ABSTRACT
Persisters are dormant variants that form a subpopulation of drug-tolerant cells largely responsible for the recalcitrance of chronic infections. However, our understanding of the genetic basis of antibiotic tolerance remains incomplete. In this study, we applied transposon sequencing (Tn-Seq) to systematically investigate the mechanism of aminoglycoside tolerance in Escherichia coli. We constructed a highly saturated transposon library that covered the majority of E. coli genes and promoter regions and exposed a stationary-phase culture to a lethal dose of gentamicin. Tn-Seq was performed to evaluate the survival of each mutant to gentamicin exposure. We found that the disruption of several distinct pathways affected gentamicin tolerance. We identified 105 disrupted gene/promoter regions with a more than 5-fold reduction in gentamicin tolerance and 37 genes with a more than 5-fold increased tolerance. Functional cluster analysis suggests that deficiency in motility and amino acid synthesis significantly diminished persisters tolerant to gentamicin, without changing the MIC. Amino acid auxotrophs, including serine, threonine, glutamine, and tryptophan auxotrophs, exhibit strongly decreased tolerance to gentamicin, which cannot be restored by supplying the corresponding amino acids to the culture. Interestingly, supplying these amino acids to wild-type E. coli sensitizes stationary-phase cells to gentamicin, possibly through the inhibition of amino acid synthesis. In addition, we found that the deletion of amino acid synthesis genes significantly increases gentamicin uptake in stationary phase, while the deletion of flagellar genes does not affect gentamicin uptake. We conclude that activation of motility and amino acid biosynthesis contributes to the formation of persisters tolerant to gentamicin.
IMPORTANCE
Persisters are responsible for the recalcitrance of chronic infections to antibiotics. The pathways of persister formation in E. coli are redundant, and our understanding of the mechanism of persister formation is incomplete. Using a highly saturated transposon insertion library, we systematically analyzed the contribution of different cellular processes to the formation of persisters tolerant to aminoglycosides. Unexpectedly, we found that activation of amino acid synthesis and motility strongly contributes to persister formation. The approach used in this study leads to an understanding of aminoglycoside tolerance and provides a general method to identify genes affecting persister formation.
INTRODUCTION
Persisters form a subpopulation of cells that can survive lethal antibiotic treatment, unlike their genetically identical kin. Persisters are tolerant to different classes of bactericidal antibiotics and are found in all bacterial species tested (1). Several studies have linked persisters to the recalcitrance of chronic disease to antibiotic therapy. We showed that late isolates of Pseudomonas aeruginosa from cystic fibrosis patients have a 100-fold increase in persister levels compared to those of early isolates from the same patients (2). In a Staphylococcus aureus deep-seated mouse biofilm infection model, a surviving persister population was observed after 48 h of treatment with a lethal dose of antibiotics (3). Upon entrance into mammalian cells, the level of persisters in Salmonella enterica serovar Typhimurium sharply increases (4). In urinary tract infection, Escherichia coli forms a dormant intracellular population tolerant to antibiotics (5).
The mechanism of persister formation has been studied for over a decade. Transcriptome analysis of isolated persisters pointed to overexpression of toxin-antitoxin (TA) modules (6, 7). Ectopic overexpression of toxins such as RelE or MazF, which inhibit protein synthesis by cleaving mRNA, increases the level of persisters. Similarly, expression of HipA, a kinase which inhibits protein synthesis by phosphorylating Glu-tRNA synthase, increases tolerance to antibiotics (8–10). However, knockouts of these toxin genes have no phenotype. Maisonneuve and colleagues showed that the deletion of five or more toxin-antitoxins decreases tolerance to both β-lactams and fluoroquinolones 100-fold (11). In some cases, a given toxin is highly expressed and can be primarily responsible for persister formation. Damage of DNA in E. coli by fluoroquinolones induces expression of the tisB toxin that forms an anion channel in the membrane, leading to a drop of proton motive force (PMF) and ATP levels (12, 13). This results in the formation of dormant, drug-tolerant persisters. The deletion of individual toxins diminishes the level of persisters in S. enterica serovar Typhimurium infecting mammalian cells (4). Apart from examining the effects of TAs, whole-genome screening for persister genes has been conducted using transposon libraries, plasmid overexpression libraries, and an open reading frame (ORF) knockout library (14–17). A selection of high persister strains of a plasmid overexpression library resulted in finding that the overexpression of glycerol metabolism genes glpD and plsB leads to increased tolerance to ampicillin and ofloxacin (16). A screen of an E. coli ORF knockout library in stationary phase identified several chaperones and global regulators that affect drug tolerance. These genes include chaperones dnaJ and dnaK, global regulators fis, hns, hnr, and dksA, and genes encoding metabolic enzymes, such as apaH (diadenosine tetraphosphatase), surA (peptidyl-prolyl cis-trans isomerase), ygfA (5-formyl-tetrahydrofolate cyclo-ligase), and yigB (flavin mononucleotide [FMN] phosphatase) (15). These data show that the mechanisms of persister formation are linked to diverse cellular pathways and are redundant.
At the same time, the surveys of persister genes are probably incomplete. There is considerable variation in the level of persister cells among biological replicates, and large-scale screens, performed by necessity with little if any repeats, are likely to miss important components. Ideally, one would want to repeat a whole-genome screen of knockout mutants multiple times. We reasoned that the newly developed transposon sequencing (Tn-Seq) method would facilitate such an approach (18). In a Tn-Seq experiment, a transposon insertion library is exposed to an antibiotic. The survivors are cultured, and deep sequencing of the transposon junctions allows for the identification of mutants that did not survive the treatment. These mutations are in candidate persister genes. In a Tn-Seq library, a given null mutation is represented by multiple transposon insertions into different positions of a gene and its promoter region. A Tn-Seq experiment is then equivalent to multiple repeats of a conventional screen. Importantly, all mutants are in the same population, minimizing errors.
The level of persisters rises sharply as the density of a culture increases, reaching 1 to 2% in stationary-phase E. coli cells, and we were interested in examining tolerance in this phase of growth. Very few antibiotics are capable of killing stationary cells. Aminoglycosides have the capability of killing both growing and nongrowing cells and can be used to investigate the tolerance of persisters in stationary state. Aminoglycosides, such as streptomycin, kanamycin, gentamicin, and tobramycin, are widely used in the clinic to treat infections caused by Gram-negative bacteria (19, 20). Aminoglycosides generally bind to the 30S ribosomal subunit and impair translational proofreading. This leads to misreading and premature termination of mRNA and causes accumulation of aberrant proteins. These proteins are then incorporated into the bacterial cell membrane and cause membrane leaks and cell death (21). Aminoglycoside uptake is dependent on the proton motive force through an unknown mechanism (22). In this study, we utilized a highly saturated Tn-Seq library to systematically study the tolerance of persisters to gentamicin in a stationary culture of E. coli.
RESULTS
Tn-Seq of gentamicin tolerance.
Persister analysis by Tn-Seq is shown schematically in Fig. 1A. A highly saturated mini-Tn10 (mTn10) insertion library in E. coli MG1655 containing ~200,000 unique mutants was generated for this study. Each mutant contains one transposon randomly inserted in the chromosome. The library was grown to stationary phase in morpholinepropanesulfonic acid (MOPS) medium supplemented with glucose and Casamino Acids. An aliquot of ~107 cells was plated, and the resulting colonies represent the input pool. The remaining stationary-phase culture was challenged with gentamicin (50 µg/ml, 500× MIC). The killing rate decreased sharply over time, indicating the existence of surviving persisters (Fig. 1B). After 6 h of incubation with gentamicin, persisters were washed and plated. Both the input and output samples were allowed to recover on plates rather than in a liquid culture to limit competition. Colonies were pooled from the input and output plates, and genomic DNA was extracted. The experiment was performed with three biological replicates to ensure reproducibility. The insertion position and frequency of each transposon mutation were determined by Illumina sequencing. The survival rate of each mutant was calculated as the frequency in the output pool versus the frequency in the input pool.
FIG 1 .
(A) Work flow of Tn-Seq for gentamicin tolerance. The E. coli MG1655 transposon library was constructed and pooled. The mutant pool was inoculated at 1:1,000 in MOPS-0.2% glucose-0.2% CAA medium and grown for 16 h to reach stationary phase. Before gentamicin challenge, an aliquot was removed and plated on LB as the input. Gentamicin was added to a stationary-phase culture at 500× MIC (50 µg/ml). After a 6-h challenge, cells were washed and plated on LB agar to recover persisters as the output. Genomic DNA was extracted from the input and output for deep sequencing. Insertion mutations in the same genes were added, and the insertion frequency for each gene was calculated. The survival index for each gene was counted as the frequency of output/input. (B) Gentamicin killing kinetics of E. coli MG1655 stationary-phase culture. (C) Insertion distribution of input. The x axis represents genome position, and the y axis represents the cumulative number of unique Tn insertions. Insertions evenly distributed throughout the genome would generate a linear pattern.
The input pool consisted of ~200,000 unique transposon insertions, yielding one insert per 20 bp on average, showing that this is a highly saturated library. The cumulative numbers of unique Tn insertions over the genome were plotted, which yielded a nearly linear relationship, indicating random distribution of insertions and the absence of large gaps or hot spots (Fig. 1C).
The insertion site of each mutant was mapped to the gene/intergenic region using custom scripts. We mapped ~200,000 insertions to 8,253 genes/intergenic regions, which would give us an average of ~25 different transposon mutants for each gene/intergenic region and thus increase the reliability of determining the survival rate for each gene. To reduce the occurrence of false negatives, only genes with 3 or more insertion sites in the input library are included in the analysis. There are ~800 genes that have less than 3 or no insertion sites in the input pool (see Data Set S1 in the supplemental material). Out of the ~800 genes, ~500 have been previously reported to be essential, encoding tRNA, ribosomal proteins, and cell wall synthesis proteins. There are also ~300 nonessential genes with less than three inserts in the input. Given the high coverage of our transposon library, these genes are likely to have an important function for growth or stationary-phase survival. Mutants with insertions in these genes could be outcompeted by other strains during growth or entry into stationary phase. Notably, several genes that we previously reported to affect persister formation, based on a screen of an E. coli knockout library, dnaKJ and hupB, have fewer than three inserts (15). Deletion of these genes does not lead to growth defects when mutants grow individually, but we speculate the mutants are outcompeted by other strains in stationary phase. These genes were excluded from further analysis.
There are 3,712 genes and 1,201 promoter regions that contain 3 or more insertion sites in the input pool. We used these genes for further analysis. We calculated predicted reads for each gene based on the length of the gene and total reads number in the library. Dval genome was defined as actual reads of each gene divided by predicted reads. The survival index (SI) for antibiotic challenge was defined as the Dval genome output divided by the Dval genome input. The survival index reflects a change in the frequency of certain mutants after gentamicin treatment. A neutral mutation gives an SI of 1. Mutants that have higher survival give an SI higher than 1, and mutants with lower survival produce an SI smaller than 1 (Fig. 1A). A full list of SI values for all genes analyzed is provided in Data Set S2 in the supplemental material.
We compared SI from independent replicates to evaluate the reproducibility of our experiment. We did pairwise linear regression on the SI of all mutants among our independent triplicates. We first calculated log2 SI, which gives positive values for mutants with increased survival and negative values for mutants with decreased survival. Mutants with the same fold change of survival have the same absolute value of log2 SI. A neutral mutant that has the same frequency in output and input has an SI equal to 1 and log2 SI equal to 0. We plotted the log2 SI of triplicate 1 on the x axis and the log2 SI of triplicate 2 on the y axis, and each dot represents one mutant (Fig. 2A). All three pairwise regressions generate a linear pattern, with the coefficient of determinations of 0.87, 0.77, and 0.73, indicating that the three independent replicates are highly consistent.
FIG 2 .

(A) Pairwise linear regression of log2 SI of each mutant between replicate 1 and 2 of the triplicates. Each dot on the plot represents one mutant. The x axis represents log2 SI (output Dval genome/input Dval genome) in triplicate 1, and the y axis represents log2 SI in triplicate 2. An ideal robust repeat would generate a linear pattern. (B) Histogram of log2 SI. x axis represents log2 SI range, and y axis represents total counts of genes/intergenic regions in the range.
To evaluate the overall distribution of survival indexes, we plotted a histogram of log2 SI with the x axis representing log2 SI and the y axis representing total counts of genes/intergenic regions (Fig. 2B). We found that the arithmetic mean of log2 SI equals 0.008, corresponding to an SI of 1.005, close to a neutral value. This indicates that most of the genes have little or no effect on persister formation, so the cumulative effect of all mutants is nearly neutral. The histogram has two tail ends starting at log2 SI equals to 2, corresponding to a 5-fold change in survival. In total, there are 142 genes/intergenic regions that have more than a 5-fold change in persister level (the tail ends on the histogram). Insertions to the majority of genes/intergenic regions (4,171) have less than 5-fold changes in persister levels. We used 5-fold as the cutoff for genes deemed to have a substantial effect on persister formation.
We used EcoCyc to annotate the 37 genes/promoter regions with an SI of >5 and 105 genes with an SI of <0.2 (see Tables S2 and S3 in the supplemental material; P value is <0.05 by Student t test) (23). We found that many of these hits clustered in the same pathways, indicating the high level of reliability of the method (see Tables S2 and S3). Among the mutants that exhibit significant increased survival, we identified the nuo operon (nuoBGLM) (see Table S2). The nuo operon encodes the NADH:ubiquinone oxidoreductase, the first coupling site of the electron transport chain. The ubi gene, which encodes a ubiquinone biosynthesis protein, was also identified among the high-tolerance group. These results demonstrate that disruption of the electron transport chain leads to increased gentamicin survival. Aminoglycoside uptake requires proton motive force (PMF); therefore, disruption of the electron transport chain (ETC) would block gentamicin uptake and increase survival. It has been reported that ubiquinone-deficient strains have an increased minimum inhibitory concentration (MIC) to gentamicin (24). We also found that mutations in the regulators of anaerobic metabolism, fnr, arcB, and appY, decreased tolerance to gentamicin. This is probably due to the inability of stationary-phase cells in a low-oxygen environment to properly switch to anaerobic respiration. As a result, the PMF will decrease, producing increased tolerance to gentamicin. Identification of these genes serves as a positive control and further validates our method.
Flagellar genes are important for gentamicin tolerance.
Strikingly, in the decreased gentamicin survival group, we found 39 genes/promoter regions related to flagellar formation, including flgABCDEFGHIJKL, flhABCD, fliAEFGHIJKLMNOPQR, and promoter regions of flgB, flgG, flhB, fliF, and flhC (Table S3). This was unexpected, as these genes have never been implicated in persister formation to our knowledge. Among these genes, flhCD is the master regulator of flagellar formation and controls the expression of all flagellar genes. fliA is the sigma factor of the flagellar regulon. The flhAB genes together with the fliOPQRHIJ operon make up the export apparatus, while fliEFGMN and flgBCFGHI form the basal body. flgA is a chaperone for P ring assembly. flgKL encode joint proteins that connect flagellin to the basal body, and flgJ and flgD participate in joint assembly. Mutations in the stator protein encoded by motAB also resulted in decreased survival. In addition, the quorum sensing response regulator qseC showed decreased tolerance. It has been reported that in enterohemorrhagic E. coli (EHEC), QseC autophosphorylates and activates flhCD in response to the autoinducer AI-3 (25). Deletion of qseC does not affect the assembly of flagella but might delay the activation of flhCD and motility. We found that the gentamicin MIC was unchanged in flagellar mutants.
To confirm these phenotypes, we used single deletion mutants in competition with a lacZ mutant. This method was chosen to best emulate the Tn-Seq conditions, where all mutants were grown to stationary phase in the same tube and challenged with antibiotic simultaneously. Strains carrying single deletions were mixed with the MG1655ΔlacZ strain at a 1:1 ratio and inoculated into fresh medium at 1:1,000. The mixed culture was grown to stationary phase and challenged with gentamicin. The survival of each strain was measured following gentamicin challenge. The ratios of the two strains before and after challenge are determined by plating on MacConkey agar containing lactose. We calculated the relative survival as the ratio of output divided by the ratio of input. In theory, the relative survival of each mutant in the competition assay should equal survival in Tn-Seq. We confirmed that the relative survival of MG1655 to survival of the MG1655ΔlacZ strain equals 1, which serves as a control for the assay (Fig. 3A). A nuoL mutant that exhibits a high SI and which is known to have high tolerance exhibits a relative survival of 30 after 24 h (Fig. 3A). This mutant served as a positive control. We confirmed the decreased tolerance of flhC, motB, fliL, and fliC by competition assay and confirmed that the deletion of flhC, motB, and fliL leads to decreased survival upon gentamicin challenge (Fig. 3A).
FIG 3 .
(A) Competition assays confirm high hits identified by Tn-Seq. Single-deletion mutants were mixed with the MG1655ΔlacZ strain at a 1:1 ratio, and the mixed culture was grown for 16 h. Gentamicin (50 µg/ml) was added to stationary-phase culture. The aliquot was washed and plated on a MacConkey-lactose plate. Pink and white survivors were enumerated, and the survival of each strain was calculated for each time point (survival after treatment/survival before treatment). The relative survival (mutant survival/wild-type survival) was graphed. Results are an average of 6 biological replicates; error bars represent standard deviation; P value between each mutant and wild type was calculated by Student's t test (*, P < 0.05; **, P < 0.01). (B) Schematic of flagellar components and their effect on tolerance to gentamicin. Genes are shown in relation to the flagellar components, and the corresponding SI is indicated. Disruption of the flagellar regulators, export machinery, basal body, and stator proteins leads to significantly decreased tolerance, in contrast to the modest change in fliCD flagellin mutants.
Next, we sought to determine whether flagellar synthesis or, rather, its rotation plays a role in antibiotic tolerance. The expression of flagellar genes is tightly coupled to the assembly process in a hierarchical order, consisting of early, middle, and late classes (26) (Fig. 3B). The deletion of early-class flagellar genes, such as those coding for the basal body and export machinery, represses the expression of other flagellar genes and stops the assembly of flagella (26). It is notable that in addition to flagellar master regulators and structural genes, insertions in motAB also decrease persisters. The motAB genes form a proton channel and serve as a stator for flagellar rotation. The motAB motor genes belong to the late class of flagellar assembly, so the deletion of motAB still allows the assembly of the flagella (27, 28). Another gene participating in the motor function is fliL. It has been suggested that FliL regulates the output of the stator and controls the flow of ions (26). Interestingly, fliL had a dramatic effect on tolerance (SI ~0.009), followed by motAB (SI ~ 0.152), compared to a more modest effect of a mutation in fliC (SI ~ 0.343). These data suggest that flagellar rotation, besides biogenesis, is important for tolerance.
Amino acid synthesis affects gentamicin tolerance.
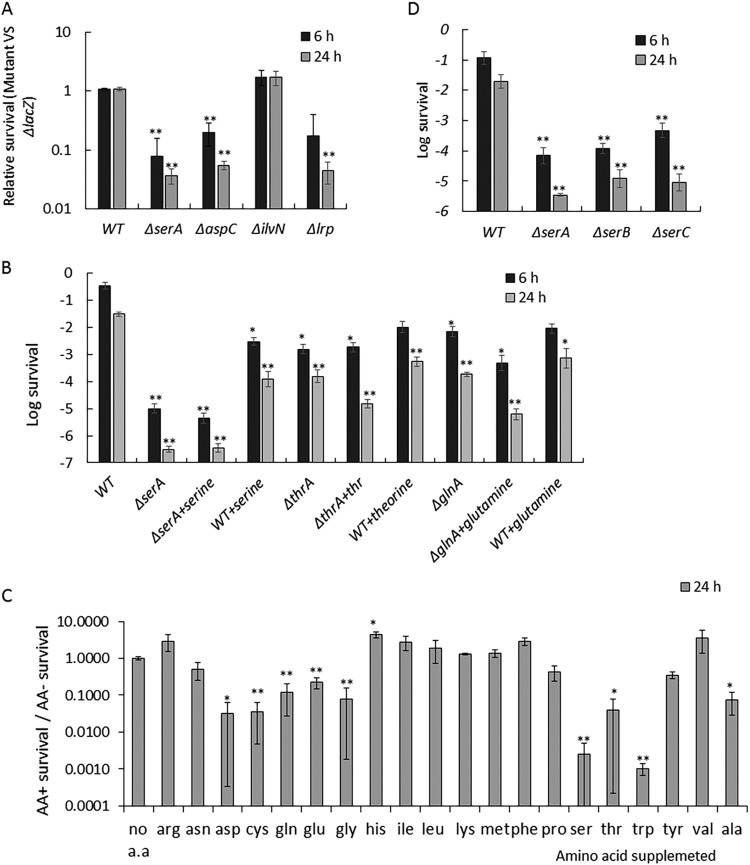
We found that 13 genes in the decreased antibiotic survival group identified by Tn-Seq encode enzymes involved in amino acid biosynthesis. These genes include serAC, which catalyze serine synthesis, aspC, which catalyzes synthesis of aspartate, phenylalanine, and tyrosine, cysK, which catalyzes cysteine biosynthesis, aroADF, which catalyze chorismate synthesis, a precursor of aromatic amino acids, tryptophan pathway trpABCDE, and tyrosine synthesis gene tyrA. Interestingly, we also found the leucine-responsive regulatory protein encoded by lrp in the decreased survival group. Lrp responds to amino acid starvation and upregulates multiple biosynthesis pathways. Decreased tolerance in the lrp mutant is consistent with the finding that deletion of amino acid synthesis genes increases gentamicin tolerance.
We compared the survival index of all amino acid biosynthesis genes. We found that many other amino acid synthesis pathways, including histidine, leucine, isoleucine, and methionine, do not significantly affect tolerance (SI ~ 1), and the ilvABCEGNY isoleucine synthesis pathways exhibit slightly higher tolerance (SI ~ 3; see Table S3 in the supplemental material). We confirmed this phenotype in the competition assay and found that serA, aspC, and lrp but not ilvN showed a decreased tolerance (Fig. 4A). This indicates that the decreased tolerance phenotype is amino acid specific, i.e., not all auxotrophs exhibit low tolerance.
FIG 4 .
Effect of amino acid synthesis on gentamicin tolerance. (A) Competition assays confirm high hits identified by Tn-Seq. Competition assay was performed as described for Fig. 3A. (B) Survival of E. coli in the presence of gentamicin. Gentamicin (50 µg/ml) was added to a stationary-phase culture. Cultures were washed and plated for CFU count. Serine, threonine, or glutamine was added to stationary-phase culture at a final concentration of 10 µg/ml 30 min before addition of gentamicin. (C) Antibiotic survival of stationary-phase cells in the presence of amino acids. The survival assay was performed as described for panel B. The relative survival rate in the presence of each amino acid, i.e., survival rate with amino acid divided by survival rate without amino acid, was graphed. (D) Survival of single deletion of genes of the serine biosynthesis pathway. Bacteria were grown in MOPS-0.2% glucose-0.2% CAA-0.1 mM pyridoxine, and the survival assay was performed as described for panel B. Results are an average of 6 biological replicates; error bars represent standard deviation; P values between each mutant/treatment and wild type without amino acid supplementation were calculated by Student's t test (*, P < 0.05; **, P < 0.01).
Based on the Tn-Seq results, we first speculated that the depletion of specific amino acids, such as serine, leads to decreased tolerance. To test this hypothesis, we separately added serine, threonine, and glutamine into a stationary-phase culture of the corresponding auxotrophs. Surprisingly, the persister level was not restored by supplementing amino acids (Fig. 4B). This suggests that the decreased tolerance in these auxotrophs is not due to a shortage of the amino acid product. In addition, we found that adding these amino acids to wild-type stationary-phase culture sensitizes the cells to gentamicin (Fig. 4B). Supplementing these amino acids would stop amino acid biosynthesis. In addition, we found that supplying serine, glycine, glutamine, tryptophan, threonine, and alanine decreases persister levels to various degrees. Supplementing the remaining amino acids (cysteine, valine, leucine, isoleucine, methionine, proline, phenylalanine, tyrosine, aspartic acid, glutamic acid, asparagine, histidine, lysine, and arginine) does not affect persister formation (Fig. 4C). These results indicate that amino acid starvation per se does not induce tolerance. However, the process of specific amino acid synthesis is important for persister formation.
We then asked the question of whether certain intermediates in the amino acid synthesis pathway are important for gentamicin tolerance. We chose the serine synthesis pathway as an example. Serine biosynthesis requires three enzymes, SerA, SerB, and SerC. We tested the survival of single-deletion serA, serB, and serC mutants. We reasoned that if a certain intermediate plays a crucial role, the downstream genes in the pathway would have a smaller effect. Since the serC mutant also exhibits pyridoxine auxotrophy, pyridoxine was added to the medium to support growth. We found that all three mutants exhibit low persister levels, suggesting that the entire biosynthetic pathway is important to maintain stationary-phase tolerance (Fig. 4D).
We then considered the alternative hypothesis that amino acid biosynthesis may affect the carbon flow and subsequently cellular energy for gentamicin uptake. To test this, we measured gentamicin uptake in the mutants by conjugating gentamicin with fluorescent Texas Red. We found that deletion of serA leads to increased gentamicin-Texas Red uptake in stationary phase (Fig. 5). This suggests that activation of serine synthesis in stationary phase decreases cellular energy, which causes gentamicin tolerance. Interestingly, we did not see a difference in gentamicin-Texas Red uptake between the flhC mutant and the wild type. Deletion of the flagellar master regulator flhC prevents activation of flagellar synthesis and leads to decreased tolerance in stationary phase. This result implies that flagellar activation causes gentamicin tolerance through a different mechanism.
FIG 5 .

Gentamicin uptake in wild type and mutants. Gentamicin-Texas Red conjugate was added to a stationary-phase culture. Uptake of Texas Red and gentamicin-Texas Red was measured by flow cytometry.
The TCA cycle and the ETC contribute to gentamicin tolerance.
We found several tricarboxylic acid (TCA) cycle enzymes in our hits. Insertions into sucC, sucD, mdh, and the intergenic region of sucB-sucC cause increased tolerance. The sucCD genes encode succinyl coenzyme A (succinyl-CoA) synthetase, which catalyzes the reaction of succinyl-CoA to succinate, and mdh encodes malate dehydrogenase, which catalyzes the reaction of malate to oxaloacetate. At the same time, insertions in another gene coding for a TCA cycle enzyme, acnB, significantly decreases gentamicin tolerance (data not shown). The acnB gene encodes aconitase, which catalyzes the conversion of citrate to isocitrate. It is surprising that disruption of different steps of the TCA cycle has opposing effects on tolerance (Fig. 6). Disrupting enzymes that catalyze reactions from oxaloacetate to 2-oxoglutarate leads to a decrease in persisters tolerant to gentamicin. Similarly, insertions which inhibit the glyoxylate cycle enzymes increase tolerance. The opposite is true when disrupting enzymes from 2-oxoglutarate to oxaloacetate (Fig. 6). It is notable that 2-oxoglutarate is a key intermediate of the TCA cycle and is linked to amino acid and other small molecule metabolism. From the heat map, it appears that accumulation of 2-oxoglutarate leads to increased tolerance.
FIG 6 .

Survival index heat map of TCA cycle and the glyoxylate bypass. The fold change of the persister level of each mutant is represented by the color code on the heat map. Disruption of acnA, acnB, and icd leads to decreased tolerance, while disruption of sucAB, sucCD, sdhABCD, fumB, and gltA leads to increased tolerance. In addition, disruption of glyoxylate bypass enzymes encoded by aceA and aceB leads to increased tolerance. Based on these data, it appears that disruption of 2-oxoglutarate synthesis decreases tolerance to gentamicin, while disruption of 2-oxoglutarate consumption increases gentamicin tolerance. The survival indexes of individual TCA enzymes can be found in Table S3 in the supplemental material.
Disruption of individual TA modules or their regulators does not affect gentamicin tolerance.
Previously, it has been reported that cumulative deletion of 10 TA modules decreases tolerance to ciprofloxacin and ampicillin, but single deletions had no effect on persister levels (11). We found that mutations in individual toxins do not have a significant effect on gentamicin survival (SI ~ 1; see Table S3 in the supplemental material), consistent with the previous report. We tested the survival of the 10-TA deletion strain (Δ10) under our screening conditions in the presence of gentamicin. We found an ~100-fold decrease in the level of persisters tolerant to gentamicin in the Δ10 strain comparing to wild type (Fig. 7).
FIG 7 .

Survival of mutants in the presence of gentamicin. Gentamicin (50 µg/ml) was added to a stationary-phase culture. Antibiotic survival assay was performed as described for Fig. 4B.
In E. coli, the activation of toxins is controlled by the degradation of antitoxins through Lon protease. Maisonneuve and coauthors proposed a model in which Lon protease degrades antitoxins and leads to the release of free toxins (29). Lon is activated by polyphosphate, which is synthesized by the kinase Ppk and degraded by the exopolyphosphatase Ppx. The polyphosphate level is in turn controlled by ppGpp through the inhibition of Ppx. It was reported that deletion of Lon protease, double deletion of ppGpp synthase genes relA and spoT, or double deletion of ppx and ppk all exhibit low tolerance to fluoroquinolone and β-lactam. However, we found that the decreased tolerance of a lon deletion resulted from a lack of degradation of sulA, encoding a cell division inhibitor induced by an SOS response. We also did not see an effect of a lon mutation on the level of tolerance to a β-lactam (30). In the current study, we found that disruption of lon does not significantly affect gentamicin tolerance (SI ~ 2), and consistently, disruption of Lon protease activator genes ppk and relA even produces a slight increase in gentamicin tolerance (SI ~ 5). These results indicate that, cumulatively, TAs have an effect on gentamicin tolerance; however, it is not controlled by a ppGpp-polyphosphate-Lon pathway.
Finally we compared the high hits we identified in this screen with the Δ10 strain. We found that in stationary phase, a deletion of serA leads to an ~1,000-fold decrease in persister level, and a deletion of fliL leads to an ~100-fold decrease in persister level (Fig. 7), comparable to the phenotype of the Δ10 strain (100-fold decrease in persister level). This indicates that TA modules are not the main component determining tolerance to gentamicin. The decreased persister levels of the ΔserA and ΔfliL strains can be restored by complementing the mutants with serA or fliL carried on a plasmid (see Fig. S1 in the supplemental material). And these mutants also exhibit a decreased level of persisters tolerant to tobramycin, another aminoglycoside antibiotic (see Fig. S2 in the supplemental material). These findings further confirmed that amino acid biosynthesis and motility play important roles on tolerance to aminoglycoside in stationary phase.
DISCUSSION
In this study, we applied Tn-Seq to systematically identify genes affecting gentamicin tolerance in E. coli. In contrast to the conventional “one pathway controlling function,” such as sporulation, persister formation appears to be highly redundant. Our Tn-Seq screen of mutants tolerant to gentamicin shows that more than 100 genes representing multiple pathways significantly affect gentamicin tolerance. This is consistent with previous findings of redundancy in mechanisms of persister formation and considerably expands the number of contributing genes and proteins. Tn-Seq allowed us to quantitatively evaluate the relative contribution of each gene to tolerance. Several cellular processes, including activation of motility and amino acid synthesis, increase tolerance to gentamicin in stationary phase.
Our most unexpected finding is that motility strongly affects gentamicin tolerance in E. coli. The interruption of flagellar structural genes, master regulator genes, or motility genes decreases tolerance. The most pronounced effect on tolerance was exhibited by mutating motAB and fliL, coding for the stator of the ion channel. These mutants are nonmotile, indicating that the rotation of flagella is important for gentamicin tolerance. Surprisingly, gentamicin uptake does not increase in these mutants. How flagellar formation and rotation affect tolerance to an antibiotic is unclear. This study is the first step toward understanding this intriguing connection. Our observations have interesting clinical implications, as motility is known to be activated during infection and biofilm formation (31). Biofilms contain substantial numbers of stationary cells. It was reported that nonmotile E. coli is outcompeted by the wild type in a mouse model of a urinary tract infection (31). These findings, together with our current study, suggest that motility contributes to both virulence and survival during antibiotic treatment.
We also found that pathways controlling the biosynthesis of amino acids affect gentamicin tolerance. The disruption of serine biosynthesis has the strongest effect on survival in the presence of gentamicin, and we investigated this case in more detail. The addition of serine had no effect on the survival of the serine biosynthesis mutants, showing that the synthesis process rather than the amino acid product is important for tolerance. Gentamicin uptake increases dramatically in the serA mutant. Gentamicin uptake is driven by the PMF; apparently, the energy level increases in the absence of serine biosynthesis, accounting for increased survival of serA mutants. Amino acid and other small molecule biosynthesis is linked to central carbon metabolism, and we speculate that amino acid synthesis changes the carbon flow in central metabolism, affecting the energy state.
Some genes identified in this screen have previously been shown to affect gentamicin sensitivity, such as genes involved in oxidative phosphorylation and respiration. Mutants with these gene deletions act as a positive control for the screen. Some of the genes we detected as hits have been previously reported to affect antibiotic susceptibility. For example, we found that disruption of the capsular polysaccharide regulator gene rcsC, which has been reported to increase β-lactam tolerance, also increases gentamicin tolerance. Disruption of the phage shock regulator gene pspA decreases gentamicin tolerance, consistent with previous reports (32, 33).
It is notable that we did not identify all genes previously reported to affect aminoglycoside tolerance. For example, interruption of the iron sulfur cluster synthesis isc operon and overexpression of dnaKJ have been reported to increase tolerance to aminoglycosides in E. coli (34, 35). We examined our input library and found that both operons do not have inserts. As we grew our transposon library into stationary phase as the input, mutants with strong growth defects would be outcompeted and therefore absent from the current screen.
TA modules play an important role in the formation of persisters (6, 36). Toxins produce a drug-tolerant, dormant state by either inhibiting protein synthesis or decreasing the energy level (13). In order to cause dormancy in a given cell, a TA module needs to be expressed and the toxin-antitoxin balance disrupted in favor of the toxin. Most antitoxins are degraded by the Lon protease in E. coli, and the activity of the protease has been proposed to be the key regulator of persister formation (11). Lon is activated by polyphosphate, which is synthesized by the kinase Ppk and degraded by the exopolyphosphatase Ppx. Polyphosphate is in turn produced under nutrient depletion by RelA/Spot (ppGpp) inhibition of Ppx. Once Ppx is inhibited, the Ppk kinase produces polyphosphate. It has been reported that deletion of lon, double deletion of relA spoT, and double deletion of ppk ppx all diminish persister formation (29). This, however, remains a topic of controversy. We did not find an effect of a lon mutation on persister formation (30). In agreement with that result, we did not find an effect of lon mutations on persister formation in this Tn-Seq study. Considering that we had 8 independent Tn inserts in the lon gene, this experiment was effectively repeated multiple times. We do see a strong drop in persisters tolerant to gentamicin in a mutant with a deletion of 10 RNA interferase (mRNA endonucleases) modules. How a given RNA interferase causes formation of persister cells requires further investigation.
In summary, we applied Tn-Seq to systematically evaluate the mechanism of gentamicin tolerance. We found that many cellular processes affect gentamicin tolerance, and we report their relative contribution by calculating the survival index of each gene. We propose that in stationary phase, the activation of motility and biosynthetic pathways decrease the cellular energy state and thus highly diminish gentamicin sensitivity. These findings have important implications for clinical aminoglycoside tolerance. We also found other genes and pathways that strongly affect gentamicin tolerance, and further study is warranted to elucidate their mechanisms. In addition, our method gives very similar survival indexes for genes in the same pathways, demonstrating that the methodology is quantitative. The platform we developed in this study can be used as a general method to study processes involving multiple pathways, such as in vivo antibiotic tolerance.
MATERIALS AND METHODS
Strains and growth conditions.
Strains used in this study are listed in Table S1 in the supplemental material. E. coli single-deletion strains were constructed in the MG1655 background by P1 transduction from the Keio collection (37) and confirmed by PCR. Cells were cultured in Luria-Bertani broth (LB) or MOPS medium supplemented with 0.2% glucose and 0.2% Casamino Acids (38) at 37°C with aeration at 220 rpm unless specified otherwise.
Construction of the E. coli transposon library.
A transposon library was constructed for E. coli MG1655 using Tn10, which randomly inserts into the sites of TA bases. Briefly, the pDL1093 plasmid containing a heat-sensitive origin of replication (ORI) with a chloramphenicol (Cam)-resistant cassette and Tn10 with a kanamycin (Kan)-resistant cassette was transformed into MG1655. Transformants were recovered at 30°C in the presence of chloramphenicol (5 µg/ml) to maintain the plasmid. The transformants were then plated on LB agar and grown at 42°C in the presence of kanamycin (50 µg/ml) for 12 h to select transposon insertion mutants. Approximately 200,000 colonies were recovered and scraped from 20 150-mm petri dishes. Each colony represents a unique insertion site in the genome, producing a coverage of 20 bp/insert. The colonies were thoroughly homogenized by vortexing and stored at −80°C in LB-30% glycerol.
Tn-Seq sample preparation.
Ten microliters of E. coli pooled transposon library glycerol stock containing ~107 CFU was inoculated into 10 ml MOPS-0.2% glucose-0.2% Casamino acids (CAA) medium in duplicate. Cultures were grown at 37°C, 220 rpm, for 16 h to stationary phase (~4 × 109 CFU). A 25-µl aliquot (~1 × 107 CFU) was removed and plated on LB agar (10 90-mm petri dishes), and this population was termed the input. Gentamicin was added to the remaining culture at a final concentration of 50 µg/ml (500× MIC) and incubated at 37°C, 220 rpm, for 6 h to kill regular cells, leaving surviving persisters. At 6 h, an aliquot of 250 µl (~1 × 107 CFU) was washed 3 times with phosphate-buffered saline (PBS) and plated onto LB agar (10 150-mm petri dishes) to recover persisters. The population was termed the output. The plates were incubated for 12 h at 37°C for both input and output. Colonies for each sample were scraped in 20 ml LB and mixed thoroughly by vortexing. Genomic DNA was extracted from a 1-ml aliquot (~5 × 109 CFU) using a DNeasy Blood & Tissue Kit (Qiagen) and subjected to deep sequencing analysis.
Library sequencing.
Illumina sequencing was carried out as previously described (39). Briefly, genomic DNA was sonicated to produce 200- to 600-bp fragments. A poly(C) tail was added to DNA fragments by using terminal deoxynucleotidyl transferase. A first round of PCR with one transposon-specific primer and one oligo(dG) primer was conducted to amplify the fragments. A second round of PCR with a nested primer was conducted to add an adaptor and bar code for Illumina sequencing. The sequencing was conducted on 8 lanes of Illumina HiSeq 2000 at The Broad Institute, Cambridge, MA.
Data analysis.
Read mapping and calculation were carried out on the Tufts University Galaxy server as described previously (39). Briefly, Illumina reads were aligned to the E. coli MG1655 genome sequence using bowtie (40). The Tufts Galaxy server custom script was applied to enumerate a read number for each insertion site. The insertion sites were then aggregated by annotated genes. In order to fully cover all genes and promoter regions, a GenBank file containing all intergenic regions was generated to allow insertion sites to aggregate to an intergenic region. The predicted number of reads for each gene was calculated based on the length of the gene and the total number of reads in the library. Dval genome for each gene was defined as actual number of reads divided by predicted number of reads. The Dval genome evaluates the frequency of the gene in a library. The survival index was defined as the Dval genome output/input.
Antibiotic survival assay.
Bacteria were inoculated at 1:1,000 in 2 ml MOPS-0.2% glucose-0.2% CAA or LB medium as indicated and grown for 16 h at 37°C, 220 rpm, to reach stationary phase. Gentamicin (50 µg/ml) was added to stationary-phase culture. For amino acid supplementation experiments, the single amino acid was added to a stationary-phase culture 30 min before antibiotics at a final concentration of 10 µg/ml. At 0 h, 6 h, and 24 h, cultures were washed with PBS twice and plated on LB agar for CFU count. The survival rate is calculated as CFU at 6 h or 24 h divided by CFU at 0 h. Results are averages from 6 biological replicates, and error bars represent standard deviations. P values were calculated by the Student's t test between the mutant and wild type. * indicates a P value of <0.05, and ** indicates a P value of <0.01. MICs were determined by the broth microdilution method, as previously reported (41).
Competition assay.
Cultures of single-deletion mutants and the MG1655ΔlacZ strain were grown separately in LB for 16 h. The MG1655ΔlacZ strain was used as the wild type and was mixed with mutant strains at a 1:1 ratio and inoculated in MOPS-0.2% glucose-0.2% CAA at 1:1,000 for 16 h before the addition of gentamicin (50 µg/ml). An aliquot was removed at 0 h, 6 h, and 24 h, washed twice with PBS, and plated on a MacConkey-1% lactose agar plate. MG1655ΔlacZ appears as white colonies and the mutant strain appears as pink colonies. Pink and white colonies were counted separately. The survival for each strain was calculated separately. Relative survival was calculated as the survival of the mutant divided by the survival of the MG1655ΔlacZ strain. Results are averages from 6 biological replicates; error bars represent standard deviations; P value was calculated by the Student t test between the mutant and wild type. * indicates a P value of <0.05, and ** indicates a P value of <0.01.
Gentamicin-Texas Red uptake.
Gentamicin-Texas Red was made as previously described (42). Texas Red-succinimidyl ester (Invitrogen) was dissolved in high-quality anhydrous N,N-dimethylformamide at 4°C at a final concentration of 20 mg/ml. Gentamicin was dissolved in 100 mM K2CO3, pH 8.5, at a final concentration of 10 mg/ml. At 4°C, 10 µl of Texas Red was slowly added to 350 µl gentamicin solution to allow a conjugation reaction. The gentamicin-Texas Red product from the reaction was used for the uptake experiment. Gentamicin uptake was measured by incubating gentamicin-Texas Red (final concentration of 50 µg/ml) with stationary-phase cells for 4 h at 37°C, 220 rpm. Five hundred microliters of culture was then washed twice with PBS and analyzed on a BD FACS Aria II flow cytometer with mCherry voltage, 10,000 recorded events per sample. Graphs were generated using FlowJo.
SUPPLEMENTAL MATERIAL
Genes that have fewer than 3 transposon inserts in the input. Download
Full list of survival indexes of all genes covered by the screen. Download
Complementing deletion strains with a plasmid copy of the gene. Gentamicin survival assay was performed in ΔserA and ΔfliL strains carrying either an empty vector or a plasmid carrying the corresponding gene coding region under an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter. Cells were grown to stationary phase in the presence of 0.1 mM IPTG. Gentamicin (50 µg/ml) was added, and antibiotic survival assay was performed as described for Fig. 4B. P values between strains carrying empty vector and gene coding regions were calculated by the Student t test. Download
Tobramycin tolerance of mutants. Tobramycin (50 µg/ml) was added to a stationary-phase culture. Antibiotic survival assay was performed as described in the legend to Fig. 4B. Download
Strains used in this study.
Mutants with increased survival. Mutants that have 5-fold-increased survival are reported in the table.
Mutants with decreased survival. Mutants that have 5-fold-decreased survival are reported in the table.
ACKNOWLEDGMENTS
The study was supported by NIH grant T-R01AI085585 to K.L. and NIH grant AI055058 to A.C.
We thank Kenn Gerdes for providing the Δ10 strain and the Broad Institute for conducting Illumina sequencing.
Footnotes
Citation Shan Y, Lazinski D, Rowe S, Camilli A, Lewis K. 2015. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. mBio 6(2):e00078-15. doi:10.1128/mBio.00078-15.
REFERENCES
- 1.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 2.Mulcahy LR, Burns JL, Lory S, Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol 192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K. 2013. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. 2014. Internalization of salmonella by macrophages induces formation of nonreplicating persisters. Science 343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 6.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol 186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Germain E, Castro-Roa D, Zenkin N, Gerdes K. 2013. Molecular mechanism of bacterial persistence by HipA. Mol Cell 52:248–254. doi: 10.1016/j.molcel.2013.08.045. [DOI] [PubMed] [Google Scholar]
- 9.Kaspy I, Rotem E, Weiss N, Ronin I, Balaban NQ, Glaser G. 2013. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat Commun 4:3001. doi: 10.1038/ncomms4001. [DOI] [PubMed] [Google Scholar]
- 10.Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. 2009. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323:396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A 108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Gurnev PA, Ortenberg R, Dörr T, Lewis K, Bezrukov SM. 2012. Persister-promoting bacterial toxin TisB produces anion-selective pores in planar lipid bilayers. FEBS Lett 586:2529–2534. doi: 10.1016/j.febslet.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dörr T, Vulić M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol 8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Y, Coates AR. 2005. Transposon mutagenesis identifies genes which control antimicrobial drug tolerance in stationary-phase Escherichia coli. FEMS Microbiol Lett 243:117–124. doi: 10.1016/j.femsle.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 15.Hansen S, Lewis K, Vulić M. 2008. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob Agents Chemother 52:2718–2726. doi: 10.1128/AAC.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spoering AL, Vulic M, Lewis K. 2006. GlpD and PlsB participate in persister cell formation in Escherichia coli. J Bacteriol 188:5136–5144. doi: 10.1128/JB.00369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernier SP, Lebeaux D, DeFrancesco AS, Valomon A, Soubigou G, Coppée JY, Ghigo JM, Beloin C. 2013. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet 9:e1003144. doi: 10.1371/journal.pgen.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Begg EJ, Barclay ML. 1995. Aminoglycosides—50 years on. Br J Clin Pharmacol 39:597–603. [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson J, Chen C, Buising K. 2013. Aminoglycosides: how should we use them in the 21st century? Curr Opin Infect Dis 26:516–525. doi: 10.1097/QCO.0000000000000012. [DOI] [PubMed] [Google Scholar]
- 21.Davis BD. 1987. Mechanism of bactericidal action of aminoglycosides. Microbiol Rev 51:341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taber HW, Mueller JP, Miller PF, Arrow AS. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev 51:439–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martínez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, Latendresse M, Muñiz-Rascado L, Ong Q, Paley S, Schröder I, Shearer AG, Subhraveti P, Travers M, Weerasinghe D, Weiss V, Collado-Vides J, Gunsalus RP, Paulsen I, Karp PD. 2013. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res 41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryan LE, Van Den Elzen HM. 1977. Effects of membrane-energy mutations and cations on streptomycin and gentamicin accumulation by bacteria: a model for entry of streptomycin and gentamicin in susceptible and resistant bacteria. Antimicrob Agents Chemother 12:163–177. doi: 10.1128/AAC.12.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. 2006. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci U S A 103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belas R. 2014. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol 22:517–527. doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Terashima H, Kojima S, Homma M. 2008. Flagellar motility in bacteria structure and function of flagellar motor. Int Rev Cell Mol Biol 270:39–85. doi: 10.1016/S1937-6448(08)01402-0. [DOI] [PubMed] [Google Scholar]
- 28.Macnab RM. 1992. Genetics and biogenesis of bacterial flagella. Annu Rev Genet 26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 29.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 30.Theodore A, Lewis K, Vulic M. 2013. Tolerance of Escherichia coli to fluoroquinolone antibiotics depends on specific components of the SOS response pathway. Genetics 195:1265–1276. doi: 10.1534/genetics.113.152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright KJ, Seed PC, Hultgren SJ. 2005. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect Immun 73:7657–7668. doi: 10.1128/IAI.73.11.7657-7668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laubacher ME, Ades SE. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J Bacteriol 190:2065–2074. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vega NM, Allison KR, Khalil AS, Collins JJ. 2012. Signaling-mediated bacterial persister formation. Nat Chem Biol 8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ezraty B, Vergnes A, Banzhaf M, Duverger Y, Huguenot A, Brochado AR, Su SY, Espinosa L, Loiseau L, Py B, Typas A, Barras F. 2013. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science 340:1583–1587. doi: 10.1126/science.1238328. [DOI] [PubMed] [Google Scholar]
- 35.Goltermann L, Good L, Bentin T. 2013. Chaperonins fight aminoglycoside-induced protein misfolding and promote short-term tolerance in Escherichia coli. J Biol Chem 288:10483–10489. doi: 10.1074/jbc.M112.420380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moyed HS, Bertrand KP. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol 155:768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, Hu LT. 2012. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics 13:578. doi: 10.1186/1471-2164-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrews JM. 2001. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48(Suppl 1):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 42.Saito H, Inui K, Hori R. 1986. Mechanisms of gentamicin transport in kidney epithelial cell line (LLC-PK1). J Pharmacol Exp Ther 238:1071–1076. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes that have fewer than 3 transposon inserts in the input. Download
Full list of survival indexes of all genes covered by the screen. Download
Complementing deletion strains with a plasmid copy of the gene. Gentamicin survival assay was performed in ΔserA and ΔfliL strains carrying either an empty vector or a plasmid carrying the corresponding gene coding region under an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter. Cells were grown to stationary phase in the presence of 0.1 mM IPTG. Gentamicin (50 µg/ml) was added, and antibiotic survival assay was performed as described for Fig. 4B. P values between strains carrying empty vector and gene coding regions were calculated by the Student t test. Download
Tobramycin tolerance of mutants. Tobramycin (50 µg/ml) was added to a stationary-phase culture. Antibiotic survival assay was performed as described in the legend to Fig. 4B. Download
Strains used in this study.
Mutants with increased survival. Mutants that have 5-fold-increased survival are reported in the table.
Mutants with decreased survival. Mutants that have 5-fold-decreased survival are reported in the table.