: 10.1093/gbe/evt108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang T-H, Lo W-S, Ku C, Chen L-L, Kuo C-H. 2014. Molecular evolution of the substrate utilization strategies and putative virulence factors in mosquito-associated Spiroplasma species. Genome Biol Evol 6:500–509. doi: 10.1093/gbe/evu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mateos M, Castrezana SJ, Nankivell BJ, Estes AM, Markow TA, Moran NA. 2006. Heritable endosymbionts of Drosophila. Genetics 174:363–376. doi: 10.1534/genetics.106.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watts T, Haselkorn TS, Moran NA, Markow TA. 2009. Variable incidence of Spiroplasma infections in natural populations of Drosophila species. PLOS ONE 4:e5703. doi: 10.1371/journal.pone.0005703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pool JE, Wong A, Aquadro CF. 2006. Finding of male-killing Spiroplasma infecting Drosophila melanogaster in Africa implies transatlantic migration of this endosymbiont. Heredity 97:27–32. doi: 10.1038/sj.hdy.6800830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herren JK, Paredes JC, Schüpfer F, Lemaitre B. 2013. Vertical transmission of a Drosophila endosymbiont via cooption of the yolk transport and internalization machinery. mBio 4:e00532-12. doi: 10.1128/mBio.00532-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haselkorn TS, Markow TA, Moran NA. 2009. Multiple introductions of the Spiroplasma bacterial endosymbiont into Drosophila. Mol Ecol 18:1294–1305. doi: 10.1111/j.1365-294X.2009.04085.x. [DOI] [PubMed] [Google Scholar]
- 23.Montenegro H, Petherwick AS, Hurst GD, Klaczko LB. 2006. Fitness effects of Wolbachia and Spiroplasma in Drosophila melanogaster. Genetica 127:207–215. doi: 10.1007/s10709-005-3766-4. [DOI] [PubMed] [Google Scholar]
- 24.Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. 2010. Adaptation via symbiosis: recent spread of a drosophila defensive symbiont. Science 329:212–215. doi: 10.1126/science.1188235. [DOI] [PubMed] [Google Scholar]
- 25.Werren JH, Skinner SW, Huger AM. 1986. Male-killing bacteria in a parasitic wasp. Science 231:990–992. doi: 10.1126/science.3945814. [DOI] [PubMed] [Google Scholar]
- 26.Werren JH, Hurst GD, Zhang W, Breeuwer JA, Stouthamer R, Majerus ME. 1994. Rickettsial relative associated with male killing in the ladybird beetle (Adalia bipunctata). J Bacteriol 176:388–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurst GD, Graf von der Schulenburg JH, Majerus TM, Bertrand D, Zakharov IA, Baungaard J, Völkl W, Stouthamer R, Majerus ME. 1999. Invasion of one insect species, Adalia bipunctata, by two different male-killing bacteria. Insect Mol Biol 8:133–139. doi: 10.1046/j.1365-2583.1999.810133.x. [DOI] [PubMed] [Google Scholar]
- 28.Jiggins FM, Hurst GD, Jiggins CD, v d Schulenburg JH, Majerus ME. 2000. The butterfly Danaus chrysippus is infected by a male-killing Spiroplasma bacterium. Parasitology 120:439–446. doi: 10.1017/S0031182099005867. [DOI] [PubMed] [Google Scholar]
- 29.Simon J-C, Boutin S, Tsuchida T, Koga R, Le Gallic J-F, Frantz A, Outreman Y, Fukatsu T. 2011. Facultative symbiont infections affect aphid reproduction. PLoS One 6:e21831. doi: 10.1371/journal.pone.0021831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duplouy A, Iturbe-Ormaetxe I, Beatson SA, Szubert JM, Brownlie JC, McMeniman CJ, McGraw EA, Hurst GD, Charlat S, O’Neill SL, Woolfit M. 2013. Draft genome sequence of the male-killing Wolbachia strain wBol1 reveals recent horizontal gene transfers from diverse sources. BMC Genomics 14:20. doi: 10.1186/1471-2164-14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie J, Vilchez I, Mateos M. 2010. Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS One 5:e12149. doi: 10.1371/journal.pone.0012149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie J, Butler S, Sanchez G, Mateos M. 2014. Male killing Spiroplasma protects Drosophila melanogaster against two parasitoid wasps. Heredity 112:399–408. doi: 10.1038/hdy.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herren JK, Paredes JC, Schüpfer F, Arafah K, Bulet P, Lemaitre B. 2014. Insect endosymbiont proliferation is limited by lipid availability. eLife 3:e02964. doi: 10.7554/eLife.02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herren JK, Lemaitre B. 2011. Spiroplasma and host immunity: activation of humoral immune responses increases endosymbiont load and susceptibility to certain gram-negative bacterial pathogens in Drosophila melanogaster. Cell Microbiol 13:1385–1396. doi: 10.1111/j.1462-5822.2011.01627.x. [DOI] [PubMed] [Google Scholar]
- 35.Hackett KJ, Lynn DE, Williamson DL, Ginsberg AS, Whitcomb RF. 1986. Cultivation of the Drosophila sex-ratio Spiroplasma. Science 232:1253–1255. doi: 10.1126/science.232.4755.1253. [DOI] [PubMed] [Google Scholar]
- 36.Duret S, Danet J-L, Garnier M, Renaudin J. 1999. Gene disruption through homologous recombination in Spiroplasma citri: an scm1-disrupted motility mutant is pathogenic. J Bacteriol 181:7449–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dally EL, Barros TS, Zhao Y, Lin S, Roe BA, Davis RE. 2006. Physical and genetic map of the Spiroplasma kunkelii CR2-3x chromosome. Can J Microbiol 52:857–867. doi: 10.1139/w06-044. [DOI] [PubMed] [Google Scholar]
- 38.Williamson DL, Sakaguchi B, Hackett KJ, Whitcomb RF, Tully JG, Carle P, Bové JM, Adams JR, Konai M, Henegar RB. 1999. Spiroplasma poulsonii sp. nov., a new species associated with male-lethality in Drosophila willistoni, a neotropical species of fruit fly. Int J Syst Bacteriol 49:611–618. doi: 10.1099/00207713-49-2-611. [DOI] [PubMed] [Google Scholar]
- 39.Ye F, Laigret F, Whitley JC, Citti C, Finch LR, Carle P, Renaudin J, Bové JM. 1992. A physical and genetic map of the Spiroplasma citri genome. Nucleic Acids Res 20:1559–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye F, Melcher U, Rascoe JE, Fletcher J. 1996. Extensive chromosome aberrations in Spiroplasma citri strain br3. Biochem Genet 34:269–286. doi: 10.1007/BF02399947. [DOI] [PubMed] [Google Scholar]
- 41.Melcher U, Sha Y, Ye F, Fletcher J. 1999. Mechanisms of spiroplasma genome variation associated with SpVl-like viral DNA inferred from sequence comparisons. Microb Comp Genomics 4:29–46. doi: 10.1089/omi.1.1999.4.29. [DOI] [PubMed] [Google Scholar]
- 42.Saillard C, Carle P, Duret-Nurbel S, Henri R, Killiny N, Carrère S, Gouzy J, Bové J-M, Renaudin J, Foissac X. 2008. The abundant extrachromosomal DNA content of the Spiroplasma citri GII3-3X genome. BMC Genomics 9:195. doi: 10.1186/1471-2164-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen AJ, Williamson DL, Oishi K. 1987. SpV3 viruses of Drosophila spiroplasmas. Isr J Med Sci 23:429–433. [PubMed] [Google Scholar]
- 44.Moran NA, Plague GR. 2004. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev 14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC, McGraw EA, Martin W, Esser C, Ahmadinejad N, Wiegand C, Madupu R, Beanan MJ, Brinkac LM, Daugherty SC, Durkin AS, Kolonay JF, Nelson WC, Mohamoud Y, Lee P. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol 2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anbutsu H, Goto S, Fukatsu T. 2008. High and low temperatures differently affect infection density and vertical transmission of male-killing Spiroplasma symbionts in Drosophila hosts. Appl Environ Microbiol 74:6053–6059. doi: 10.1128/AEM.01503-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brüssow H, Canchaya C, Hardt W-D. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68:560–602–table of contents. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marais A, Bové JM, Renaudin J. 1996. Characterization of the recA gene regions of Spiroplasma citri and Spiroplasma melliferum. J Bacteriol 178:7003–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ku C, Lo W, Kuo C. 2014. Molecular evolution of the actin-like MreB protein gene family in wall-less bacteria. Biochem Biophys Res Commun 446:927–932. doi: 10.1016/j.bbrc.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 50.Kürner J, Frangakis AS, Baumeister W. 2005. Cryo-electron tomography reveals the cytoskeletal structure of Spiroplasma melliferum. Science 307:436–438. doi: 10.1126/science.1104031. [DOI] [PubMed] [Google Scholar]
- 51.Bedford JJ. 1977. The carbohydrate levels of insect haemolymph. Comp Biochem Physiol A Physiol 57:83–86. doi: 10.1016/0300-9629(77)90354-1. [DOI] [Google Scholar]
- 52.Evans JD, Schwarz RS. 2011. Bees brought to their knees: microbes affecting honey bee health. Trends Microbiol 19:614–620. doi: 10.1016/j.tim.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 53.Gaurivaud P, Danet JL, Laigret F, Garnier M, Bové JM. 2000. Fructose utilization and phytopathogenicity of Spiroplasma citri. Mol Plant Microbe Interact 13:1145–1155. doi: 10.1094/MPMI.2000.13.10.1145. [DOI] [PubMed] [Google Scholar]
- 54.Bokma E, van Koningsveld GA, Jeronimus-Stratingh M, Beintema JJ. 1997. Hevamine, a chitinase from the rubber tree Hevea brasiliensis, cleaves peptidoglycan between the C-1 of N-acetylglucosamine and C-4 of N-acetylmuramic acid and therefore is not a lysozyme. FEBS Lett 411:161–163. doi: 10.1016/S0014-5793(97)00682-0. [DOI] [PubMed] [Google Scholar]
- 55.Duret S, André A, Renaudin J. 2005. Specific gene targeting in Spiroplasma citri: improved vectors and production of unmarked mutations using site-specific recombination. Microbiology 151(Pt 8):2793–2803. doi: 10.1099/mic.0.28123-0. [DOI] [PubMed] [Google Scholar]
- 56.Freeman BA, Sissenstein R, McManus TT, Woodward JE, Lee IM, Mudd JB. 1976. Lipid composition and lipid metabolism of Spiroplasma citri. J Bacteriol 125:946–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang KC, Mukhopadhyay R, Wingreen NS. 2006. A curvature-mediated mechanism for localization of lipids to bacterial poles. PLoS Comput Biol 2:e151. doi: 10.1371/journal.pcbi.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carvalho M, Sampaio JL, Palm W, Brankatschk M, Eaton S, Shevchenko A. 2012. Effects of diet and development on the Drosophila lipidome. Mol Syst Biol 8:600. doi: 10.1038/msb.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Béven L, Wróblewski H. 1997. Effect of natural amphipathic peptides on viability, membrane potential, cell shape and motility of Mollicutes. Res Microbiol 148:163–175. doi: 10.1016/S0923-2508(97)87647-4. [DOI] [PubMed] [Google Scholar]
- 60.Pilo P, Vilei EM, Peterhans E, Bonvin-Klotz L, Stoffel MH, Dobbelaere D, Frey J. 2005. A metabolic enzyme as a primary virulence factor of Mycoplasma mycoides subsp. mycoides small colony. J Bacteriol 187:6824–6831. doi: 10.1128/JB.187.19.6824-6831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nappi A, Poirié M, Carton Y.. 2009. The role of melanization and cytotoxic by-products in the cellular immune response of Drosophila against parasitic wasps. Adv Parasitol 70:99–121. doi: 10.1016/S0065-308X(09)70004-1. [DOI] [PubMed] [Google Scholar]
- 62.Gonzalvez F, Gottlieb E. 2007. Cardiolipin: setting the beat of apoptosis. Apoptosis 12:877–885. doi: 10.1007/s10495-007-0718-8. [DOI] [PubMed] [Google Scholar]
- 63.Cleavinger CM, Kim MF, Im JH, Wise KS. 1995. Identification of mycoplasma membrane proteins by systematic TnphoA mutagenesis of a recombinant library. Mol Microbiol 18:283–293. doi: 10.1111/j.1365-2958.1995.mmi_18020283.x. [DOI] [PubMed] [Google Scholar]
- 64.Wróblewski H, Johansson K-E, Hjérten S. 1977. Purification and characterization of spiralin, the main protein of the Spiroplasma citri membrane. Biochim Biophys Acta 465:275–289. doi: 10.1016/0005-2736(77)90079-7. [DOI] [PubMed] [Google Scholar]
- 65.Wróblewski H, Robic D, Thomas D, Blanchard A. 1984. Comparison of the amino acid compositions and antigenic properties of spiralins purified from the plasma membranes of different spiroplasmas. Microbiol, Ann (Paris) 135A:73–82. doi: 10.1016/S0769-2609(84)80061-7. [DOI] [PubMed] [Google Scholar]
- 66.Duret S, Batailler B, Dubrana M-P, Saillard C, Renaudin J, Béven L, Arricau-Bouvery N. 2014. Invasion of insect cells by Spiroplasma citri involves spiralin relocalization and lectin/glycoconjugate-type interactions. Cell Microbiol 16:1119–1132. doi: 10.1111/cmi.12265. [DOI] [PubMed] [Google Scholar]
- 67.Béven L, Duret S, Batailler B, Dubrana M-P, Saillard C, Renaudin J, Arricau-Bouvery N. 2012. The repetitive domain of ScARP3d triggers entry of Spiroplasma citri into cultured cells of the vector Circulifer haematoceps. PLoS One 7:e48606. doi: 10.1371/journal.pone.0048606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Be NA, Bishai WR, Jain SK. 2012. Role of Mycobacterium tuberculosis pknD in the pathogenesis of central nervous system tuberculosis. BMC Microbiol 12:7. doi: 10.1186/1471-2180-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toh H, Weiss BL, Perkin SA, Yamashita A, Oshima K, Hattori M, Aksoy S. 2006. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res 16:149–156. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burke GR, Moran NA. 2011. Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol 3:195–208. doi: 10.1093/gbe/evr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 72.Hamilton PT, Leong JS, Koop BF, Perlman SJ. 2014. Transcriptional responses in a Drosophila defensive symbiosis. Mol Ecol 23:1558–1570. doi: 10.1111/mec.12603. [DOI] [PubMed] [Google Scholar]
- 73.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download
Extensive genome rearrangements among closely related Spiroplasma species. The color blocks represent regions of homologous backbone sequences without rearrangement among the genomes compared (top, S. citri GII-3X; middle, S. poulsonii MRSO; bottom, S. melliferum IPMB4A). The vertical red bars indicate the boundaries of individual contigs. The average nucleotide sequence identities were calculated on the basis of single-copy genes that were conserved among the three genomes compared in each group. Download
Phylogenetic tree of spiralin and ARP amino acid sequences. Phylogenetic analysis of whole protein sequences with the MUSCLE algorithm (multiple-sequence comparison by log expectation). Shown are the spiralin (A) and ARP (B) sequences of closely related Spiroplasma species. The number in brackets indicates the contig position. The only functional ARP found in the S. poulsonii genome is ARP S. p. (B); ARP S. p (6) is truncated by a viral insertion; the N-terminal and C-terminal portions were put together for the sequence alignment. GenBank accession numbers are listed in Materials and Methods. Download
List of annotated features.
List of S. poulsonii peptides and proteins identified by proteomics.
List of shared and genome-specific homologous gene clusters. Taps legends: all, all genes; Sch, S. chrysopicola; Sci, S. citri GII-3X; Sme, S. melliferum IPMB4A; Spo, S. poulsonii MSRO; +, presence.