Abstract
Background:
Epidermal growth factor receptor (EGFR) is overexpressed in a subset of human epidermal growth factor receptor 2 (HER2)-positive breast cancers, and coexpression of HER2 and EGFR has been reported to be associated with poor clinical outcome. Moreover, interaction between HER2 and EGFR has been suggested to be a possible basis for trastuzumab resistance.
Methods:
We analysed the clinical significance of EGFR overexpression and EGFR gene copy number alterations in 242 HER2-positive primary breast cancers. In addition, we examined the correlations between EGFR overexpression, trastuzumab response and clinical outcome in 447 primary, and 112 metastatic HER2-positive breast cancer patients treated by trastuzumab.
Results:
Of the 242 primary cases, the level of EGFR overexpression was 2+ in 12.7% and 3+ in 11.8%. High EGFR gene copy number was detected in 10.3%. Epidermal growth factor receptor overexpression was associated with hormone receptor negativity and high Ki-67 proliferation index. In survival analyses, EGFR overexpression, but not high EGFR copy number, was associated with poor disease-free survival in all patients, and in the subgroup not receiving adjuvant trastuzumab. In 447 HER2-positive primary breast cancer patients treated with adjuvant trastuzumab, EGFR overexpression was also an independent poor prognostic factor. However, EGFR overexpression was not associated with trastuzumab response, progression-free survival or overall survival in the metastatic setting.
Conclusions:
Epidermal growth factor receptor overexpression, but not high EGFR copy number, is a poor prognostic factor in HER2-positive primary breast cancer. Epidermal growth factor receptor overexpression is a predictive factor for trastuzumab response in HER2-positive primary breast cancer, but not in metastatic breast cancer.
Keywords: breast carcinoma, gene amplification, EGFR, HER2, trastuzumab, immunohistochemistry
Epidermal growth factor receptor (EGFR, ERBB1) expression has been widely studied in breast cancer by immunohistochemistry (IHC), and the frequency of EGFR overexpression in breast cancer is quite variable, ranging from 7 to 43% (Wrba et al, 1988; Suo et al, 2002; Bhargava et al, 2005; DiGiovanna et al, 2005; Nieto et al, 2007; Koletsa et al, 2010; Lv et al, 2011b; Hwangbo et al, 2013). Epidermal growth factor receptor overexpression is associated with hormone receptor negativity, large tumour size, high histologic grade and poor clinical outcome (DiGiovanna et al, 2005; Nieto et al, 2007; Koletsa et al, 2010; Lv et al, 2011b). In particular, coexpression of human epidermal growth factor receptor 2 (HER2) and EGFR is associated with worse survival in patients with HER2-positive breast cancer (Suo et al, 2002; DiGiovanna et al, 2005; Nieto et al, 2007).
Interaction between HER2 and other ERBB coreceptors, such as EGFR and HER3 (ERBB3), has been suggested as a possible mechanism of resistance to trastuzumab, a humanised monoclonal antibody against the extracellular portion of the HER2 protein (Franklin et al, 2004; Diermeier et al, 2005). Upon ligand binding, ERBB receptors form homo- or heterodimers, and these phosphorylate their cytoplasmic tyrosine kinase domains and activate intracellular signalling, leading to cell division, motility, survival and angiogenesis (Yarden and Sliwkowski, 2001). Ritter et al (2007)generated trastuzumab-resistant BT-474 cells in vivo; these contained higher levels of phosphorylated EGFR and EGFR/HER2 heterodimers. Application of the EGFR tyrosine kinase inhibitors, erlotinib and gefitinib, was associated with decreased phosphorylation of HER2, suggesting that amplification of ligand-induced activation of ERBB receptors through heterodimerisation is a plausible mechanism for resistance to trastuzumab (Ritter et al, 2007). Actually, it was shown that trastuzumab is not able to block ligand-induced EGFR/HER2 and HER2/HER3 heterodimers (Cho et al, 2003; Franklin et al, 2004). However, the significance of EGFR overexpression for trastuzumab response is not clear in clinical settings. Gori et al (2009) evaluated the expression of EGFR in 45 HER2-positive metastatic breast cancer patients treated with trastuzumab, and found that EGFR overexpression was not associated with response to trastuzumab, time to progression or overall survival. In contrast, in another study, EGFR expression was associated with decreased overall survival of HER2-positive metastatic breast cancer patients treated with trastuzumab (Gallardo et al, 2012).
Epidermal growth factor receptor copy number alteration, one of the mechanisms of EGFR overexpression, is also highly variable in breast cancer, with amplification frequencies up to 24% in triple-negative breast cancer (Bhargava et al, 2005; Gumuskaya et al, 2010; Koletsa et al, 2010; Shao et al, 2011; Lv et al, 2011a; Martin et al, 2012). Recently, we reported that high EGFR copy number because of EGFR amplification or high polysomy is an independent prognostic factor for poor disease-free survival in patients with triple-negative breast cancer (Park et al, 2014). However, the prognostic significance of EGFR copy number in HER2-positive breast cancer is not clear.
In this study, we assessed the clinicopathologic significance of EGFR overexpression and EGFR copy number alteration in a large series of HER2-positive primary breast cancers from two institutions. In addition, we examined the correlation of EGFR expression with trastuzumab response and clinical outcome in HER2-positive primary and metastatic breast cancer patients treated by trastuzumab.
Materials and methods
Patients and tissue samples
We used three different sets of breast cancer samples in this study. First, we retrospectively examined 242 cases of HER2-positive primary breast cancers, which comprised 105 cases operated on at Seoul National University Bundang Hospital (SNUBH) between 2003 and 2009, and 137 cases operated on at Asan Medical Center (AMC) between 2003 and 2004. Expression of HER2 was scored according to the 2007 ASCO/CAP guidelines: 0, no staining; 1+, weak and incomplete membranous staining in ⩾10% of the tumour cells; 2+, weak to moderate, complete membranous staining in ⩾10% of the tumour cells and 3+, strong, complete membranous staining in ⩾30% of the tumour cells (Wolff et al, 2007). Human epidermal growth factor receptor2 positivity was defined as a score of 3+ in the IHC test, or amplification shown by fluorescence in situ hybridisation. Baseline characteristics of the patients are summarised in Supplementary Table S1.
The second set comprised 447 cases of HER2-positive primary breast cancer treated with chemotherapy and with adjuvant trastuzumab for 1 year at Asan Medical Center from 2006 to 2011, which were used for a previous study (Lee et al, 2014a). Of the 450 cases analysed in the previous study (Lee et al, 2014a), three cases were occult breast cancers, and thus were excluded for this study. The third set was 112 cases of HER2-positive metastatic breast cancer treated with trastuzumab at Seoul National University Bundang Hospital, Seoul National University Hospital and Asan Medical Center from 2001 to 2011 (Lee et al, 2014b). In 80 of the 112 metastatic cases, the tissue originated from the primary cancer site, and in the remaining cases it was obtained from metastatic sites. Trastuzumab was administered as the first-line treatment for metastatic breast cancer in 99 patients (88.4%). Taxane was used most frequently used in trastuzumab-based combination chemotherapy (in 107 patients, 95.5%), and other agents (i.e. gemcitabine, capecitabine, and vinorelbine) were combined with trastuzumab in three cases.
All cases were independently reviewed by two breast pathologists (SYP and HJL), and the following histopathologic variables were examined: histologic subtype, T stage, nodal status, Nottingham combined histologic grade and lymphvascular invasion. This study was approved by the institutional review boards of Seoul National University Bundang Hospital, Asan Medical Center and Seoul National University Hospital; the requirement for informed consent was waived.
Tissue microarray construction
For the first set, tissue microarrays (TMAs) were constructed from archival formalin-fixed, paraffin-embedded tissue blocks. Briefly, a representative tumour area was carefully selected for each tumour from haematoxylin- and eosin-stained sections. The designated zone of each donor block was punched with a 2 mm diameter tissue cylinder, and the sample was transferred to a recipient block. Each sample was arrayed in triplicate. For the second set, we used TMAs that were constructed in a previous study (Lee et al, 2014a). For metastatic cases, TMAs were constructed from archival formalin-fixed, paraffin-embedded tissue blocks from resected cases. After selection of a representative tumour area, each donor block was punched with a tissue cylinder 4 mm in diameter. For biopsy specimen, whole sections were evaluated.
Immunohistochemistry and scoring
Formalin-fixed and paraffin-embedded tissue sections were cut, dried, dewaxed in xylene and rehydrated through graded alcohol. Epidermal growth factor receptor expression was detected using EGFR pharmDx Kit (Dako, Capinteria, CA, USA) and scored as follows: 0, no staining, or weak membranous staining in <10% of the tumour cells; 1+, weak membranous staining in ⩾10% of the tumour cells; 2+, moderate, membranous staining in ⩾10% of the tumour cells; 3+, strong membranous staining in ⩾10% of the tumour cells. Both complete and incomplete membranous staining was accepted. If the triplicate TMA cores yielded different scores, the highest score was used. In the first set, we classified tumours into two groups according to the staining pattern of EGFR: homogeneous expression (0/1+ or 2+/3+ in all TMA cores) and heterogeneous expression (mixed pattern of 0/1+ and 2+/3+ in three TMA cores).
Oestrogen and progesterone receptors were regarded as positive if there were at least 1% positive tumour nuclei (Hammond et al, 2010). Cases with 10% or more positive staining were grouped as positive for p53. The Ki-67 proliferation index was defined as the percentage of tumour cells showing nuclear positivity.
Fluorescence in situ hybridisation assays for EGFR
Fluorescence in situ hybridisation (FISH) was performed on the TMA samples with commercially available locus-specific and chromosome enumeration probes (CEPs) (LSI EGFR SpectrumOrange probe (7p12) and CEP 7 SpectrumGreen probe (7p11.1–q11.1); Abbott Molecular, Des Plaines, IL, USA). Briefly, 4-μm deparaffinised TMA sections were incubated in the pretreatment solution (Abbott Molecular) at 80 °C for 30 min, and then in the protease solution (Abbott Molecular) for 25 min at 37 °C. Probes were diluted in tDen-Hyb-2 hybridisation buffer (InSitus Biotechnologies, Albuquerque, NM, USA). Codenaturation of the probes and DNA was achieved by incubating at 75 °C for 5 min in a HYBrite hybridisation chamber (Abbott Molecular) followed by 16-h hybridisation at 37 °C. Posthybridisation washes were performed according to the supplier protocols. Slides were mounted in 4′,6-diamidino-2-phenylindole/antifade and viewed with a fluorescence microscope.
A total of 50 tumour cells were evaluated for each core, and the genetic variables evaluated were: EGFR gene copy number, chromosome 7 copy number and average EGFR gene : chromosome 7 ratio. The University of Colorado Cancer Center criteria for the EGFR gene were used as follows: disomy (⩽2 copies in ⩾90% of cells), low trisomy (⩽2 copies in ⩾40% of cells, 3 copies in 10–40% of cells, ⩾4 copies in <10% of cells), high trisomy (⩽2 copies in ⩾40% of cells, 3 copies in ⩾40% of cells, ⩾4 copies in <10% of cells), low polysomy (⩾4 copies in 10–40% of cells) and high polysomy (⩾4 copies in ⩾40% of cells). For gene amplification, the criterion was a ratio of EGFR to chromosome 7 of ⩾2 or ⩾15 copies of EGFR per cell in ⩾10% of the analysed cells (Cappuzzo et al, 2005). For further analysis, the patients were divided into two groups according to EGFR copy number as follows: low EGFR copy number (disomy, low trisomy, high trisomy and low polysomy) and high EGFR copy number (high polysomy and gene amplification).
Assessment of trastuzumab response for HER2-positive metastatic breast cancer
Responses to trastuzumab-based therapy were evaluated every 8 to 12 weeks using Response Evaluation Criteria in Solid Tumours criteria version 1.1 (Eisenhauer et al, 2009). Clinical benefit was defined as complete response, partial response or stable disease over at least 6 months. Time to progression was defined as the time from the initiation of trastuzumab treatment to disease progression, and overall survival as the time from initiation of trastuzumab treatment to patient's death from any cause.
Statistical analysis
Statistical significance was assessed using Statistical Package, SPSS version 18.0 for Windows (SPSS, Chicago, IL, USA). The correlation between EGFR overexpression and EGFR gene : chromosome 7 ratio was determined by Spearman's correlation analysis. The associations of EGFR overexpression or high EGFR copy number with clinicopathological characteristics of the tumours were analysed using Student's t-test, Fisher's exact test or the χ2 test, depending on the test conditions. Survival curves were estimated using the Kaplan–Meier method, and the significance of differences between survival curves was determined using the log-rank test. Covariates that were statistically significant in the univariate analysis were included in a multivariate analysis using a Cox proportional hazards regression model, and the hazard ratio (HR) and its 95% confidence interval (CI) were assessed for each factor. P-values of <0.05 were considered statistically significant. All P-values reported are two-sided.
Results
Correlation between EGFR overexpression and EGFR copy number alteration in HER2-positive primary breast cancer
The EGFR IHC and FISH results obtained in the 242 HER2-positive primary breast cancers are shown in Supplementary Table S2. Epidermal growth factor receptor protein expression was not evaluable in 5 of the 242 cases, because of the loss of TMA cores. Of the other 237 cases, 149 (62.9%) did not express EGFR protein (IHC score 0); 30 (12.7%) were scored as 1+, another 30 (12.7%) as 2+ and 28 cases (11.8%) were scored as 3+. Of the 58 cases with EGFR expression of 2+ or more, 27 showed heterogeneous EGFR expression in three TMA cores. Epidermal growth factor receptor copy number data were available in 232 cases; they were unavailable in the remainder because of the detachment of TMA cores or inadequate hybridisation. Epidermal growth factor receptor gene amplification and high polysomy were detected in 11 (4.7%) and 13 cases (5.6%), respectively. The distribution of EGFR IHC scores and EGFR copy number did not differ between the two institutions (χ2 square test, P=0.723, P=0.664, respectively).
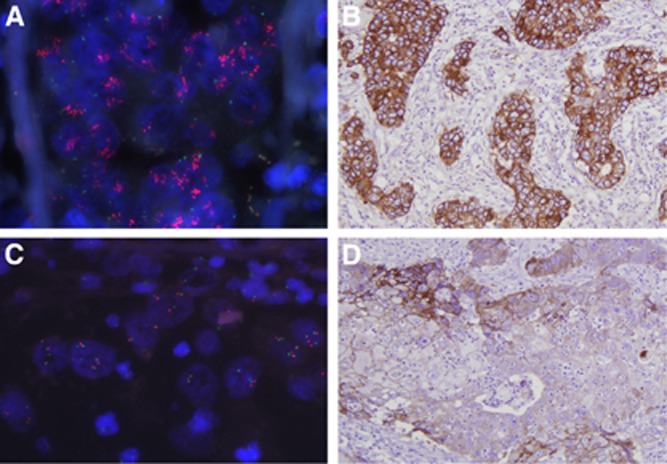
We compared the results of EGFR IHC and EGFR FISH in each TMA core. Epidermal growth factor receptor protein overexpression of 3+ and 2+ or more was associated with high EGFR copy number (all P<0.001; Table 1). In a correlation analysis, a weak but significant positive correlation was found between EGFR IHC score and EGFR: chromosome 7 ratio (ρ=0.282, P<0.001). Of the 11 cases in which EGFR was amplified, five scored as 3+, three as 2+ and three as 0 (Supplementary Table S3 and Figure 1).
Table 1. Correlation between EGFR protein expression and EGFR copy number alteration in tissue microarray cores.
|
EGFR
FISH |
|||
|---|---|---|---|
| Low gene copy numbera (%) | High polysomy (%) | Gene amplification (%) | |
|
EGFR IHC | |||
| 0 | 354 (70.5) | 10 (45.5) | 3 (14.3) |
| 1+ | 68 (13.5) | 2 (9.1) | 1 (4.8) |
| 2+ | 52 (10.4) | 6 (27.3) | 4 (19.0) |
| 3+ | 28 (5.6) | 4 (18.2) | 13 (61.9) |
Abbreviations: EGFR=epidermal growth factor receptor; FISH=fluorescence in situ hybridization; IHC=immunohistochemistry; TMA=tissue microarray.
Include disomy, trisomy and low polysomy.; Of the 726 TMA cores (three cores for 242 cases), data of 545 cores were available because of tissue loss, inadequate hybridisation or failure of FISH.
Figure 1.
EGFR fluorescence in situ hybridisation and EGFR immunohistochemistry. An example of EGFR amplification (A) showing strong (3+) EGFR overexpression (B). An example of EGFR high polysomy (C) showing strong (3+) but focal EGFR overexpression (D).
Clinicopathologic characteristics of cases with EGFR overexpression or high EGFR copy number
We examined the clinicopathologic significance of EGFR overexpression by two different criteria (3+ 2+ or more), because there is no standardised cutoff point for EGFR overexpression in breast cancer. Clinicopathologic characteristics of tumours showing EGFR overexpression or high EGFR copy number are summarised in Table 2. Epidermal growth factor receptor overexpression was significantly correlated with hormone receptor negativity, and higher Ki-67 proliferation index, irrespective of the criteria for positivity (EGFR overexpression of 3+, P<0.001, P=0.014, respectively; EGFR overexpression of 2+ or more, P<0.001, P=0.003, respectively). High EGFR copy number tended to be associated with hormone receptor negativity, but this did not reach statistical significance (P=0.078).
Table 2. Clinicopathologic characteristics of HER2-positive primary breast cancers with EGFR overexpression or high EGFR copy number in the first set.
|
EGFR overexpression (3+) |
EGFR overexpression (2+ or more) |
High
EGFR
copy number |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Clinicopathological characteristics | Absent (n=209) | Present (n=28) | P-value | Absent (n=179) | Present (n=58) | P-value | Absent (n=208) | Present (n=24) | P-value |
| pT stage | 0.070 | 0.130 | 0.830 | ||||||
| T1 | 99 (47.4) | 8 (28.6) | 86 (48.0) | 21 (36.2) | 95 (45.7) | 10 (41.7) | |||
| T2–T4 | 110 (52.6) | 20 (71.4) | 93 (52.0) | 37 (63.8) | 113 (54.3) | 14 (58.3) | |||
| pN stage | 0.322 | 0.230 | 1.000 | ||||||
| N0 | 111 (53.1) | 12 (42.9) | 97 (54.2) | 26 (44.8) | 108 (51.9) | 13 (54.2) | |||
| N1–N3 | 98 (46.9) | 16 (57.1) | 82 (45.8) | 32 (55.2) | 100 (48.1) | 11 (45.8) | |||
| Stage | 0.356 | 0.119 | 1.000 | ||||||
| I and II | 159 (76.1) | 19 (67.9) | 139 (77.7) | 39 (67.2) | 158 (76.0) | 18 (75.0) | |||
| III | 50 (23.9) | 9 (32.1) | 40 (22.3) | 19 (32.8) | 50 (24.0) | 6 (25.0) | |||
| Histologic grade | 0.385 | 0.067 | 0.234 | ||||||
| I and II | 63 (30.1) | 6 (21.4) | 58 (32.4) | 11 (19.0) | 62 (29.8) | 4 (16.7) | |||
| III | 146 (69.9) | 22 (78.6) | 121 (67.6) | 47 (81.0) | 146 (70.2) | 20 (83.3) | |||
| Lymphovascular invasion | 0.838 | 0.282 | 1.000 | ||||||
| Negative | 126 (60.3) | 16 (57.1) | 111 (62.0) | 31 (53.4) | 126 (60.6) | 15 (62.5) | |||
| Positive | 83 (39.7) | 12 (42.9) | 68 (38.0) | 27 (46.6) | 82 (39.4) | 9 (37.5) | |||
| Hormone receptor | <0.001 | <0.001 | 0.078 | ||||||
| Negative | 123 (58.9) | 26 (92.9) | 94 (52.5) | 55 (94.8) | 125 (60.1) | 19 (79.2) | |||
| Positive | 86 (41.1) | 2 (7.1) | 85 (47.5) | 3 (5.2) | 83 (39.9) | 5 (20.8) | |||
| p53 overexpression | 0.227 | 0.098 | 0.130 | ||||||
| Negative | 117 (56.0) | 12 (42.9) | 103 (57.5) | 26 (44.8) | 115 (55.3) | 9 (37.5) | |||
| Positive | 92 (44.0) | 16 (57.1) | 76 (42.5) | 32 (55.2) | 93 (44.7) | 15 (62.5) | |||
| Ki-67 index | 0.014 | 0.003 | 0.879 | ||||||
| 25.1±14.3 | 32.3±15.1 | 24.4±13.3 | 30.8±17.1 | 26.1±14.2 | 26.6±16.9 | ||||
Abbreviations: EGFR=epidermal growth factor receptor; HER2=human epidermal growth factor receptor 2.
Prognostic significance of EGFR overexpression and EGFR copy number increase
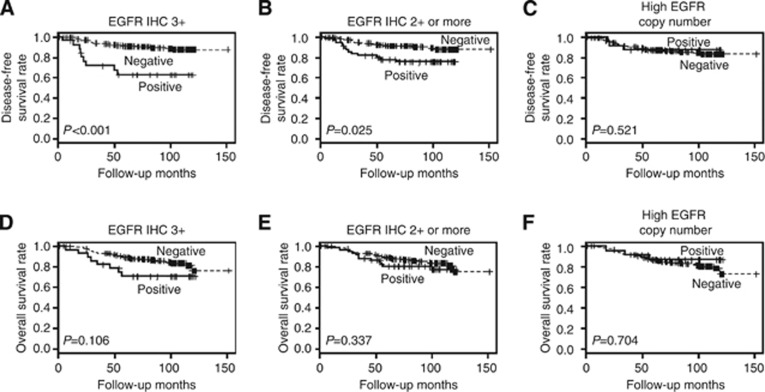
At the time of the analysis, the median follow-up for patients was 98.3 months (range, 1–152 months). There were 41 (16.9%) recurrences and 1 (0.4%) cancer-related death as the first event. In survival analysis, EGFR overexpression defined as 3+ (P<0.001; Figure 2A) and 2+ or more (P=0.025; Figure 2B) was associated with worse disease-free survival of the patients. However, high EGFR copy number was not associated with disease-free survival (P=0.521; Figure 2C). The overall survival of the patients was not affected by EGFR expression or EGFR copy number alteration (Figures 2D–F). Pattern of EGFR overexpression (heterogeneous or homogeneous) was not associated with survival of the patients, either.
Figure 2.
Disease-free and overall survival according to EGFR overexpression and EGFR copy number alteration in HER2-positive primary breast cancer. EGFR overexpression defined as 3+ (A) and EGFR overexpression defined as 2+ or more (B) were associated with poor disease-free survival, whereas high EGFR copy number (C) was not associated with disease-free survival. EGFR overexpression defined as 3+ (D) or as 2+ or more (E), or high EGFR copy number (F) was not associated with overall survival.
Besides EGFR overexpression (3+, 2+ or more), high T stage and lymph node metastasis were associated with poor disease-free survival (Table 3). In multivariate analysis including EGFR overexpression (3+), high T stage and lymph node metastasis, both lymph node metastasis (pN0 vs pN1∼pN3; HR, 5.611; 95% CI, 2.262–13.357; P<0.001) and EGFR overexpression (0∼2+ vs 3+ HR, 3.013; 95% CI, 1.481–6.131; P=0.002) were found to be independent prognostic factors of poor disease-free survival. However, in multivariate analysis including EGFR overexpression (2+ or more), only lymph node metastasis (HR, 5.465; 95% CI, 2.235–13.363; P<0.001) was an independent prognostic indicator of poor disease-free survival.
Table 3. Univariate anaylsis of factors associated with disease-free and overall survival for HER2-positive primary breast cancer patients in the first set.
|
Disease-free survival |
Overall survival |
|||||
|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|
EGFR overexpression (3+) | ||||||
| Negative | — | — | ||||
| Positive | 3.593 | 1.774–7.278 | <0.001 | 1.928 | 0.870–4.273 | 0.106 |
|
EGFR overexpression (2+ or more) | ||||||
| Negative | — | — | ||||
| Positive | 2.121 | 1.100–4.089 | 0.025 | 1.403 | 0.703–2.799 | 0.337 |
|
High EGFR copy number | ||||||
| Negative | — | — | ||||
| Positive | 0.68 | 0.209–2.208 | 0.521 | 0.798 | 0.244–2.594 | 0.704 |
|
Age (years) | ||||||
| <50 | — | — | ||||
| ⩾50 | 1.284 | 0.690–2.388 | 0.431 | 1.256 | 0.662–2.385 | 0.485 |
|
pT stage | ||||||
| T1 | — | — | ||||
| T2–T4 | 2.214 | 1.105–4.436 | 0.025 | 1.387 | 0.716–2.684 | 0.332 |
|
pN stage | ||||||
| N0 | — | — | ||||
| N1–N3 | 5.832 | 2.579–13.191 | <0.001 | 3.654 | 1.727–7.732 | 0.001 |
|
Histologic grade | ||||||
| I and II | — | — | ||||
| III | 1.492 | 0.707–3.148 | 0.294 | 1.264 | 0.613–2.607 | 0.526 |
|
Lymphovascular invasion | ||||||
| Negative | — | — | ||||
| Positive | 1.756 | 0.944–3.269 | 0.076 | 1.668 | 0.903–3.081 | 0.102 |
|
Hormone receptor status | ||||||
| Negative | — | — | ||||
| Positive | 0.672 | 0.342–1.318 | 0.247 | 0.751 | 0.379–1.489 | 0.413 |
|
p53 expression | ||||||
| Negative | — | — | ||||
| Positive | 0.829 | 0.440–1.562 | 0.562 | 0.962 | 0.535–1.930 | 1.016 |
|
Adjuvant trastuzumab | ||||||
| Not received | — | — | ||||
| Received | 0.804 | 0.212–3.048 | 0.748 | 1.289 | 0.318–5.225 | 0.723 |
|
Adjuvant chemotherapy | ||||||
| Not received | — | — | ||||
| Received | 0.582 | 0.257–1.317 | 0.194 | 0.561 | 0.246–1.279 | 0.169 |
Abbreviations: CI=confidence interval; EGFR=epidermal growth factor receptor; HR=hazard ratio.
In the subgroup analyses, according to adjuvant trastuzumab therapy, EGFR overexpression of 3+ was also an independent prognostic factor of poor disease-free survival (0∼2+ vs 3+ HR, 3.065; 95% CI, 1.480–6.348; P=0.003) in the subgroup not receiving adjuvant trastuzumab (n=209), but EGFR overexpression of 2+ or more and high EGFR copy number were not associated with disease-free survival. However, as only a small number of patients (n=33) were treated with adjuvant trastuzumab, we could not determine the prognostic and predictive significance of EGFR overexpression in this subgroup. Therefore, we additionally analysed EGFR overexpression in an independent set of HER2-positive primary breast cancer patients treated with adjuvant trastuzumab.
Correlation between EGFR overexpression and clinical outcome in HER2-positive primary breast cancer patients treated with adjuvant trastuzumab
In the second set, which comprised 447 HER2-positive primary breast cancer patients treated with adjuvant trastuzumab for 1 year, EGFR expression data were not available in five cases, because of the loss of TMA cores. Of the other 442 cases, 42 (9.5%) were scored as 1+, 46 (10.4%) as 2+ and 30 cases (6.8%) as 3+. Epidermal growth factor receptor overexpression also showed correlations with high histologic grade, hormone receptor negativity and high Ki-67 labelling index (Supplementary Table S4).
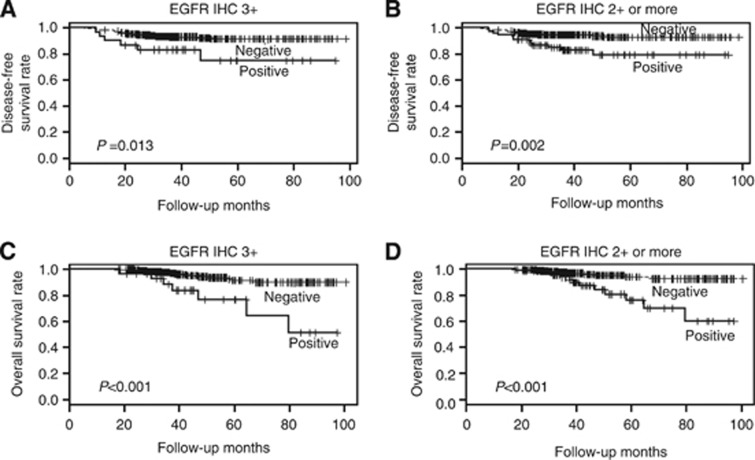
At the time of the analysis, the median follow-up for patients was 49 months (range, 18–104 months). There were 34 (7.6%) recurrences. In survival analysis, the patients with EGFR-overexpressing tumour had shorter disease-free survival time than those without it (3+, P=0.013; 2+ or more, P=0.002; Figures 3A and B). The overall survival of the patients was also significantly affected by EGFR overexpression, showing poor overall survival (3+, P<0.001; 2+ or more, P<0.001; Figures 3C and D). In this second set, high T stage, lymph node metastasis and lymphovascular invasion were also associated with poor disease-free or overall survival of the patients (Supplementary Table S5). In multivariate analysis, both EGFR overexpression (3+ 2+ or more) and lymphovascular invasion were found to be independent prognostic factors for poor disease-free and overall survival of the patients (Table 4).
Figure 3.
Disease-free and overall survival according to EGFR overexpression in HER2-positive primary breast cancer patients treated with adjuvant trastuzumab. EGFR overexpression defined as 3+ (A and C) and EGFR overexpression defined as 2+ or more (B and D) were associated with poor disease-free and overall survival.
Table 4. Multivariate survival analyses for HER2-positive primary breast cancer patients treated with adjuvant trastuzumab in the second set.
|
Multivariate analysis |
|||||
|---|---|---|---|---|---|
| Model | Variable | Category | HR | 95% CI | P-value |
|
Disease-free survival | |||||
| 1 | EGFR overexpression (3+) | Positive vs negative | 2.837 | 1.172–6.863 | 0.021 |
| pT stage | T2–T3 vs T1 | 2.000 | 0.928–4.310 | 0.077 | |
| pN stage | N1–N3 vs N0 | 1.608 | 0.637–4.063 | 0.315 | |
| Lymphovascular invasion | Positive vs negative | 2.368 | 1.113–5.038 | 0.025 | |
| 2 | EGFR overexpression (2+ or more) | Positive vs negative | 3.177 | 1.587–6.359 | 0.001 |
| pT stage | T2–T3 vs T1 | 2.106 | 0.980–4.522 | 0.056 | |
| pN stage | N1–N3 vs N0 | 1.673 | 0.667–4.196 | 0.273 | |
| Lymphovascular invasion | Positive vs negative | 2.487 | 1.175–5.264 | 0.017 | |
|
Overall survival | |||||
| 1 | EGFR overexpression (3+) | Positive vs negative | 4.697 | 1.958–11.266 | 0.001 |
| pT stage | T2–T3 vs T1 | 2.640 | 0.987–7.061 | 0.053 | |
| Lymphovascular invasion | Positive vs negative | 2.870 | 1.190–6.924 | 0.019 | |
| 2 | EGFR overexpression (2+ or more) | Positive vs negative | 4.582 | 2.075–10.119 | <0.001 |
| pT stage | T2–T3 vs T1 | 2.808 | 1.051–7.498 | 0.039 | |
| Lymphovascular invasion | Positive vs negative | 3.143 | 1.303–7.582 | 0.011 | |
Abbreviations: CI=confidence interval; EGFR=epidermal growth factor receptor; HR=hazard ratio.
Correlation between EGFR overexpression and response to trastuzumab-based therapy in HER2-positive metastatic breast cancer in the third set
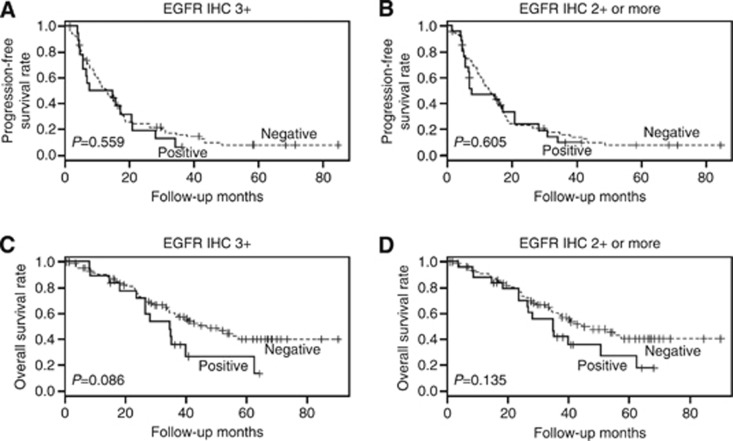
We also examined EGFR overexpression in HER2-positive metastatic breast cancers treated with trastuzumab-based chemotherapy to evaluate the predictive significance of EGFR overexpression, as well as its prognostic significance. Epidermal growth factor receptor IHC failed in 5 of the 112 cases. Epidermal growth factor receptor expression was 2+ in 7.5% (8 out of 107) and 3+ in 17.8% (19 out of 107). Epidermal growth factor receptor overexpression was also associated with hormone receptor negativity (3+, P=0.002; 2+ or more, P<0.001; Supplementary Table S6). The clinical benefit of trastuzumab treatment did not differ between the cases in which there was EGFR overexpression and those in which there was not (3+, 84.2% vs 84.3%, P=1.000; 2+ or more, 84.6% vs 84.2%, P=1.000). Epidermal growth factor receptor overexpression was not associated with progression-free survival irrespective of the criterion used (3+ HR, 1.177; 95% CI, 0.682–2.033; P=0.559 (Figure 4A); 2+ or more, HR, 1.140; 95% CI, 0.694–1.870; P=0.605 (Figure 4B)). Although EGFR overexpression tended to be associated with decreased overall survival, the effect did not reach statistical significance (3+ HR, 1.733; 95% CI, 0.925–3.244; P=0.086 (Figure 4C); 2+ or more; HR, 1.561; 95% CI, 0.870–2.800; P=0.135 (Figure 4D)).
Figure 4.
Progression-free and overall survival according to EGFR expression in HER2-positive metastatic breast cancer. EGFR overexpression defined as 3+ (A) and 2+ or more (B) were not associated with progression-free survival. EGFR overexpression defined as 3+ (C) and 2+ or more (D) tends to be associated with decreased overall survival.
Discussion
Epidermal growth factor receptor alterations occur at an advanced stage of malignancy characterised by metastatic competence, and EGFR is thought to promote cancer cell migration and invasion (Masuda et al, 2012). In the present study, we have shown that EGFR overexpression is a poor prognostic factor in two independent sets of HER2-positive primary breast cancer patients. In the first set, patients with EGFR-overexpressing tumours showed poor disease-free survival, and the poor prognostic impact of EGFR overexpression (3+) was also applied to the subgroup not receiving trastuzumab. Furthermore, in the second set, EGFR overexpression was associated with poor disease-free and overall survival of the patients treated with adjuvant trastuzumab. Therefore, these results indicate that EGFR overexpression has a prognostic and predictive value in HER2-positive primary breast cancers.
However, there are, at present, no validated scoring systems for EGFR protein overexpression assessed by IHC. We therefore analysed the clinical significance of EGFR overexpression using two different cutoff values (3+, 2+ or more). We found that prognostic impact of EGFR overexpression using two different criteria was similar, although not same. Previous studies that showed prognostic significance of EGFR overexpression in HER2-positive breast cancer also used different cutoff values for EGFR expression (Suo et al, 2002; DiGiovanna et al, 2005; Nieto et al, 2007), and even any membranous staining was found to be significant (DiGiovanna et al, 2005). However, they used different clones and different staining methods for EGFR IHC. In this study, we used EGFR pharmDx Kit, which is approved for the identification of colorectal cancer patients eligible for treatment with cetuximab or panitumumab, but any membranous staining or expression of 1+ or more had no prognostic impact on the survival of the patients. Thus, most reliable cutoff values for EGFR overexpression and its standard staining method should be determined to predict the prognosis of HER2-positive breast cancer patients, but, in the study, EGFR overexpression of 3+ was found to be most predictive.
We also revealed that the clinicopathologic characteristics of EGFR-overexpressing HER2-positive breast cancers were same irrespective of the cutoff value used. Especially, the association of EGFR overexpression with hormone receptor negativity was clearly demonstrated in the three different sets of HER2-positive breast cancers. Recently, the Cancer Genome Atlas (TCGA) Network analysed primary breast cancers with various platforms, and showed that there were at least two types of clinically HER2-positive breast cancers (Cancer Genome Atlas N, 2012). One corresponded to the subtype which is enriched for HER2 mRNA and in which there is high expression of receptor tyrosine kinases including EGFR, HER2 and FGFR4. The other is the luminal mRNA subtype with high expression of GATA3, BCL2 and ESR1, the so-called luminal cluster of genes. In our study, the correlation between EGFR overexpression and hormone receptor negativity is consistent with the results of TCGA analysis, and suggests that HER2-positive breast cancers have different features depending on their hormone receptor status.
The relationship between EGFR overexpression and high EGFR copy number has not been clearly defined. In this study, analysis of TMA cores revealed a positive correlation between EGFR gene status and EGFR protein overexpression. In particular, cases with high levels of EGFR amplification (case nos. 196 and 224; Supplementary Table S3) yielded 3+ EGFR IHC results in all three cores evaluated. However, our data also revealed that EGFR overexpression had low specificity and low positive predictive value for high EGFR copy number as not all tumours with high EGFR copy number overexpressed EGFR and not all EGFR-overexpressing tumours had high EGFR copy numbers. Furthermore, while EGFR overexpression was associated with poor prognosis in HER2-positive primary breast cancer patients, high EGFR copy number did not have prognostic significance. These results suggest that besides increased EGFR gene copy number, EGFR overexpression may be induced by other mechanisms, such as mutation, aberrant transcription or translational modification.
Epidermal growth factor receptor overexpression did not affect the response to trastuzumab and progression-free survival in patients with HER2-positive metastatic breast cancers, although EGFR overexpression tended to be associated with poor overall survival. In this study, predictive value of EGFR overexpression for trastuzumab response in metastatic setting seems to be different from that in an adjuvant setting. However, the third metastatic breast cancer set has some limitations; not all patients were treated by the same protocol, and most of the tissues used for analysis were primary tumours even though there were possibilities of change in EGFR status during tumour progression. In HER2-positive primary breast cancer, EGFR overexpression was associated with worse survival in the patients not treated with trastuzumab, as well as those treated with adjuvant trastuzuamb. Therefore, the association of EGFR overexpression with poor clinical outcome in patients receiving adjuvant trastuzuamb cannot be solely explained by resistance to trastuzumab caused by EGFR overexpression. The utility of EGFR overexpression as a predictive biomarker for traststuzumab response should be validated carefully in further large-scale studies.
In the light of the known interplay between EGFR and HER2, EGFR inhibitor gefitinib has been tried in combination with trastuzuamb to treat patients with HER2-positive metastatic breast cancer (Arteaga et al, 2008; Somlo et al, 2012). However, this combination of gefitinib and trastuzumab was not found to be more effective than traststuzumab alone. Moreover, EGFR expression was not associated with response to treatment. However, in those studies, the number of enrolled patients was too small to permit meaningful statistical analysis. Hence, the utility of EGFR overexpression as a predictive biomarker for EGFR-targeted therapy and as a prognostic factor for HER2-positive metastatic breast cancer should be validated in large studies.
There are treatments that may improve the clinical outcome of patients with HER2-positive breast cancers overexpressing EGFR. A dual EGFR/HER2 tyrosine kinase inhibitor, lapatinib, has been shown to be useful after previous treatments such as trastuzumab in cases with progressive metastatic HER2-positive disease (Geyer et al, 2006). Fabi et al (2013) evaluated EGFR gene copy number and responses to lapatinib, and identified increased EGFR copy number as a positive predictive factor for lapatinib response. In addition to lapatinib, other small-molecule inhibitors targeting ERBB family members, such as afatinib and neratinib, are being investigated in ongoing trials in the context of trastuzumab-resistant metastatic HER2-positive breast cancer (Gradishar, 2013).
In conclusion, our study showed that EGFR protein overexpression is an independent poor prognostic factor in HER2-positive primary breast cancer, and it is also a predictive factor for trastuzumab response in HER2-positive primary breast cancer, but not in metastatic breast cancer.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (Grant No. NRF-2012R1A1B4000557) and by Seoul National University Bundang Hospital Research Fund (Grant No. 11-2012-007).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Arteaga CL, O'Neill A, Moulder SL, Pins M, Sparano JA, Sledge GW, Davidson NE. A phase I–II study of combined blockade of the ErbB receptor network with trastuzumab and gefitinib in patients with HER2 (ErbB2)-overexpressing metastatic breast cancer. Clin Cancer Res. 2008;14:6277–6283. doi: 10.1158/1078-0432.CCR-08-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava R, Gerald WL, Li AR, Pan Q, Lal P, Ladanyi M, Chen B. EGFR gene amplification in breast cancer: correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations. Mod Pathol. 2005;18:1027–1033. doi: 10.1038/modpathol.3800438. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, Haney J, Witta S, Danenberg K, Domenichini I, Ludovini V, Magrini E, Gregorc V, Doglioni C, Sidoni A, Tonato M, Franklin WA, Crino L, Bunn PA, Jr, Varella-Garcia M. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Jr, Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- Diermeier S, Horvath G, Knuechel-Clarke R, Hofstaedter F, Szollosi J, Brockhoff G. Epidermal growth factor receptor coexpression modulates susceptibility to Herceptin in HER2/neu overexpressing breast cancer cells via specific erbB-receptor interaction and activation. Exp Cell Res. 2005;304:604–619. doi: 10.1016/j.yexcr.2004.12.008. [DOI] [PubMed] [Google Scholar]
- DiGiovanna MP, Stern DF, Edgerton SM, Whalen SG, Moore D, 2nd, Thor AD. Relationship of epidermal growth factor receptor expression to ErbB-2 signaling activity and prognosis in breast cancer patients. J Clin Oncol. 2005;23:1152–1160. doi: 10.1200/JCO.2005.09.055. [DOI] [PubMed] [Google Scholar]
- Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Fabi A, Merola R, Ferretti G, Di Benedetto A, Antoniani B, Ercolani C, Nistico C, Papaldo P, Ciccarese M, Sperduti I, Vici P, Marino M, Gori S, Botti C, Malaguti P, Cognetti F, Mottolese M. Epidermal growth factor receptor gene copy number may predict lapatinib sensitivity in HER2-positive metastatic breast cancer. Expert Opin Pharmacother. 2013;14:699–706. doi: 10.1517/14656566.2013.779672. [DOI] [PubMed] [Google Scholar]
- Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2–pertuzumab complex. Cancer Cell. 2004;5:317–328. doi: 10.1016/s1535-6108(04)00083-2. [DOI] [PubMed] [Google Scholar]
- Gallardo A, Lerma E, Escuin D, Tibau A, Munoz J, Ojeda B, Barnadas A, Adrover E, Sanchez-Tejada L, Giner D, Ortiz-Martinez F, Peiro G. Increased signalling of EGFR and IGF1R, and deregulation of PTEN/PI3K/Akt pathway are related with trastuzumab resistance in HER2 breast carcinomas. Br J Cancer. 2012;106:1367–1373. doi: 10.1038/bjc.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, Pienkowski T, Jagiello-Gruszfeld A, Crown J, Chan A, Kaufman B, Skarlos D, Campone M, Davidson N, Berger M, Oliva C, Rubin SD, Stein S, Cameron D. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355:2733–2743. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- Gori S, Sidoni A, Colozza M, Ferri I, Mameli MG, Fenocchio D, Stocchi L, Foglietta J, Ludovini V, Minenza E, De Angelis V, Crino L. EGFR, pMAPK, pAkt and PTEN status by immunohistochemistry: correlation with clinical outcome in HER2-positive metastatic breast cancer patients treated with trastuzumab. Ann Oncol. 2009;20:648–654. doi: 10.1093/annonc/mdn681. [DOI] [PubMed] [Google Scholar]
- Gradishar WJ. Emerging approaches for treating HER2-positive metastatic breast cancer beyond trastuzumab. Ann Oncol. 2013;24:2492–2500. doi: 10.1093/annonc/mdt217. [DOI] [PubMed] [Google Scholar]
- Gumuskaya B, Alper M, Hucumenoglu S, Altundag K, Uner A, Guler G. EGFR expression and gene copy number in triple-negative breast carcinoma. Cancer Genet Cytogenet. 2010;203:222–229. doi: 10.1016/j.cancergencyto.2010.07.118. [DOI] [PubMed] [Google Scholar]
- Hammond ME, Hayes DF, Wolff AC, Mangu PB, Temin S. American society of clinical oncology/college of american pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Oncol Pract. 2010;6:195–197. doi: 10.1200/JOP.777003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwangbo W, Lee JH, Ahn S, Kim S, Park KH, Kim CH, Kim I. EGFR gene amplification and protein expression in invasive ductal carcinoma of the breast. Korean J Pathol. 2013;47:107–115. doi: 10.4132/KoreanJPathol.2013.47.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koletsa T, Kotoula V, Karayannopoulou G, Nenopoulou E, Karkavelas G, Papadimitriou CS, Kostopoulos I. EGFR expression and activation are common in HER2 positive and triple-negative breast tumours. Histol Histopathol. 2010;25:1171–1179. doi: 10.14670/HH-25.1171. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Park IA, Park SY, Seo AN, Lim B, Chai Y, Song IH, Kim NE, Kim JY, Yu JH, Ahn JH, Gong G. Two histopathologically different diseases: hormone receptor-positive and hormone receptor-negative tumors in HER2-positive breast cancer. Breast Cancer Res Treat. 2014;145:615–623. doi: 10.1007/s10549-014-2983-x. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Seo AN, Kim EJ, Jang MH, Suh KJ, Ryu HS, Kim YJ, Kim JH, Im S, Gong G, Jung KH, Park IA, Park SY.2014HER2 heterogeneity affects trastuzumab responses and survival in patients with HER2-positive metastatic breast cancer Am J Clin Patholin press. [DOI] [PubMed]
- Lv N, Xie X, Ge Q, Lin S, Wang X, Kong Y, Shi H, Wei W. Epidermal growth factor receptor in breast carcinoma: association between gene copy number and mutations. Diagn Pathol. 2011;6:118. doi: 10.1186/1746-1596-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv N, Xie X, Ge Q, Lin S, Wang X, Kong Y, Shi H, Xie X, Wei W. Epidermal growth factor receptor in breast carcinoma: association between gene copy number and mutations. Diagn Pathol. 2011;6:118. doi: 10.1186/1746-1596-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V, Botta F, Zanellato E, Molinari F, Crippa S, Mazzucchelli L, Frattini M. Molecular characterization of EGFR and EGFR-downstream pathways in triple negative breast carcinomas with basal like features. Histol Histopathol. 2012;27:785–792. doi: 10.14670/HH-27.785. [DOI] [PubMed] [Google Scholar]
- Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136:331–345. doi: 10.1007/s10549-012-2289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto Y, Nawaz F, Jones RB, Shpall EJ, Nawaz S. Prognostic significance of overexpression and phosphorylation of epidermal growth factor receptor (EGFR) and the presence of truncated EGFRvIII in locoregionally advanced breast cancer. J Clin Oncol. 2007;25:4405–4413. doi: 10.1200/JCO.2006.09.8822. [DOI] [PubMed] [Google Scholar]
- Park HS, Jang MH, Kim EJ, Kim HJ, Lee HJ, Kim YJ, Kim JH, Kang E, Kim SW, Kim IA, Park SY. High EGFR gene copy number predicts poor outcome in triple-negative breast cancer. Mod Pathol. 2014;27:1212–1222. doi: 10.1038/modpathol.2013.251. [DOI] [PubMed] [Google Scholar]
- Ritter CA, Perez-Torres M, Rinehart C, Guix M, Dugger T, Engelman JA, Arteaga CL. Human breast cancer cells selected for resistance to trastuzumab in vivo overexpress epidermal growth factor receptor and ErbB ligands and remain dependent on the ErbB receptor network. Clin Cancer Res. 2007;13:4909–4919. doi: 10.1158/1078-0432.CCR-07-0701. [DOI] [PubMed] [Google Scholar]
- Shao MM, Zhang F, Meng G, Wang XX, Xu H, Yu XW, Chen LY, Tse GM. Epidermal growth factor receptor gene amplification and protein overexpression in basal-like carcinoma of the breast. Histopathology. 2011;59:264–273. doi: 10.1111/j.1365-2559.2011.03921.x. [DOI] [PubMed] [Google Scholar]
- Somlo G, Martel CL, Lau SK, Frankel P, Ruel C, Gu L, Hurria A, Chung C, Luu T, Morgan R, Jr., Leong L, Koczywas M, McNamara M, Russell CA, Kane SE. A phase I/II prospective, single arm trial of gefitinib, trastuzumab, and docetaxel in patients with stage IV HER-2 positive metastatic breast cancer. Breast Cancer Res Treat. 2012;131:899–906. doi: 10.1007/s10549-011-1850-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo Z, Risberg B, Kalsson MG, Willman K, Tierens A, Skovlund E, Nesland JM. EGFR family expression in breast carcinomas. c-erbB-2 and c-erbB-4 receptors have different effects on survival. J Pathol. 2002;196:17–25. doi: 10.1002/path.1003. [DOI] [PubMed] [Google Scholar]
- Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- Wrba F, Reiner A, Ritzinger E, Holzner JH, Reiner G. Expression of epidermal growth factor receptors (EGFR) on breast carcinomas in relation to growth fractions, estrogen receptor status and morphological criteria. An immunohistochemical study. Pathol Res Pract. 1988;183:25–29. [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.