ABSTRACT
The pathways involved in aromatic compound oxidation under perchlorate and chlorate [collectively known as (per)chlorate]-reducing conditions are poorly understood. Previous studies suggest that these are oxygenase-dependent pathways involving O2 biogenically produced during (per)chlorate respiration. Recently, we described Sedimenticola selenatireducens CUZ and Dechloromarinus chlorophilus NSS, which oxidized phenylacetate and benzoate, two key intermediates in aromatic compound catabolism, coupled to the reduction of perchlorate or chlorate, respectively, and nitrate. While strain CUZ also oxidized benzoate and phenylacetate with oxygen as an electron acceptor, strain NSS oxidized only the latter, even at a very low oxygen concentration (1%, vol/vol). Strains CUZ and NSS contain similar genes for both the anaerobic and aerobic-hybrid pathways of benzoate and phenylacetate degradation; however, the key genes (paaABCD) encoding the epoxidase of the aerobic-hybrid phenylacetate pathway were not found in either genome. By using transcriptomics and proteomics, as well as by monitoring metabolic intermediates, we investigated the utilization of the anaerobic and aerobic-hybrid pathways on different electron acceptors. For strain CUZ, the results indicated utilization of the anaerobic pathways with perchlorate and nitrate as electron acceptors and of the aerobic-hybrid pathways in the presence of oxygen. In contrast, proteomic results suggest that strain NSS may use a combination of the anaerobic and aerobic-hybrid pathways when growing on phenylacetate with chlorate. Though microbial (per)chlorate reduction produces molecular oxygen through the dismutation of chlorite (ClO2−), this study demonstrates that anaerobic pathways for the degradation of aromatics can still be utilized by these novel organisms.
IMPORTANCE
S. selenatireducens CUZ and D. chlorophilus NSS are (per)chlorate- and chlorate-reducing bacteria, respectively, whose genomes encode both anaerobic and aerobic-hybrid pathways for the degradation of phenylacetate and benzoate. Previous studies have shown that (per)chlorate-reducing bacteria and chlorate-reducing bacteria (CRB) can use aerobic pathways to oxidize aromatic compounds in otherwise anoxic environments by capturing the oxygen produced from chlorite dismutation. In contrast, we demonstrate that S. selenatireducens CUZ is the first perchlorate reducer known to utilize anaerobic aromatic degradation pathways with perchlorate as an electron acceptor and that it does so in preference over the aerobic-hybrid pathways, regardless of any oxygen produced from chlorite dismutation. D. chlorophilus NSS, on the other hand, may be carrying out anaerobic and aerobic-hybrid processes simultaneously. Concurrent use of anaerobic and aerobic pathways has not been previously reported for other CRB or any microorganisms that encode similar pathways of phenylacetate or benzoate degradation and may be advantageous in low-oxygen environments.
INTRODUCTION
After carbohydrates, aromatic compounds are the most abundant class of organic compounds found in nature (1) and occur naturally in lignin, flavonoids, quinones, and some amino acids. Many aromatic compounds, including components of crude oil and fossil fuels, are considered major environmental pollutants (1, 2), and therefore, their detection and removal are of interest. Despite the high stability conferred by the resonance energy of the aromatic ring (150 kJ·mol benzene−1), microorganisms have evolved that can degrade most naturally occurring aromatic compounds in both oxic and anoxic environments (3). Under oxic conditions, microorganisms utilize oxygen as both a terminal electron acceptor and a cosubstrate for oxygenases to activate and cleave the aromatic ring (3, 4). In anoxic environments, aromatic degradation proceeds via coenzyme A (CoA) activation, reductive dearomatization of the ring, and hydrolytic cleavage (3, 4). A third, novel, pathway that combines aspects of both the aerobic and anaerobic catabolic routes has been recently elucidated, and its use under low or fluctuating oxygen conditions was postulated (3–5). In this pathway, known as the aerobic-hybrid pathway, intermediates are processed as CoA thioesters (similar to anaerobic pathway intermediates), but dearomatization of the aromatic ring involves an epoxidation reaction that requires molecular oxygen (5). Finally, the ring is hydrolytically cleaved (3–5).
Phenylacetate is found in the environment as a common carbon source and is a central intermediate in the degradation of many aromatic compounds such as phenylalanine, phenylacetaldehyde, 2-phenylethylamine, phenylacetyl esters, lignin-related phenylpropane units, phenylalkanoic acids with an even number of carbon atoms, and environmental contaminants like styrene and ethylbenzene (5–7). Although the anaerobic pathway of phenylacetate degradation in bacteria is well characterized (1, 4, 8, 9), the aerobic pathway has only recently been discovered (3–5). Unlike aerobic phenylacetate degradation in fungi, in which hydroxylases convert phenylacetate to homogentisate (10–12), the novel bacterial aerobic-hybrid pathway proceeds through CoA-dependent activation, epoxidation of the aromatic ring, and hydrolytic ring cleavage (4, 5). To date, this hybrid pathway is the only known aerobic pathway used by bacteria in the degradation of phenylacetate (4, 5). The production of phenylacetyl-CoA as an intermediate in both the anaerobic and aerobic-hybrid pathways is an efficient and rapid way to respond to oxygen fluctuations in the environment, as the phenylacetyl-CoA intermediate can be routed to either pathway, depending on the concentration of oxygen (4, 13). This is also true of the anaerobic and aerobic-hybrid pathways of benzoate degradation, both of which produce benzoyl-CoA as a key intermediate (4).
Perchlorate-reducing bacteria (PRB) and chlorate-reducing bacteria (CRB) are microorganisms that can utilize perchlorate (ClO4−) or chlorate (ClO3−) as a terminal electron acceptor. While PRB can reduce both perchlorate and chlorate via the perchlorate reductase enzyme (Pcr), CRB only reduce the latter with the chlorate reductase (Clr) (14). Pcr and Clr reduce (per)chlorate and chlorate, respectively, to chlorite (ClO2−), which is subsequently dismutated into molecular O2 and Cl− by the chlorite dismutase (Cld) enzyme present in both PRB and CRB (15). Chlorite dismutation is the only chemotrophic microbial metabolism shown to produce significant amounts of free O2 (15). While chemotrophic nitrous oxide disproportionation could putatively produce molecular O2 and this reaction has been invoked in anaerobic methane oxidation coupled to nitrate reduction (16), no enzyme is known that can carry out this reaction.
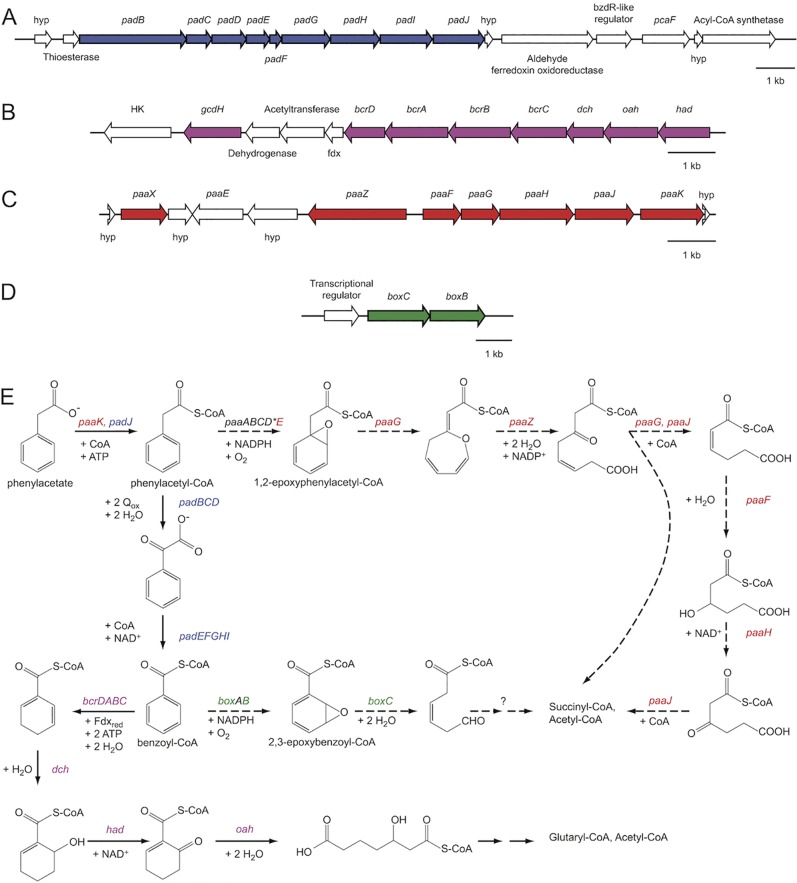
The degradation of aromatic compounds coupled to the electron acceptor (per)chlorate has only been shown to occur via aerobic pathways that exploit the biogenic O2 from CLO2− dismutation not only for respiration but also as a cosubstrate for oxygenases in anoxic environments (15, 17–19). However, the recently described PRB Sedimenticola selenatireducens CUZ and the closely related CRB Dechloromarinus chlorophilus NSS possess genes that encode both anaerobic and aerobic-hybrid pathways of phenylacetate and benzoate degradation (C. I. Carlström et al., in press) (Fig. 1; see Fig. S1 in the supplemental material). The anaerobic phenylacetate pathway is encoded by two gene clusters, the pad cluster (Fig. 1A; see Fig. S1A), which converts phenylacetate to benzoyl-CoA (Fig. 1E), and the bcr, oah, dch, and had cluster (here referred to as the bcr cluster) (Fig. 1B; see Fig. S1B), which degrades benzoyl-CoA under anoxic conditions (Fig. 1E) (1, 4). Genes involved in the aerobic-hybrid pathways of phenylacetate (paa) and benzoate (box) degradation (Fig. 1E) (1, 5) were also present in the genomes of both strains (Fig. 1C; see Fig. S1C). However, in the case of phenylacetate, all of the genes encoding the key oxygenase of the system, phenylacetyl-CoA epoxidase (paaABCDE), were missing except paaE, which likely functions as an iron-sulfur oxidoreductase that transfers electrons from NADPH to the active center of the oxygenase complex (5). Detailed information about these genes is provided in the legend to Fig. 1; for the locus tags, see Table S1 in the supplemental material.
FIG 1 .
Genes and pathways of phenylacetate and benzoate degradation in S. selenatireducens CUZ. (A) pad gene cluster. (B) bcr gene cluster. (C) paa gene cluster. (D) box gene cluster. (E) Pathways of phenylacetate and benzoate degradation in strain CUZ. Red, paa gene cluster involved in the aerobic-hybrid pathway of phenylacetate degradation; blue, pad gene cluster involved in anaerobic phenylacetate degradation; purple, bcr gene cluster involved in anaerobic benzoate and phenylacetate degradation; green, box gene cluster involved in the aerobic-hybrid pathway of benzoate degradation; *, gene not found in strain CUZ. The anaerobic phenylacetate pathway is encoded by two gene clusters, the pad cluster (A; see Fig. S1A in the supplemental material) that converts phenylacetate to benzoyl-CoA (E), and the bcr cluster (B; see Fig. S1B) that degrades benzoyl-CoA to glutaryl-CoA and acetyl-CoA under anoxic conditions (E) (1, 4). The pad cluster (A) includes genes for the phenylacetate-CoA ligase (padJ), the phenylacetyl-CoA:acceptor oxidoreductase (padBCD), and the phenylglyoxylate:NAD+ oxidoreductase (padEFGHI). The bcr cluster (B) contains genes encoding a benzoyl-CoA reductase (bcrCBAD), an enoyl-CoA hydratase (dch), an oxoacyl-CoA hydrolase (oah), and a hydroxyacyl-CoA dehydrogenase (had). The box (D; see Fig. S1D) cluster of aerobic-hybrid benzoate degradation encodes the beta subunit of the benzoyl-CoA oxygenase (boxB) and the 2,3-epoxybenzoyl-CoA dihydrolase (boxC). Genes involved in the aerobic-hybrid pathway of phenylacetate degradation (E) (1, 5) encode an oxepin-CoA hydrolase (paaZ), a 2,3-dehydroadipyl-CoA hydratase (paaF), a 1,2-epoxyphenylacetyl-CoA isomerase (paaG), a 3-hydroxyadipyl-CoA dehydrogenase (paaH), a 3-oxoadipyl-CoA thiolase (paaJ), and a phenylacetate-CoA ligase (paaK) (C; see Fig. S1C). Block arrows indicate the direction of transcription. Thin solid arrows, anaerobic pathways; thin dashed arrows, aerobic-hybrid pathways.
Since the (per)chlorate reducer S. selenatireducens CUZ and the chlorate reducer D. chlorophilus NSS possess genes that encode both anaerobic and aerobic-hybrid pathways of phenylacetate and benzoate degradation (Carlström et al., in press), they provide an unusual opportunity to study anaerobic versus aerobic pathway preference in bacteria capable of internally generating oxygen. At present, the degradation of aromatic compounds coupled to perchlorate or chlorate reduction has been shown to occur only by recycling the oxygen produced from chlorite dismutation to activate aerobic pathways (15, 17–19). This study aimed to test whether strains CUZ and NSS utilize aerobic-hybrid, anaerobic, or both pathways when degrading aromatic compounds in the presence of perchlorate and chlorate. Using transcriptomics, proteomics, and chemical analysis, we demonstrate that strain CUZ not only uses but unexpectedly prefers the anaerobic catabolic pathways when degrading phenylacetate and benzoate coupled to perchlorate reduction. Strain NSS also utilizes anaerobic pathways, but this study indicates that aerobic-hybrid pathways are also expressed in the presence of chlorate, suggesting that strain NSS may use both pathways concomitantly.
RESULTS
Growth with phenylacetate.
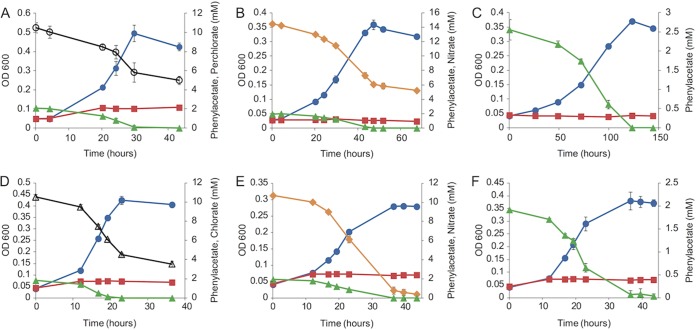
S. selenatireducens CUZ and D. chlorophilus NSS grew by the oxidation of phenylacetate coupled to the reduction of perchlorate (strain CUZ) and chlorate (strain NSS), nitrate, or oxygen (Fig. 2). The doubling time of both strains was shortest on (per)chlorate and longest on oxygen, and the percentage of phenylacetate assimilated into biomass varied from 26 to 41% (Table 1). While strain CUZ grew fully aerobically (20%, vol/vol, oxygen in the headspace) with phenylacetate as the electron donor, strain NSS could grow only with 10% (vol/vol) oxygen in sealed bottles (Fig. 2C and F; Table 1).
FIG 2 .
Growth curves of S. selenatireducens strain CUZ (A to C) and D. chlorophilus strain NSS (D to F) with 2 mM phenylacetate in the presence of perchlorate (A), chlorate (D), nitrate (B and E), or oxygen (C and F) as the electron acceptor. Blue circles, cell density; red squares, cell density of the no-acceptor control; green triangles, phenylacetate concentration; black open circles, perchlorate concentration; black open triangles, chlorate concentration; orange diamonds, nitrate concentration. Points and error bars represent the average and standard deviation of triplicate samples. OD 600, optical density at 600 nm.
TABLE 1 .
Doubling times and percentages of biomass assimilation of strains CUZ and NSS with different electron acceptors
| Strain and electron acceptor | Equation | Avg doubling time (h) ± SD | Avg biomass assimilation (%) ± SD |
|---|---|---|---|
| CUZ | |||
| ClO4 | 2 C8H7O2− + 9 ClO4− + 2 H+ → 16 CO2 + 9 Cl− + 8 H2O | 7 ± 2 | 41 ± 1 |
| NO3 | 5 C8H7O2− + 36 NO3− + 41 H+ → 40 CO2 + 18 N2 + 38 H2O | 10 ± 2 | 34 ± 3 |
| O2 | C8H7O2− + 9 O2 + H+ → 8 CO2 + 4 H2O | 29.7 ± 0.4 | NDa |
| NSS | |||
| ClO3− | C8H7O2− + 6 ClO3− + H+ → 6 Cl− + 8 CO2 + 4 H2O | 4.3 ± 0.3 | 36 |
| NO3− | 5 C8H7O2− + 36 NO3− + 41 H+ → 40 CO2 + 18 N2 + 38 H2O | 7.4 ± 1.1 | 26 |
| O2 | C8H7O2− + 9 O2 + H+ → 8 CO2 + 4 H2O | 6 | ND |
ND, not determined.
Both strains also grew by benzoate oxidation coupled to the reduction of (per)chlorate and nitrate (see Fig. S2 in the supplemental material). Additionally, strain CUZ could oxidize benzoate aerobically (10%, vol/vol, oxygen in the headspace; see Fig. S2) while strain NSS was unable to do so even at very low (1%, vol/vol) oxygen concentrations (data not shown).
Transcriptomics.
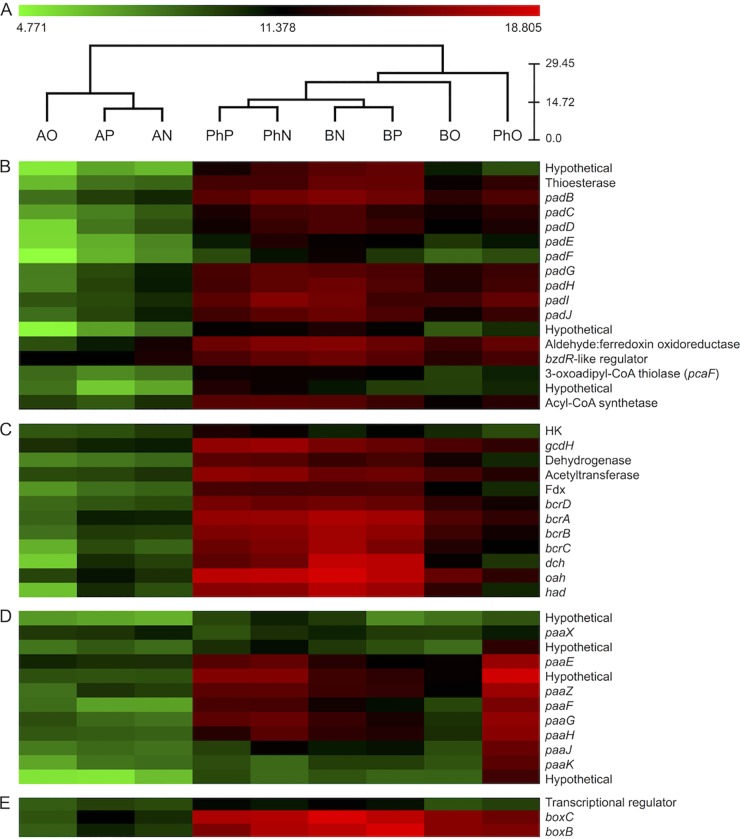
Transcriptome sequencing (RNA-seq) experiments were carried out with strain CUZ to investigate which pathways of phenylacetate degradation are transcribed when perchlorate is used as the electron acceptor. Hierarchical clustering analysis of the pad, bcr, paa, and box clusters grouped samples by both electron donor and degree of aerobicity (Fig. 3A). Acetate samples grouped together, as did samples containing phenylacetate and benzoate (Fig. 3A). The phenylacetate-oxygen and benzoate-oxygen samples clustered distinctly from their anaerobic counterparts according to the amount of oxygen used (see Materials and Methods).
FIG 3 .
Hierarchical clustering (A) and heat maps (B to E) of RNA-seq data from S. selenatireducens strain CUZ. Counts were normalized with DeSeq2 and then transformed via log2 (normalized counts + 1) prior to clustering and visualization. (B) pad gene cluster involved in anaerobic phenylacetate degradation. (C) bcr gene cluster involved in anaerobic phenylacetate and benzoate degradation. (D) paa gene cluster involved in the aerobic-hybrid pathway of phenylacetate degradation. (E) box gene cluster involved in the aerobic-hybrid pathway of benzoate degradation. Conditions: AP, acetate and perchlorate; AN, acetate and nitrate; AO, acetate and 10% (vol/vol) oxygen; PhP, phenylacetate and perchlorate; PhN, phenylacetate and nitrate; PhO, phenylacetate and 20% (vol/vol) oxygen; BP, benzoate and perchlorate; BN, benzoate and nitrate; BO, benzoate and 10% (vol/vol) oxygen. The scale bar represents Euclidean distance. For locus tags, see Table S1 in the supplemental material.
Genes in the pad cluster (Fig. 1A and E), which anaerobically converts phenylacetate into benzoyl-CoA, and genes in the bcr cluster (Fig. 1B and E), which is responsible for the downstream processing of benzoyl-CoA under anoxic conditions, were upregulated in the presence of benzoate or phenylacetate, regardless of the electron acceptor (Fig. 3B and C). However, genes in these clusters were more highly transcribed during growth with nitrate and perchlorate than during growth with oxygen (Fig. 3B and C). Transcription of genes in the bcr cluster (Fig. 3C) was higher on benzoate-oxygen than on phenylacetate-oxygen (Fig. 3C), presumably because growth on phenylacetate-oxygen does not generate benzoyl-CoA, a known inducer of anaerobic benzoate degradation pathways (1, 20).
The paa genes, which are involved in phenylacetate degradation under oxic conditions (Fig. 1C and E) (and do not produce benzoyl-CoA as an intermediate), were found to be most highly expressed under the phenylacetate-oxygen condition, although transcription in the presence of phenylacetate-perchlorate and phenylacetate-nitrate was also observed (Fig. 3D). Expression of genes in this cluster was also seen under benzoate conditions (Fig. 3D); however, it was lower than in any of the phenylacetate samples, probably because this pathway is unnecessary for the degradation of benzoate. Genes encoding the enzymes for the aerobic degradation of benzoate (box pathway) were found to be transcribed in the presence of both benzoate and phenylacetate, regardless of the electron acceptor utilized (Fig. 3E). Surprisingly, expression was lower in the oxygen samples than the perchlorate or nitrate samples (Fig. 3E). Overall, the phenylacetate-perchlorate condition more closely resembled the phenylacetate-nitrate condition than the phenylacetate-oxygen condition, suggesting that the degradation of phenylacetate with perchlorate as an electron acceptor favored the anaerobic pathway. However, because both the anaerobic and aerobic-hybrid pathways were transcribed with perchlorate, transcriptomic data alone could not rule out the possibility that both types of pathways were active under this condition.
Proteomics.
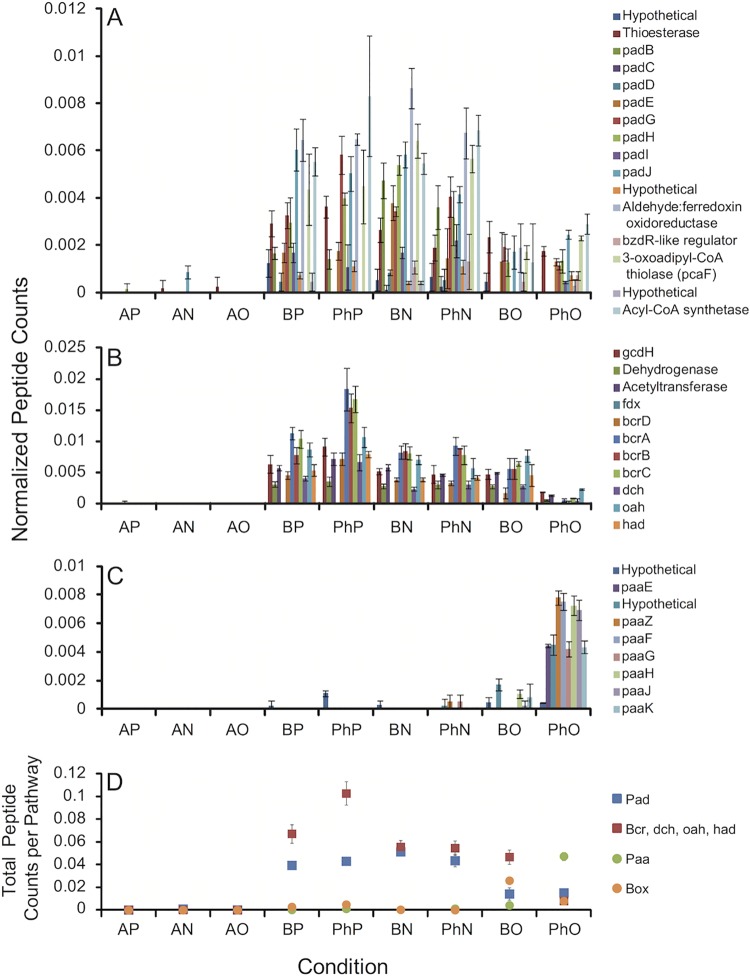
Proteomic experiments were carried out with strains CUZ and NSS. All of the conditions tested were identical, except that strain NSS was tested with chlorate, rather than perchlorate, as the electron acceptor and that strain NSS did not grow with benzoate and oxygen (data not shown). Additionally, while strain CUZ was tested with 20% (vol/vol) oxygen in the headspace with phenylacetate as the electron donor, strain NSS was only able to grow with 10% (vol/vol) oxygen in the headspace in the presence of phenylacetate. Nonparametric multidimensional scaling (nMDS) analysis indicated that all triplicate samples (except one acetate and nitrate replicate of strain NSS) clustered with each other with at least 68% similarity between the two strains (see Fig. S3 in the supplemental material).
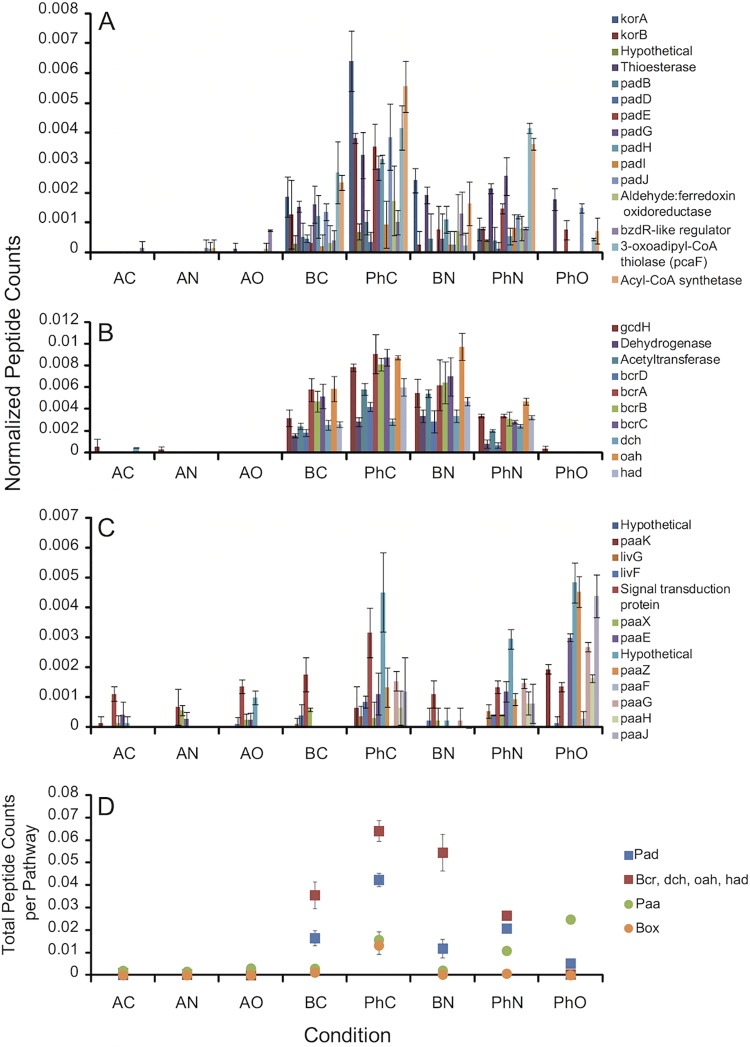
During growth with phenylacetate, peptide counts for gene products of the PAD and BCR clusters of strain CUZ were higher in the presence of perchlorate and nitrate than in the presence of oxygen (Fig. 4A and B). With either perchlorate or nitrate, the cumulative peptide counts for the PAD cluster were 2.8 times as high as with oxygen (Fig. 4D). A two-tailed heteroscedastic Student t test applied to biological replicate data estimated this increase as significant at a P value of <0.011 for nitrate and a P value of <0.003 for perchlorate. The BCR pathway was minimally expressed on phenylacetate-oxygen, probably because under these conditions the PAA pathway predominates, benzoyl-CoA is not produced, and the BCR pathway is not induced (20, 21). Cumulative peptide counts for the BCR cluster were 12.8 times higher on perchlorate than on oxygen (P < 0.004) and 6.8 times higher on nitrate than on oxygen (P < 0.007) (Fig. 4D). Peptide counts for the PAA pathway of strain CUZ were very low or absent under all conditions, except for phenylacetate-oxygen (Fig. 4C), which is consistent with the paa genes encoding an aerobic pathway of phenylacetate degradation (Fig. 1E). The cumulative peptide counts for PAA with phenylacetate-oxygen were 11 times as high as for benzoate-oxygen, which was qualitatively low despite being the second most expressed condition (P < 0.002) (Fig. 4D).
FIG 4 .
Normalized peptide counts of proteins from S. selenatireducens strain CUZ. (A) PAD cluster (anaerobic phenylacetate degradation). (B) BCR cluster (anaerobic phenylacetate-benzoate degradation). (C) PAA cluster (aerobic-hybrid phenylacetate degradation). (D) Sum of all the normalized peptide counts per pathway (PAD, BCR, PAA, and BOX). Error bars represent the standard deviation of triplicate samples. For locus tags, see Table S1 in the supplemental material. For definitions of conditions, see the legend to Fig. 3.
During growth with benzoate, the PAD pathway of strain CUZ was also more expressed in the presence of perchlorate and nitrate than in the presence of oxygen (Fig. 4A). Cumulative peptide counts for PAD were higher with nitrate (3.6 times, P < 0.007) and perchlorate (2.8 times, P < 0.015) than with oxygen (Fig. 4D). Protein expression for BCR on benzoate was similar for all electron acceptors (Fig. 4B), with only a low 1.4 times greater expression on perchlorate than on oxygen (P < 0.03) and no statistically discernible difference between nitrate and either perchlorate or oxygen.
In strain CUZ, protein expression of the Box pathway of aerobic-hybrid benzoate degradation was highest for the benzoate-oxygen samples (Fig. 4D), although a small number of peptides from BoxC and BoxB were measurable under the phenylacetate-oxygen, phenylacetate-perchlorate, and benzoate-perchlorate conditions (data not shown). Total peptide counts for the BOX pathway were nine times as high for benzoate-oxygen as those for benzoate-perchlorate (P < 0.002; Fig. 4D), while benzoate-perchlorate- and benzoate-nitrate-grown cells were not statistically distinguishable from each other. This reinforces the idea that benzoate-perchlorate and benzoate-nitrate rely mainly on the anaerobic BCR pathway with little or no contribution from the aerobic-hybrid BOX pathway (Fig. 4D). Although cells grown on benzoate-oxygen produce peptides for both the BCR pathway and the Box pathway (Fig. 4D), the benzoyl-CoA reductase of the BCR pathway is highly oxygen sensitive (22), suggesting that the BOX pathway is more likely to be actually used under aerobic conditions.
Proteomic results of strain NSS were similar to those of strain CUZ with respect to the PAD and BCR clusters (Fig. 5A and B). Protein expression of the PAD and BCR pathways was much higher on phenylacetate-chlorate or phenylacetate-nitrate than on phenylacetate-oxygen (Fig. 5A and B). With phenylacetate, the total peptide counts for the PAD pathway were higher with nitrate (4.0 times, P < 0.004) and chlorate (8.1 times, P < 0.002) than with oxygen (Fig. 5D), while the BCR peptides were nearly absent on oxygen (Fig. 5B and D). In contrast, proteins of the PAA pathway were apparently more abundant on phenylacetate-oxygen than under any other condition, with a statistically significant P value of at least < 0.04 for all comparisons (Fig. 5C and D). In contrast to strain CUZ, peptides from the PAA pathway were still detected in phenylacetate-chlorate and phenylacetate-nitrate samples of strain NSS (Fig. 5C and D). This creates the possibility that strain NSS may use a combination of aerobic and anaerobic pathways to degrade phenylacetate with chlorate as the electron acceptor. Finally, proteins of the Box pathway were more abundant in the presence of phenylacetate-chlorate than under any other condition (at least P < 0.04 for all comparisons) (Fig. 5D). This is consistent with benzoyl-CoA (an anaerobic phenylacetate degradation intermediate) and oxygen acting as BOX inducers (20, 21). Cells grown with benzoate-nitrate or benzoate-chlorate expressed much lower levels of BOX peptides, suggesting that they rely on the BCR pathway (Fig. 5D).
FIG 5 .
Normalized peptide counts of proteins from D. chlorophilus strain NSS. (A) PAD cluster (anaerobic phenylacetate degradation). (B) BCR cluster (anaerobic phenylacetate-benzoate degradation). (C) PAA cluster (aerobic-hybrid phenylacetate degradation). (D) Sum of all the normalized peptide counts per pathway (PAD, BCR, PAA, and BOX). Error bars represent the standard deviation of triplicate samples. For locus tags, see Table S1 in the supplemental material. For definitions of conditions, see the legend to Fig. 3.
Measurements of pathway intermediates.
Samples of strain CUZ with 2 mM phenylacetate as the electron donor and perchlorate, nitrate, or oxygen as the electron acceptor were analyzed for benzoyl-CoA and phenylacetyl-CoA concentrations in mid-log phase. The concentration of benzoyl-CoA, a key intermediate produced only in the anaerobic pathway of phenylacetate degradation, was measured as a diagnostic for the activity of the anaerobic (PAD) pathway with perchlorate, nitrate, and oxygen as electron acceptors. As shown in Fig. 6A, the pools of benzoyl-CoA in the perchlorate and nitrate samples were very similar (1.61 ± 0.3 and 1.8 ± 0.6 μM, respectively) and 14 to 15 times larger (P < 4 × 10−5) than when oxygen was used as the electron acceptor (0.119 ± 0.02 μM). On the other hand, the pool of phenylacetyl-CoA, which is the first intermediate produced by both the anaerobic and the aerobic-hybrid pathways, was 87 times as large (P < 2 × 10−9) under the oxic condition (54 ± 9 μM) than it was with perchlorate (0.6 ± 0.5 μM) or nitrate (0.6 ± 0.6 μM) as the electron acceptor (Fig. 6B).
FIG 6 .
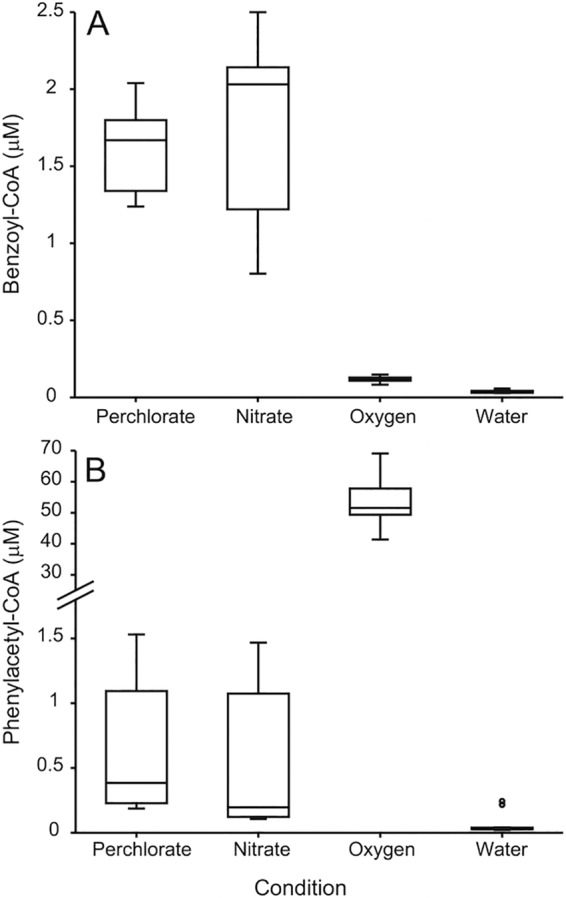
Metabolites extracted from mid-log-phase samples of S. selenatireducens strain CUZ with phenylacetate as the electron donor and perchlorate, nitrate, or oxygen as the electron acceptor. Water was run between sets as a negative control. (A) Concentration of benzoyl-CoA pool. (B) Concentration of phenylacetyl-CoA pool. Three biological replicates were sampled two to four times each, giving 12 perchlorate, 9 nitrate, and 11 oxygen samples. Data are represented in a Tukey box plot with the box extending to the upper and lower quartiles and the horizontal line indicating the median. Whiskers extend beyond the box to the highest and lowest measured concentrations within 1.5 times the interquartile range of the edge of the box. Outlying points that exceed 1.5 times the interquartile range are indicated by open circles.
DISCUSSION
We compared phenylacetate and benzoate degradation under aerobic and anaerobic (per)chlorate-reducing and nitrate-reducing conditions in the PRB S. selenatireducens strain CUZ and the CRB D. chlorophilus NSS. Previous work has shown that PRB and CRB can utilize oxygen generated from (per)chlorate reduction to activate oxygenase-dependent pathways (17, 18, 23). In contrast, this study demonstrated that PRB and CRB can also degrade aromatic compounds coupled to (per)chlorate respiration by utilizing fully anaerobic pathways that do not rely on oxygen produced from chlorite dismutation. The PRB S. selenatireducens strain CUZ and the CRB D. chlorophilus strain NSS degraded phenylacetate both aerobically and anaerobically coupled to nitrate reduction (Fig. 2), and their genomes encoded aerobic-hybrid and anaerobic pathways, allowing for the investigation of pathway activity in the presence of different electron acceptors. However, genes (except paaE) encoding the epoxidase (paaABCDE) of the aerobic-hybrid pathway were not found in either genome, opening the possibility that the aerobic-hybrid pathway may differ somewhat from the previously described process.
When strain CUZ was grown with phenylacetate, the genes for the anaerobic (pad, bcr) and aerobic-hybrid (paa) pathways both appeared to be transcribed in the presence of nitrate, perchlorate, and oxygen (Fig. 3). However, proteomics indicated that the aerobic-hybrid pathways of phenylacetate (PAA) and benzoate (BOX) degradation were used predominantly in the presence of oxygen and not in the presence of perchlorate or nitrate (Fig. 4), while the proteins of the PAD and BCR pathways were abundant in the presence of nitrate and perchlorate. In the case of phenylacetate, this was further supported by 14 times higher intracellular concentrations of the anaerobic intermediate benzoyl-CoA during growth with perchlorate and nitrate than during growth with oxygen (Fig. 6A). Given these results, we conclude that with perchlorate as the sole electron acceptor, strain CUZ oxidizes phenylacetate and benzoate via anaerobic pathways.
In strain CUZ, very high levels of phenylacetyl-CoA in the presence of phenylacetate-oxygen (Fig. 6B) suggested a bottleneck in the aerobic-hybrid pathway, which may be a result of the absence of genes for the key epoxidase (paaABCDE) of this system that would ordinarily be expected to act on phenylacetyl-CoA. While strain CUZ grew on phenylacetate-oxygen and clearly transcribed other aerobic-hybrid pathway genes, the aerobic-hybrid pathway may rely on a yet-to-be-identified oxygenase-epoxidase that could be less efficient than the canonical 1,2-phenylacetyl-CoA epoxidase. BoxB, the key oxygenase of the aerobic-hybrid benzoate degradation pathway, is the closest relative of the oxygenase of the aerobic-hybrid phenylacetate degradation pathway (PaaA) and may thus appear to be a likely candidate; however, previous studies have shown that the BoxB enzyme of Azoarcus evansii cannot process phenylacetyl-CoA (24). Similarity percentage analysis of samples used for proteomic analysis revealed a cluster of genes (CUZ_02282 to CUZ_02289; NSS_00000690 to NSS_00000740) that may be important for growth with phenylacetate-oxygen. These genes are likely involved in the degradation of 4-hydroxyphenylpyruvate to acetoacetate and fumarate (25). Of particular interest is the presence of three genes encoding oxygenases-hydroxylases (CUZ_02283, CUZ_02285, CUZ_02288; NSS_00000700, NSS_00000720, NSS_00000750) that may be relevant to the aerobic-hybrid phenylacetate degradation pathway in strains CUZ and NSS. Genetic studies are necessary to fully understand this pathway and the necessary genes.
Strain NSS grew anaerobically with phenylacetate (Fig. 2) and benzoate (see Fig. S2 in the supplemental material) but was unable to oxidize benzoate aerobically under the conditions tested. As with strain CUZ, proteomics demonstrated that the anaerobic phenylacetate and benzoate pathways (PAD, BCR) were upregulated in the presence of nitrate or chlorate but not in the presence of oxygen (Fig. 5). Unexpectedly, the aerobic-hybrid phenylacetate (PAA) pathway genes were expressed under all of the conditions where phenylacetate was present. The total PAA peptide count was only about 2.3 times as high in the presence of oxygen as it was in the presence of nitrate (P < 0.004), which is much less distinctive than the >35-fold increase in strain CUZ when PAA expression was compared under the same conditions. Phenylacetate-chlorate in strain NSS produced intermediate PAA peptide counts that could not be statistically distinguished from phenylacetate-nitrate. On the basis of these data, it is possible that phenylacetate degradation coupled to chlorate reduction in strain NSS uses both the aerobic-hybrid and anaerobic pathways simultaneously. It is also possible that phenylacetate activates the expression of PAA even in the absence of oxygen. In strain NSS, the benzoate degradation pathway is also unclear. Since strain NSS cannot grow with benzoate in the presence of oxygen (even when it is reduced to 1%, vol/vol) there is no baseline level to use as a positive control for aerobic growth. The aerobic-hybrid benzoate pathway (box) was never activated in the presence of benzoate; hence, it appears that benzoate is degraded exclusively by the anaerobic pathway (bcr) under all of the conditions tested. However, peptides of the box pathway were observed in the presence of phenylacetate and chlorate. If oxygen is present, the box pathway could act on the benzoyl-CoA intermediate of the anaerobic phenylacetate pathway, though it is unknown if this is occurring.
These results indicate that PRB, and likely CRB, can couple (per)chlorate reduction to aromatic degradation using anaerobic pathways. This is in contrast to previous studies showing PRB and CRB using (per)chlorate to facilitate the aerobic degradation of compounds such as benzene, toluene, naphthalene, phenol, and catechol (17, 18, 23, 26, 27). Even though both pathways are present, strain CUZ preferred the anaerobic pathway. This preference is somewhat unexpected, as key enzymes in the anaerobic pathways, benzoyl-CoA reductase and phenylglyoxylate:NAD+ oxidoreductase, are highly oxygen sensitive (22, 28). However, because chlorite dismutase is a periplasmic enzyme (29, 30), the O2 produced from chlorite dismutation is physically separated from the cytoplasm; thus, it is likely that the cytoplasmic concentration of O2 was low enough to prevent significant inhibition of these enzymes. One possible reason to preferentially utilize the anaerobic pathways is to conserve O2 for other processes such as respiration. It may be more energetically favorable to respire the O2 produced than to use it for ring cleavage. It was observed that both strains CUZ and NSS grew more rapidly on perchlorate and chlorate, respectively, than on nitrate, suggesting that the extra oxygen does convey a metabolic advantage (Fig. 1). Thus, bacteria in microaerophilic environments may apply a mixture of anaerobic and aerobic strategies that allow them to make the best possible use of limited oxygen.
MATERIALS AND METHODS
Culture conditions for S. selenatireducens CUZ.
The medium used to culture S. selenatireducens CUZ contained NaCl (30 g/liter), KCl (0.67 g/liter), NaHCO3 (2.5 g/liter), 10 ml of vitamins, 10 ml of minerals, and 20 ml of RST minerals (31). The RST minerals contained NaCl (40 g/liter), NH4Cl (50 g/liter), KCl (5 g/liter), KH2PO4 (5 g/liter), MgSO4 ⋅ 7H2O (10 g/liter), and CaCl2 ⋅ 2H2O (1 g/liter). The medium was boiled and then simultaneously cooled on ice and degassed under a N2-CO2 headspace (80/20). The pH was ~6.8. Each anaerobic tube (Bellco, Vineland, NJ) received 9 ml of this medium and was autoclaved. All growth experiments were carried out at 30°C.
Culture conditions for D. chlorophilus NSS.
The medium used to culture D. chlorophilus NSS contained NaCl (20 g/liter), KCl (0.67 g/liter), monosodium piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; 1.6220 g/liter), disodium PIPES (1.7317 g/liter), 10 ml of vitamins, 10 ml of minerals, and 20 ml of RST minerals (31 g/liter). The medium was boiled and then simultaneously cooled on ice and degassed under a N2 headspace. The pH was ~7.5. Each anaerobic tube received 8 ml of this medium and was autoclaved. After autoclaving, 0.5 ml of MgCl2 ⋅ 6H2O (21.2 g/100 ml of distilled H2O [dH2O]) and 0.5 ml of CaCl2 ⋅ 2H2O (3.04 g/100 ml of dH2O) were added to each tube from sterile, aqueous stock solutions that were kept under an anoxic headspace. All growth experiments were carried out at 37°C.
Growth with phenylacetate, benzoate, and acetate.
To test growth on phenylacetate coupled to the reduction of perchlorate and chlorate (corresponding to strains CUZ and NSS, respectively) or nitrate, 2 mM phenylacetate and 10 mM perchlorate or chlorate or 15 mM nitrate were added to triplicate samples. Negative controls included no-donor and no-acceptor controls. Samples were taken for optical density and anion concentration measurements as described below.
For strain CUZ, growth in sealed 2.8-liter Fernbach flasks with a total liquid volume of 500 ml with phenylacetate (2 mM) and oxygen (20%, vol/vol) was carried out in triplicate, thus allowing for a large oxic headspace. A no-acceptor control was included. The Fernbach flasks were shaken at 200 rpm. The medium was made by adding all of the components except bicarbonate to the flasks and autoclaving it aerobically. After autoclaving, the flasks were sealed and bicarbonate from an aqueous, sterile stock was added. HCl was used to adjust the pH of the medium to ~6.8. In the case of strain NSS, 160-ml sealed bottles were used with a 50-ml liquid volume. Ten milliliters of oxygen was injected into triplicate cultures twice, once at inoculation, and a second time in early log phase. A no-acceptor control was included. Bottles were shaken horizontally at 200 rpm.
For both strains, growth with 10 mM acetate and (per)chlorate and 1 mM benzoate with 5 mM (per)chlorate or 10 mM nitrate was used as a control for RNA-seq and proteomic experiments. Growth with 1 mM benzoate and oxygen was also used as a control with strain CUZ but not strain NSS. Cultures with 1 mM benzoate and 10% (vol/vol) oxygen in the headspace were grown in 160-ml bottles shaken horizontally at 200 rpm.
Cell growth profiles.
Cell growth was measured spectrophotometrically at 600 nm. All experimental analyses were performed in triplicate, and the results are expressed as the average of these determinations.
Analytical methods.
Perchlorate, nitrate, benzoate, and phenylacetate concentrations were measured via ion chromatography (ICS-2100; Thermo Fisher Scientific, Sunnyvale, CA). The system utilized an IonPac AG16 (4 by 50 mm) guard column, an IonPac AS16 (4 by 250 mm) analytical column, an ASRS-300 4-mm suppressor system, and a DS6 heated conductivity cell. A KOH gradient was generated with an EGC III KOH generator at an isocratic flow rate of 1.5 ml/min. The KOH gradient was 1.5 mM from 0 to 7 min, increased to 10 mM from min 7 to 13, held at 10 mM from min 13 to 16, increased to 35 mM during min 16 to 17, held at 35 mM from min 17 to 27, and decreased to 1.5 mM from min 27 to 30. The suppressor controller was set at 130 mA for the analysis. The injection volume was 25 µl.
Chlorate concentration was measured by ion chromatography with a Dionex ICS 1500 equipped with a Dionex Ion Pac AS 25 column (4 by 250 mm). The mobile phase was 36 mM NaOH at a flow rate of 1.0 ml/min. Background conductivity was suppressed with a Dionex ASRS-300 (4 mm) in recycle mode. The suppressor controller was set at 90 mA for analysis, and the injection volume was 10 µl.
RNA extraction and RNA-seq.
Triplicate cultures (40 ml each) were harvested in mid-log phase, centrifuged at 7,000 × g for 10 min at 4°C, and resuspended in 1 ml of TRIzol (Life Technologies). To keep samples as anoxic as possible, N2-CO2 or N2 gas (depending on the medium) was blown into 50-ml Falcon tubes prior to centrifugation of the sample. RNA was extracted according to the manufacturer’s protocol, and biological triplicates were combined into one sample per condition at the end of the extraction. Each sample was then treated with DNase to remove DNA contamination (Turbo DNA-free; Life Technologies). The RNA was tested for DNA contamination with the universal 16S rRNA amplification primers 27F (5′ AGAGTTTGATCMTGGCTCAG 3′) and 1492R (5′ GGTTACCTTGTTACGACTT 3′). The Vincent J. Coates Genomics Sequencing Laboratory (University of California, Berkeley) performed rRNA removal (Ribo-Zero rRNA removal kit; Epicentre), cDNA synthesis, library preparation (Apollo 324; WaferGen Biosystems), and 50-bp single-end sequencing with the Illumina Hiseq2000 system. Scythe (https://github.com/ucdavis-bioinformatics/scythe), seqtk (https://github.com/lh3/seqtk/), and sickle (https://github.com/ucdavis-bioinformatics/sickle) were used to remove adapter contamination, cut 5 bp on the 5′ end, and perform quality trimming, respectively. Bowtie2 (32) was used to map processed reads to the S. selenatireducens CUZ genome. The GenomicRanges package (33) in R (R Core Team, 2013) was used to count reads mapped to coding regions by using summarizeOverlaps in “union” mode. The DESeq2 package (34) in R was used to normalize counts and perform differential-expression analysis. Data were log2 transformed. MeV 4.9 (http://www.tm4.org/) was used for data visualization and hierarchical clustering analysis.
Protein extraction and LC-MS/MS.
Samples used for proteomic analysis were prepared as described in previous protocols with modifications (35, 36). Triplicate cultures (40 ml) were harvested in mid-log phase and centrifuged at 7,000 × g for 10 min at 4°C. To keep samples as anoxic as possible, N2-CO2 or N2 gas (depending on the medium) was blown into 50-ml Falcon tubes prior to centrifugation of the sample. The pellet was then washed with 1 ml of 100 mM NH4HCO3, centrifuged for 2 min at 10,000 × g, and resuspended in 0.5 ml of 100 mM NH4HCO3. Cells were lysed at 4°C with a 550 Sonic Dismembrator (Fisher Scientific, Waltham, MA) at a power of 1.5. Each sample was subject to three rounds of 30 s each, with a 30-s rest on ice between rounds. Five hundred nanograms of trypsin was then added, and the samples were incubated for 3 h at 37°C. Samples were then centrifuged at 14,000 × g for 5 min, and the supernatant was transferred to new tubes. Five microliters of 100 mM dithiothreitol and 5 µl of 100 mM iodoacetamide were then added in two sequential 30-min incubations at room temperature. Another 500 ng of trypsin was then added in an overnight incubation at 37°C. Twenty microliters of 2.5% (vol/vol) trifluoroacetic acid (TFA) was added; this was followed by a 5-min centrifugation at 14,000 × g. The supernatant was transferred to a new tube. C18 Zip Tips (Millipore, Billerica, MA) were used to concentrate peptides and remove salts. Eighty-five percent (vol/vol) acetonitrile–0.1% TFA was used to elute the peptides, and vacuum centrifugation was used to remove acetonitrile for a final volume of 30 µl.
Trypsin-digested proteins were analyzed with a Thermo Dionex UltiMate 3000 RSLCnano liquid chromatograph that was connected in line with an LTQ Orbitrap XL mass spectrometer equipped with a nanoelectrospray ionization (nanoESI) source (Thermo Fisher Scientific, Waltham, MA). The liquid chromatograph was equipped with a C18 analytical column (Acclaim PepMap RSLC, 150-mm length by 0.075-mm inner diameter, 2-µm particles, 100 Å pores; Thermo) and a 1-µl sample loop. Acetonitrile (Fisher Optima grade, 99.9%), formic acid (1-ml ampules, >99% pure; Thermo Pierce), and water purified to a resistivity of 18.2 MΩ × cm (at 25°C) with a Milli-Q Gradient ultrapure water purification system (Millipore, Billerica, MA) were used to prepare mobile-phase solvents for liquid chromatography (LC)-tandem mass spectrometry (MS/MS). Solvent A was 99.9% (vol/vol) water–0.1% (vol/vol) formic acid, and solvent B was 99.9% (vol/vol) acetonitrile–0.1% (vol/vol) formic acid. Samples contained in polypropylene autosampler vials with septa caps (Wheaton, Millville, NJ) were loaded into the autosampler compartment prior to analysis. The autosampler compartment was temperature controlled at 4°C. The elution program consisted of isocratic flow at 5% solvent B for 4 min, a linear gradient to 30% solvent B over 128 min, isocratic flow at 95% solvent B for 6 min, and isocratic flow at 5% solvent B for 12 min at a flow rate of 300 nl/min.
The column exit was connected to the nanoESI emitter in the nanoESI source of the mass spectrometer with polyimide-coated fused-silica tubing (20-µm inner diameter, 280-µm outer diameter; Thermo). Full-scan mass spectra were acquired in the positive-ion mode over a range of m/z = 350 to 1,600 with the Orbitrap mass analyzer in profile format at a mass resolution setting of 60,000 (at m/z = 400, measured at full width at half-maximum peak height).
In the data-dependent mode, the eight most intense ions exceeding an intensity threshold of 30,000 counts were selected from each full-scan mass spectrum for MS/MS analysis by collision-induced dissociation (CID). MS/MS spectra were acquired with the linear ion trap in centroid format with the following parameters: isolation width, 3 U; normalized collision energy, 28%; default charge state, 2+; activation Q, 0.25; activation time, 30 ms. Real-time charge state screening was enabled to exclude singly charged ions and unassigned charge states from MS/MS analysis. To avoid the occurrence of redundant MS/MS measurements, real-time dynamic exclusion was enabled to preclude reselection of previously analyzed precursor ions, with the following parameters: repeat count, 2; repeat duration, 10 s; exclusion list size, 500; exclusion duration, 90 s; exclusion mass width, 20 ppm. Data acquisition was controlled with Xcalibur software (version 2.0.7 SP1; Thermo). Raw data files were searched against the S. selenatireducens strain CUZ or D. chlorophilus NSS protein database with Proteome Discoverer software (version 1.3, SEQUEST algorithm; Thermo) for tryptic peptides (i.e., peptides resulting from cleavage at the C-terminal end of lysine and arginine residues) with up to three missed cleavages, carbamidomethylcysteine as a static posttranslational modification, and methionine sulfoxide as a variable posttranslational modification. A decoy database was used to characterize the false-discovery rate, and the target false-discovery rate was 0.01 (i.e., 1%).
Peptide data were normalized (dividing by the total number of peptides per sample) and square root transformed. The hierarchical clustering and nMDS functions in Primer 6 (Primer-E Ltd., Plymouth, United Kingdom) were used to determine the clustering patterns among samples. Similarity percentage (SIMPER) analysis was used to identify genes contributing to the top 50% of the differences between groups. Bray-Curtis dissimilarity matrices were used in all cases. P values for assessing differences in protein expression were calculated with a two-tailed heteroscedastic Student t test.
Metabolite extraction and LC-MS/MS.
For growth on phenylacetate and perchlorate and phenylacetate and nitrate, cells (500 ml) were grown in triplicate in 1-liter Pyrex bottles sealed anoxically. For growth with phenylacetate and oxygen, cells (500 ml) were grown in Fernbach flasks as described above. For the nitrate and perchlorate conditions, all of the work except centrifugation was performed inside an anaerobic chamber. Samples for metabolite extraction were collected at mid-log phase and centrifuged at 10,000 × g for 10 min at 4°C. The volume of cells collected was normalized across different conditions by the total optical density (OD) (OD of 125). The pellet was resuspended in 500 µl of sterile, degassed, acidic acetonitrile (32 ml of acetonitrile, 7.7 ml of dH2O, 314.6 µl of formic acid) and incubated on ice for 15 min. Samples were then centrifuged at 14,000 × g for 3 min, and the supernatant was filtered into LC-MS vials with a 0.2-µm filter. Benzoyl-CoA and phenylacetyl-CoA standards (Sigma Aldrich) were run at concentrations ranging from 10 to 5,000 nM.
Compounds were separated with a 1200 Series LC instrument (Agilent Technologies, Santa Clara, CA). A 40-µl aliquot of each sample was injected onto an Agilent Zorbax SB-C18 column (2.1-mm inside diameter, 30-mm length, 3.5-µm particle size) and eluted at 30°C at a flow rate of 0.5 ml/min with the following gradient: 2 min with isocratic 99% buffer A (40 mM ammonium acetate)–1% buffer B (methanol), 8 min with a linear gradient to 100% buffer B, 2 min with isocratic 100% buffer B, and then 3 min of equilibration with 99% buffer A and 1% buffer B. The eluent from the column was introduced into a mass spectrometer for 11 min after the first minute. Water was run between sets as a negative control.
MS was performed with an LTQ XL ion trap instrument (Thermo, Fisher Scientific, San Jose, CA) with an ESI source operated in positive-ion mode. The MS settings were a capillary temperature of 350°C, an ion spray voltage of 3.5 kV, a sheath gas flow rate of 60 arbitrary units, an auxiliary gas flow rate of 10 arbitrary units, and a sweep gas flow rate of 5 arbitrary units.
For MS/MS product ion scanning, the scan range was m/z 245 to 1,000. The compounds at m/z 872.1 (benzoyl-CoA) and m/z 886.2 (phenylacetyl-CoA) were isolated with a 2 isolation width and fragmented with a normalized CID energy setting of 35%, an activation time of 30 ms, and an activation Q of 0.250. P values for assessing differences between metabolite concentrations were calculated with a two-tailed heteroscedastic Student t test.
Nucleotide sequence accession numbers.
The genomes of S. selenatireducens CUZ and D. chlorophilus NSS are available in the Integrated Microbial Genomes (IMG) system of the Joint Genome Institute, and their 16S rRNA gene sequences are available in the GenBank database (accession no. KM192219 and AF170359, respectively).
SUPPLEMENTAL MATERIAL
Genes involved in phenylacetate and benzoate degradation in D. chlorophilus NSS. (A) pad gene cluster. (B) bcr gene cluster. (C) paa gene cluster. (D) box gene cluster. Gene colors correspond to the pathways shown in Fig. 2E. Red, paa gene cluster involved in the aerobic-hybrid pathway of phenylacetate degradation; blue, pad gene cluster involved in anaerobic phenylacetate degradation; purple, bcr gene cluster involved in anaerobic benzoate and phenylacetate degradation; green, box gene cluster involved in the aerobic-hybrid pathway of benzoate degradation. Bar, 1 kb. Arrows indicate the direction of transcription. Download
Growth curves of S. selenatireducens strain CUZ (A to C) and D. chlorophilus strain NSS (D and E) on 1 mM benzoate with perchlorate or chlorate (A and D), nitrate (B and E), or oxygen (C) as the electron acceptor. Blue circles, cell density; red squares, cell density of the no-acceptor control; green triangles, phenylacetate concentration; black open circles, perchlorate concentration; black open triangles, chlorate concentration; orange diamonds, nitrate concentration. Points and error bars represent the average and standard deviation of triplicate samples. Download
nMDS plot of samples used for proteomic analysis of S. selenatireducens strain CUZ (A) and D. chlorophilus strain NSS (B) based on a Bray-Curtis similarity matrix. Circles represent percent similarity based on hierarchical clustering. Colored symbols represent different electron donors. Letters represent different electron acceptors. N, nitrate; P, perchlorate; O, oxygen; C, chlorate. Download
IMG locus tags for genes in the paa, pad, bcr, and box clusters of strains CUZ and NSS.
ACKNOWLEDGMENTS
Funding supporting research on microbial (per)chlorate metabolism in the lab of J. D. Coates is provided by the Energy Biosciences Institute, Berkeley, CA. C. I. Carlström is grateful for funding support from the National Science Foundation Graduate Research Fellowship Program. This work used the Vincent J. Coates Genomics Sequencing Laboratory at University of California Berkeley, supported by NIH S10 instrumentation grants S10RR029668 and S10RR027303.
Footnotes
Citation Carlström CI, Loutey D, Bauer S, Clark IC, Rohde RA, Iavarone AT, Lucas L, Coates JD. 2015. (Per)chlorate-reducing bacteria can utilize aerobic and anaerobic pathways of aromatic degradation with (per)chlorate as an electron acceptor. mBio 6(2):e02287-14. doi:10.1128/mBio.02287-14.
Contributor Information
Victor de Lorenzo, Centro National de Biotecnologia.
Sang Yup Lee, Korea Advanced Institute of Science and Technology.
REFERENCES
- 1.Carmona M, Zamarro MT, Blázquez B, Durante-Rodríguez G, Juárez JF, Valderrama JA, Barragán MJ, García JL, Díaz E. 2009. Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol Mol Biol Rev 73:71–133. doi: 10.1128/MMBR.00021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Díaz E, Jiménez JI, Nogales J. 2013. Aerobic degradation of aromatic compounds. Curr Opin Biotechnol 24:431–442. doi: 10.1016/j.copbio.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Ismail W, Gescher J. 2012. Epoxy coenzyme A thioester pathways for degradation of aromatic compounds. Appl Environ Microbiol 78:5043–5051. doi: 10.1128/AEM.00633-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuchs G, Boll M, Heider J. 2011. Microbial degradation of aromatic compounds—from one strategy to four. Nat Rev Microbiol 9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- 5.Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, Eisenreich W, Haehnel W, Fuchs G. 2010. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc Natl Acad Sci U S A 107:14390–14395. doi: 10.1073/pnas.1005399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navarro-Llorens JM, Patrauchan MA, Stewart GR, Davies JE, Eltis LD, Mohn WW. 2005. Phenylacetate catabolism in Rhodococcus sp. strain RHA1: a central pathway for degradation of aromatic compounds. J Bacteriol 187:4497–4504. doi: 10.1128/JB.187.13.4497-4504.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luengo JM, García JL, Olivera ER. 2001. The phenylacetyl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Mol Microbiol 39:1434–1442. doi: 10.1046/j.1365-2958.2001.02344.x. [DOI] [PubMed] [Google Scholar]
- 8.Rhee SK, Fuchs G. 1999. Phenylacetyl-CoA: acceptor oxidoreductase, a membrane-bound molybdenum-iron-sulfur enzyme involved in anaerobic metabolism of phenylalanine in the denitrifying bacterium Thauera aromatica. Eur J Biochem 262:507–515. doi: 10.1046/j.1432-1327.1999.00399.x. [DOI] [PubMed] [Google Scholar]
- 9.Wöhlbrand L, Kallerhoff B, Lange D, Hufnagel P, Thiermann J, Reinhardt R, Rabus R. 2007. Functional proteomic view of metabolic regulation in “Aromatoleum aromaticum” strain EbN1. Proteomics 7:2222–2239. doi: 10.1002/pmic.200600987. [DOI] [PubMed] [Google Scholar]
- 10.Mingot JM, Peñalva MA, Fernández-Cañón JM. 1999. Disruption of phacA, an Aspergillus nidulans gene encoding a novel cytochrome P450 monooxygenase catalyzing phenylacetate 2-hydroxylation, results in penicillin overproduction. J Biol Chem 274:14545–14550. doi: 10.1074/jbc.274.21.14545. [DOI] [PubMed] [Google Scholar]
- 11.Cox HH, Faber BW, Van Heiningen WN, Radhoe H, Doddema HJ, Harder W. 1996. Styrene metabolism in Exophiala jeanselmei and involvement of a cytochrome P-450-dependent styrene monooxygenase. Appl Environ Microbiol 62:1471–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kishore G, Sugumaran M, Vaidyanathan CS. 1976. Metabolism of dl-(+/−)-phenylalanine by Aspergillus niger. J Bacteriol 128:182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs G. 2008. Anaerobic metabolism of aromatic compounds, p 82–99. In Wiegel J, Maier RJ, Adams MWW (ed), Incredible anaerobes: from physiology to genomics to fuels, volume 1125. Blackwell Publishing, Oxford, United Kingdom. [Google Scholar]
- 14.Clark IC, Melnyk RA, Engelbrektson A, Coates JD. 2013. Structure and evolution of chlorate reduction composite transposons. mBio 4:e00379-13. doi: 10.1128/mBio.00379-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coates JD, Achenbach LA. 2004. Microbial perchlorate reduction: rocket-fueled metabolism. Nat Rev Microbiol 2:569–580. doi: 10.1038/nrmicro926. [DOI] [PubMed] [Google Scholar]
- 16.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJ, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJ, Janssen-Megens EM, Francoijs KJ, Stunnenberg H, Weissenbach J, Jetten MS, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 17.Weelink SA, Tan NC, ten Broeke H, van den Kieboom C, van Doesburg W, Langenhoff AA, Gerritse J, Junca H, Stams AJ. 2008. Isolation and characterization of Alicycliphilus denitrificans strain BC, which grows on benzene with chlorate as the electron acceptor. Appl Environ Microbiol 74:6672–6681. doi: 10.1128/AEM.00835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlström CI, Wang O, Melnyk RA, Bauer S, Lee J, Engelbrektson A, Coates JD. 2013. Physiological and genetic description of dissimilatory perchlorate reduction by the novel marine bacterium Arcobacter sp. strain CAB. mBio 4:e00217-13. doi: 10.1128/mBio.00217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motzer WE. 2001. Perchlorate: problems, detection, and solutions. Environ Forensics 2:301–311. doi: 10.1006/enfo.2001.0059. [DOI] [Google Scholar]
- 20.Schühle K, Gescher J, Feil U, Paul M, Jahn M, Schägger H, Fuchs G. 2003. Benzoate-coenzyme A ligase from Thauera aromatica: an enzyme acting in anaerobic and aerobic pathways. J Bacteriol 185:4920–4929. doi: 10.1128/JB.185.16.4920-4929.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valderrama JA, Durante-Rodríguez G, Blázquez B, García JL, Carmona M, Díaz E. 2012. Bacterial degradation of benzoate: cross-regulation between aerobic and anaerobic pathways. J Biol Chem 287:10494–10508. doi: 10.1074/jbc.M111.309005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boll M, Fuchs G. 1995. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain k172. Eur J Biochem 234:921–933. doi: 10.1111/j.1432-1033.1995.921_a.x. [DOI] [PubMed] [Google Scholar]
- 23.Coates JD, Bruce RA, Patrick J, Achenbach LA. 1999. Hydrocarbon bioremediative potential of (per)chlorate-reducing bacteria. Bioremediat J 3:323–334. doi: 10.1080/10889869991219415. [DOI] [Google Scholar]
- 24.Zaar A, Gescher J, Eisenreich W, Bacher A, Fuchs G. 2004. New enzymes involved in aerobic benzoate metabolism in Azoarcus evansii. Mol Microbiol 54:223–238. doi: 10.1111/j.1365-2958.2004.04263.x. [DOI] [PubMed] [Google Scholar]
- 25.Arias-Barrau E, Olivera ER, Luengo JM, Fernández C, Galán B, García JL, Díaz E, Miñambres B. 2004. The homogentisate pathway: a central catabolic pathway involved in the degradation of l-phenylalanine, l-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J Bacteriol 186:5062–5077. doi: 10.1128/JB.186.15.5062-5077.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oosterkamp MJ, Veuskens T, Plugge CM, Langenhoff AA, Gerritse J, van Berkel WJ, Pieper DH, Junca H, Goodwin LA, Daligault HE, Bruce DC, Detter JC, Tapia R, Han CS, Land ML, Hauser LJ, Smidt H, Stams AJ. 2011. Genome sequences of Alicycliphilus denitrificans strains BC and K601T. J Bacteriol 193:5028–5029. doi: 10.1128/JB.00365-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coates JD, Bruce RA, Haddock JD. 1998. Anoxic bioremediation of hydrocarbons. Nature 396:730. doi: 10.1038/25470. [DOI] [PubMed] [Google Scholar]
- 28.Hirsch W, Schägger H, Fuchs G. 1998. Phenylglyoxylate: NAD+ oxidoreductase (CoA benzoylating), a new enzyme of anaerobic phenylalanine metabolism in the denitrifying bacterium Azoarcus evansii. Eur J Biochem 251:907–915. doi: 10.1046/j.1432-1327.1998.2510907.x. [DOI] [PubMed] [Google Scholar]
- 29.Mehboob F, Wolterink AF, Vermeulen AJ, Jiang B, Hagedoorn P-L, Stams AJ, Kengen SW. 2009. Purification and characterization of a chlorite dismutase from Pseudomonas chloritidismutans. FEMS Microbiol Lett 293:115–121. doi: 10.1111/j.1574-6968.2009.01517.x. [DOI] [PubMed] [Google Scholar]
- 30.Stenklo K, Thorell HD, Bergius H, Aasa R, Nilsson T. 2001. Chlorite dismutase from Ideonella dechloratans. J Biol Inorg Chem 6:601–607. doi: 10.1007/s007750100237. [DOI] [PubMed] [Google Scholar]
- 31.Bruce RA, Achenbach LA, Coates JD. 1999. Reduction of (per)chlorate by a novel organism isolated from paper mill waste. Environ Microbiol 1:319–329. doi: 10.1046/j.1462-2920.1999.00042.x. [DOI] [PubMed] [Google Scholar]
- 32.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–U354. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLOS Comput Biol 9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlson HK, Iavarone AT, Gorur A, Yeo BS, Tran R, Melnyk RA, Mathies RA, Auer M, Coates JD. 2012. Surface multiheme c-type cytochromes from Thermincola potens and implications for respiratory metal reduction by Gram-positive bacteria. Proc Natl Acad Sci U S A 109:1702–1707. doi: 10.1073/pnas.1112905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark IC, Carlson HK, Iavarone AT, Coates JD. 2012. Bioelectrical redox cycling of anthraquinone-2,6-disulfonate coupled to perchlorate reduction. Energy Environ Sci 5:7970–7978. doi: 10.1039/c2ee21594b. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes involved in phenylacetate and benzoate degradation in D. chlorophilus NSS. (A) pad gene cluster. (B) bcr gene cluster. (C) paa gene cluster. (D) box gene cluster. Gene colors correspond to the pathways shown in Fig. 2E. Red, paa gene cluster involved in the aerobic-hybrid pathway of phenylacetate degradation; blue, pad gene cluster involved in anaerobic phenylacetate degradation; purple, bcr gene cluster involved in anaerobic benzoate and phenylacetate degradation; green, box gene cluster involved in the aerobic-hybrid pathway of benzoate degradation. Bar, 1 kb. Arrows indicate the direction of transcription. Download
Growth curves of S. selenatireducens strain CUZ (A to C) and D. chlorophilus strain NSS (D and E) on 1 mM benzoate with perchlorate or chlorate (A and D), nitrate (B and E), or oxygen (C) as the electron acceptor. Blue circles, cell density; red squares, cell density of the no-acceptor control; green triangles, phenylacetate concentration; black open circles, perchlorate concentration; black open triangles, chlorate concentration; orange diamonds, nitrate concentration. Points and error bars represent the average and standard deviation of triplicate samples. Download
nMDS plot of samples used for proteomic analysis of S. selenatireducens strain CUZ (A) and D. chlorophilus strain NSS (B) based on a Bray-Curtis similarity matrix. Circles represent percent similarity based on hierarchical clustering. Colored symbols represent different electron donors. Letters represent different electron acceptors. N, nitrate; P, perchlorate; O, oxygen; C, chlorate. Download
IMG locus tags for genes in the paa, pad, bcr, and box clusters of strains CUZ and NSS.