Abstract
Background:
Aberrant expression of microRNAs (miRNAs) is associated with cancer progression, initiation and metastasis. MiR34a is a miRNA that has been previously described as a tumour suppressor. Herein, we assess the expression of miR34a in three independent breast cancer cohorts using a quantitative in situ hybridisation assay (qISH) and determined its association with disease-specific death in breast cancer.
Methods:
The qISH method was applied to three independent primary breast cancer cohorts (Cohort 1 with 461, Cohort 2 with 279 and Cohort 3 with 795 patients) using 5′ and 3′ double DIG-labelled LNA-modified probe against miR34a using the protocol described previously. Level of expression measured as automated quantitative analysis (AQUA) score for miR34a was determined for each patient and assessed for association with risk of disease-specific death. An optimal cutpoint was determined using the X-tile software for disease-specific survival in Cohort 1 and this cutpoint was then applied to the other two cohorts after median normalisation of AQUA scores.
Results:
Loss of miR34a is associated with poor outcome in three independent breast cancer cohorts (uncorrected log-rank P=0.0188 for Cohort 1, log-rank P=0.0024 for Cohort 2 and log-rank P=0.0455 for Cohort 3). In all three cohorts, loss of miR34a is able to stratify patients with poor disease-specific survival among node-negative patients, but not in node-positive population. Multivariate Cox proportional hazards analysis in Cohort 1 (P=0.0381) and Cohort 2 (P=0.0468) revealed that loss of miR34a is associated with poor outcome, independent of age, node status, receptor status and tumour size.
Conclusion:
Loss of the tumour suppressor, miR34a, identifies a subgroup of breast cancer patients with poor disease-specific survival. This study is consistent with the well-established preclinical observations for miR34a as a tumour suppressor and suggests that miR34a may have future value as a biomarker in breast cancer.
Keywords: miR34a, breast cancer, qISH
MicroRNAs (miRNAs) are a class of short non-coding RNAs expressed as primary miRNA transcripts that undergo Drosha–Dicer–Argonaut-initiated processing to yield 18- to 25-nucleotide long mature miRs (Bartel, 2004; Kim, 2005; Lee and Dutta, 2009). The coding sequence of miRNAs in human genome resides both within the intronic and exonic regions of genes (Pencheva and Tavazoie, 2013). MicroRNAs elicit their transcriptional regulation by binding to the 3′ untranslated regions of the target mRNAs leading to either degradation or disruption of their translation. Each miRNA has multiple different target mRNAs and each mRNA is regulated by several different miRNAs causing a very complex web of tight regulation. It has been estimated that miRNAs regulate 30–60% of all mRNAs and thus have key roles in the regulation of proliferation, differentiation and virtually all cellular processes (Lewis et al, 2005; Friedman et al, 2009; Kasinski and Slack, 2011). Furthermore, aberrant expression of miRNAs has a major role in the cancer initiation, progression and metastasis, and they can function as oncogenes or tumor suppressors (Croce, 2009; Garzon et al, 2009; Cortez et al, 2010; Mukherji et al, 2011; Luo et al, 2013).
miR34a is one such miRNA often reported as a putative tumour suppressor that is downregulated or lost during transformation of normal epithelial cells to cancer cells (Welch et al, 2007; Chen and Hu, 2012). Studies in cell lines and animals have identified miR34a functions in regulating somatic cell reprogramming, cell cycle, differentiation, apoptosis, cancer cell progression and metastasis (Kaller et al, 2011; Mackiewicz et al, 2011; Pramanik et al, 2011a; Kasinski and Slack, 2012; Kumar et al, 2012; Li et al, 2012, 2013; Yang et al, 2013a).
In fact it has been found to be downregulated in most cancer types including breast, lung, pancreas, colon and leukaemia (Welch et al, 2007; Gallardo et al, 2009; Yao et al, 2009; Liu et al, 2011; Genovese et al, 2012; Javeri et al, 2013; Li et al, 2013; Yang et al, 2013b; Li et al, 2014). It has also been shown to affect cell cycle, differentiation and apoptosis by targeting a variety of genes. Through cell line data it has been well established that overexpression of miR34a induces apoptosis and cell cycle arrest via p53-dependent tumour suppressor network and regulation of miR34a expression (Chen and Hu, 2012). MiR34a belongs to a family of three miRs that are homologues; miR34a is expressed in all cell types except in the lung tissue, whereas miR34b/c are expressed almost exclusively in the lungs (Bommer et al, 2007).
Mir34a expression can be assessed with miRNA microarray, qPCR or in situ hybridisation (ISH) methods (Peurala et al, 2011; Jamieson et al, 2012; Javeri et al, 2013). Each method has specific advantages and limitations. Use of miRNA microarray and qPCR methods requires RNA extraction and thus cannot assign signals to cell type. Traditional ISH methods allow the visualisation of miR34a expression with cells; however, quantification has been limited because of DAB staining. Herein, we utilised our qISH approach (Hanna et al, 2012) to quantitatively assess the level of expression of miR34a within tumour epithelia and correlate it with the disease-free survival outcome in three independent breast cancer cohorts.
Patients and methods
Cohorts
The study was conducted using data from three independent cohorts of breast cancer patients. The first cohort consists of 461 patients who underwent surgery at the Yale University Cancer Center/Yale New Haven hospital between years 1962 and 1982 and had formalin-fixed, paraffin-embedded (FFPE) primary invasive breast tumours available for study. Cohort 2 consists of 279 patients, nonoverlapping with cohort 1, who had surgery for breast cancer at the Yale University Cancer Center/Yale New Haven hospital between years 1976 and 2005 and for whom FFPE tissue was available. Tissue microarrays were constructed and assessed in two-fold redundancy for each cohort. Both cohorts have been described previously (Giltnane et al, 2009; Welsh et al, 2011). The follow-up information on cases was obtained from the Yale New Haven Tumor Registry, the Yale-New Haven Hospital medical records and the Connecticut Death Records. Use of these resources was approved by the Yale Institutional Review Board and the tissues were collected in accordance with the consent guidelines using protocol 8219 issued to DLR from the Yale Institutional Review Board. Cohort 3 consists of 795 patients who were diagnosed with invasive breast cancer and underwent surgery between years 2003 and 2005 in Poland. Signed informed consents were obtained from each patient for Cohort 3 in accordance with the National Cancer Institute and Poland Institutional Review Boards (Horne et al, 2014). Clinicopathological characteristics of all three cohorts are presented in Table 1.
Table 1. Clinicopathological characteristics of all three cohorts.
| Parameter | Cohort 1, N (%) | Cohort 2, N (%) | Cohort 3, N (%) |
|---|---|---|---|
| All patients | 461 | 279 | 795 |
| Year of diagnosis | 1962–1982 | 1976–2005 | 2000–2003 |
|
Age (years) | |||
| <50 | 127 (27.5) | 94 (33.7) | 270 (34) |
| ⩾50 | 334 (72.5) | 185 (66.3) | 525 (66) |
| Unknown | 0 | 0 | 0 |
|
Nodal status | |||
| Positive | 243 (52.7) | 59 (21.1) | 304 (38.2) |
| Negative | 201 (43.6) | 157 (56.3) | 469 (59) |
| Unknown | 17 (3.7) | 63 (22.6) | 22 (2.78) |
|
Tumour size | |||
| ⩽2 cm | 193 (41.9) | 187 (56.9) | 412 (51.8) |
| >2–5 cm | 177 (38.4) | 72 (35.9) | 343 (43.1) |
| >5 cm | 53 (11.5) | 0 (0) | 26 (3.3) |
| Unknown | 38 (8.2) | 19 (7.2) | 14 (1.8) |
|
ERα (path) | |||
| Positive (1–3) | 239 (51.8) | 142 (50.9) | 488 (61.4) |
| Negative (0) | 209 (45.3) | 97 (34.8) | 285 (35.8) |
| Unknown | 13 (2.8) | 40 (14.3) | 22 (2.8) |
|
PR (path) | |||
| Positive (1–3) | 221 (47.9) | 127 (45.5) | 403 (50.6) |
| Negative (0) | 211 (45.8) | 102 (36.6) | 371 (46.6) |
| Unknown | 29 (6.3) | 50 (17.9) | 22 (2.8) |
|
HER2 (IHC) | |||
| Positive (3) | 45 (9.8) | 17 (6.1) | 72 (9.1) |
| Negative (0–2) | 397 (86.1) | 216 (77.4) | 566 (71.2) |
| Unknown | 19 (4.1) | 46 (16.5) | 157 (19.7) |
|
Follow-up (m) | |||
| Median (range) | 105.46 (2.39–498.03 ) | 121 (7–385) | 114.84 (3.6–147.96) |
Abbreviations: ERα=estrogen receptor alpha; Her2=human epidermal growth factor receptor 2; IHC=immunohistochemistry; m=months; N=number; PR=progesterone receptor.
miRNA quantitative in situ hybridisation
The miRNA qISH assays were performed on TMA slides as described previously (Hanna et al, 2012). Briefly, the arrays were deparaffinised first by melting at 60 °C in an oven equipped with a fan for 20 min followed by 2 × xylene treatment for 20 min each. Slides were then rehydrated with an ethanol gradient followed by Proteinase K digestion for 10 min at 37 °C, and fixed with 4% formaldehyde followed by another fixation step with EDC. Endogenous peroxidase was blocked with 1% hydrogen peroxide in PBS for 30 min. Slides were then pre-hybridised at 50 °C in a hybridisation oven for 30 min followed by hybridisation with 200 nM double DIG-labelled LNA-modified probes for an hour at 50 °C. Slides were washed with 2 × SSC buffer once at hybridisation temperature and twice at room temperature for 5 min each. Slides were blocked with 2% BSA for 30 min followed by 1 h incubation at room temperature with anti-DIG antibody (Roche, Indianapolis, IN, USA) along with rabbit anti-pan cytokeratin antibody. After washing away the primary antibodies, slides were then incubated with CY5-conjugated tyramide (TSA plus CY5 system) for 10 min to visualise the miRNA. After washing, slides were incubated with goat anti-rabbit secondary antibody conjugated to Alexa546 to visualise cytokeratin (Molecular Probes, Grand Island, NY, USA) for an hour at room temperature. After washing, slides were mounted with ProLong gold mixed with DAPI (Molecular Probes). Serial sections of the index array used for assay standardisation were stained alongside each cohort to assess the reproducibility of assay. An additional serial section of the index array was stained with each experiment with a DIG-labelled scramble probe (5′-GTGTAACACGTCTATACGCCCA-3′) as a negative control. The miR34a-unlabelled blocking oligo (800 nM; 5′-TGGCAGTGTCTTAGCTGGTTGT-3′) was pre-incubated with the DIG-labelled miR34a probe (200 nM; 5′-ACAACCAGCTAAGACACTGCCA-3′) for an hour at hybridisation temperature followed by hybridisation of the mixed probes on the TMA.
Automated quantitative analysis (AQUA)
The AQUA technology allows quantitative measurement of biomolecules in subcellular compartments as described previously (Camp et al, 2002; Moeder et al, 2009). Briefly, a series of monochromatic images for each histospot was captured using PM-2000 microscope equipped with automated stage. A binary ‘tumour mask' was created using cytokeratin staining of the histospot representing only epithelial cells and excluding stromal features. AQUA scores for miR34a were calculated by dividing the signal intensity (scored on a scale from 0 to 255) to the area of the specific compartment (in this case within the tumour mask area).
Statistical analysis
Pearson's correlation coefficient (R) was used to assess the reproducibility of the assay between near-serial sections of the index array. On the basis of the extensive experience in our laboratory, an R2 value greater than 0.4 was considered acceptable for both inter- and intra-array reproducibility for miRNA. For both Yale Cohorts (1 and 2), miR34a AQUA scores from two independent cores for each histospot were averaged and the averages were used for final analysis. AQUA scores from Cohorts 2 and 3 were median normalised to the Cohort 1 to compare a single cutpoint on all three cohorts. An optimal cutpoint was determined for Cohort 1 using the X-tile software (Yale University, New Haven, CT, USA) for disease-specific survival differences (Camp et al, 2004). This cutpoint was then applied to the two validation cohorts (Cohorts 2 and 3). A multivariate Cox proportional hazards analysis was used to examine prognostic value of miR34a while controlling for other common prognostic factors as well as potential confounders, including age, receptor status, node status and tumour size. Log-rank P-value<0.05 was considered to be statistically significant. All statistical analyses were carried out using SAS 9.2 (SAS Institute, Cary, NC, USA).
Results
Reproducibility, specificity and heterogeneity of miR34a expression in breast cancer cohorts
We developed and validated the qISH assay to quantitatively assess the level of expression for miR34a using TMA and the IF-based AQUA technology. Pan-cytokeratin staining was used to define the epithelial component of tumours in which the signal intensity of miR34a was assessed. As expected, a cytoplasmic localisation pattern was observed for miR34a (Figure 1A). In order to assess specificity we performed the qISH assay on serially sectioned index array with either the DIG-labelled miR34a probe or a scrambled probe. As shown in Figure 1B (panels a–c for miR34a probe and g–i for scrambled probe) for the same histospot, there is a high signal for miR34a probe, but no specific signal for the scrambled probe. Furthermore, we pre-incubated a miR34a-specific-unlabelled blocking oligo (the same sequence as the endogenous miR34a) with the labelled miR34a probe to compete with endogenous miR34a-specific probe hybridisation. As shown in Figure 1B (panels d–f), no specific signal was detected for the same histospot when the blocking oligo was used.
Figure 1.
Development of a robust and specific qISH assay to assess miR34a. (A) Representative tumour histospot with Pan cytokeratin staining used to define the tumour mask (white), Pan cytokeratin (green) and miR34a (red), showing a cytoplasmic staining pattern. Panels a–c represent a histospot with high expression of miR34a, whereas d–f represent a histospot with low expression of miR34a. (B) Specificity of qISH assay for miR34a. Panels a–c, d–f and g–i represent the same histospot from a serially sectioned index TMA hybridised with either DIG-labelled miR34a (a–c), pre-incubated unlabelled blocking oligo with labelled miR34a probe (d–f) or labelled scramble probe (g–i). Arrowhead in c indicates expression of miR34a in a stromal compartment.
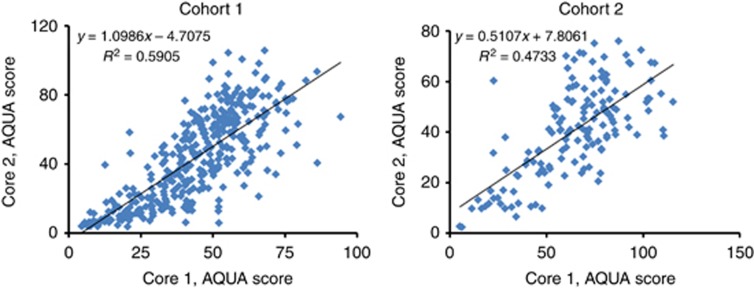
Intra-array reproducibility of the qISH assay was evaluated by staining serial sections of an index array TMA consisting of a small subgroup of breast cancer patients used as control array for each experiment. This yielded Pearson's R2 values of 0.646 and 0.514 performed as four individual assays indicative of good reproducibility (Supplementary Figure 1). Tumour heterogeneity (inter-array reproducibility) was evaluated by comparing miR34a AQUA scores in two different cores (histospots) from the same tumour block. This showed significant correlation between histospots (R2=0.5905 for Cohort 1 and 0.4733 for Cohort 2), as shown in Figure 2.
Figure 2.
Assessment of tumour heterogeneity for miR34a expression in two independent primary breast cancer cohorts. Linear regression between AQUA scores obtained from two independent assays on two different cores (builds) of each patient on two Yale Cohorts, 1 (461 patients) and 2 (279 patients).
Association of miR34a with disease-specific survival
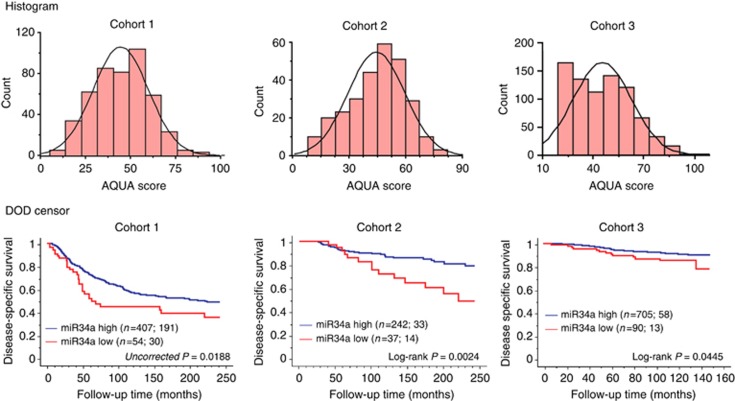
Expression levels of miR34a were analysed using averaged AQUA scores of two independent cores for the two Yale cohorts (Cohorts 1 and 2) and single AQUA scores for Cohort 3. Distribution of AQUA scores for all three cohorts were similar to each other as shown in Figure 3A. As there are no previous data or biological hypotheses on which we can divide the data to determine the prognostic value of miR34a, we used Cohort 1 as a training set for the X-tile method to determine an optimal cutpoint with 20-year disease-specific survival. Low expression of miR34a was strongly associated with poor outcome (uncorrected log-rank P=0.0188, Figure 3B). The X-tile cutpoint was then applied to Cohorts 2 and 3, consisting of 279 and 795 patients, respectively. This cutpoint showed that decreased levels of miR34 was associated with worse outcome (log-rank P=0.0024 and 0.0445, respectively).
Figure 3.
Distribution of AQUA scores and Kaplan–Meier analysis for miR34a in three independent primary breast cancer cohorts. (A) Distribution of AQUA scores for each cohort. (B) An optimal cutpoint was determined for cohort 1 using the X-tile software. This cutpoint was then applied to the median normalised AQUA scores of the validation cohorts 2 and 3. In parenthesis the number of patients in each group is shown followed by the number of events due to disease. Log-rank P-values are shown for 20 year follow-up for cohorts 1 and 2 and maximum time (148 months) for cohort 3.
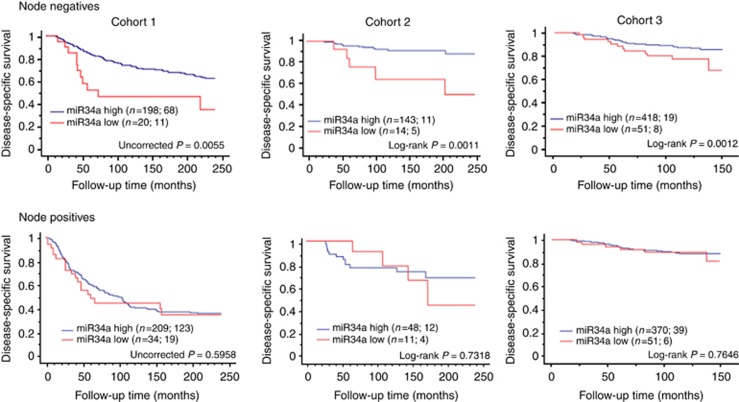
In addition, we assessed the impact of miR34a on survival within node-negative and -positive subgroups of all three cohorts. The miR34a expression was prognostic in the node-negative subset with a log-rank P-value of 0.0055 for Cohort 1; 0.0011 for Cohort 2 and 0.0012 for Cohort 3, but not in node-positive patients using the same cutpoint as for whole population (Figure 4A and B). Further, box and whisker plots demonstrated no significant differences in the distribution of AQUA scores for miR34a between node-negative and node-positive populations (Supplementary Figure 2) in all three breast cancer cohorts. Thus, the prognostic value of miR34a among node negatives appears to be associated with tumour biology within this subpopulation group. Assessment of miR34a in other subclasses (hormone receptor-positive or -negative, HER2-enriched and triple negative) showed no relationships with outcome.
Figure 4.
Kaplan–Meier analysis for miR34a in subpopulations based on nodal status in all three primary breast cancer cohorts. Each cohort was divided into low and high miR34a groups and disease-specific survival of low miR34a expression was compared with the high miR34a expression. The number of patients in each group is shown followed by the number of patient deaths due to disease. Log-rank P-values are shown for 20 year follow-up for two Yale cohorts and maximum years for the third cohort. Only miR34a scores in node-negative subpopulations are statistically significant in all three cohorts (P=0.0055 for cohort 1; P=0.0011 for cohort 2 and P=0.0012 for cohort 3).
Univariate and multivariate analyses of all three cohorts
A univariate Cox proportional hazard analysis was carried out to assess the potential of miR34a as a prognostic marker in all three cohorts. As shown in Table 2, high miR34a demonstrated lower risk for disease-specific survival in all three cohorts (hazard ratio (HR)=0.632, P=0.0199 for Cohort 1; HR=0.393, P=0.0034 for Cohort 2; and HR=0.545, P=0.0479 for Cohort 3). In addition, in univariate analysis, nodal status and tumour size were also associated with poor disease-specific survival in all three cohorts (Table 2).
Table 2. Univariate and multivariate Cox proportional hazards analyses for disease-specific survival in association with miR34a expression for all three cohorts.
|
Cohort 1 (n=394; 189)a |
Cohort 2 (n=182; 22)a |
Cohort 3 (n=621; 53)a |
||||
|---|---|---|---|---|---|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
|
Age | ||||||
| Univariate | ||||||
| <50 | 0.834 (0.62–1.12) | 0.2335 | 1.451 (0.82–2.58) | 0.2059 | 0.817 (0.49–1.35) | 0.435 |
| Multivariate | ||||||
| <50 | 0.807 (0.57–1.14) | 0.2268 | 1.402 (0.51–3.86) | 0.5131 | 0.900 (0.49–1.63) | 0.7261 |
| >50 | 1.00 | 1.00 | 1.00 | |||
|
Tumour size | ||||||
| Univariate | ||||||
| 2–5 cm | 1.756 (1.29–2.40) | 0.0014 | 3.781 (2.04–7.00) | <0.0001 | 2.2026 (1.22–3.38) | 0.0068 |
| >5 cm | 3.191 (2.16–4.71) | <0.0001 | 8.402 (3.9–18.09) | <0.0001 | ||
| Multivariate | ||||||
| <2 cm | 1 | <0.0001 | 1 | 0.0022 | 1 | 0.0006 |
| >2–5 cm | 1.772 (1.28–2.47) | 4.709 (1.74–12.73) | 1.517 (0.81–2.86) | |||
| >5 cm | 2.959 (1.97–4.44) | 5.608 (2.31–13.62) | ||||
|
Nodal status | ||||||
| Univariate | ||||||
| Negative | 0.438 (0.33–0.58) | <0.0001 | 0.352 (0.18–0.70) | 0.0031 | 0.350 (0.22–0.57) | <0.0001 |
| Multivariate | ||||||
| Positive | 1.00 | 1.00 | 1.00 | |||
| Negative | 0.549 (0.40–0.75) | 0.0002 | 0.640 (0.25–1.66) | 0.3597 | 0.295 (0.16–0.54) | <0.0001 |
|
ERα | ||||||
| Univariate | ||||||
| Negative | 1.512 (1.16–1.97) | 0.0024 | 1.391 (0.70–2.76) | 0.345 | 1.895 (1.17–3.06) | 0.009 |
| Multivariate | ||||||
| Positive | 1.00 | 1.00 | 1.00 | |||
| Negative | 1.396 (1.00–1.93) | 0.0439 | 1.247 (0.38–4.05) | 0.7139 | 1.259 (0.66–2.42) | 0.4889 |
|
PR | ||||||
| Univariate | ||||||
| Negative | 1.409 (1.07–1.85) | 0.0133 | 1.610 (0.81–3.19) | 0.1736 | 2.666 (1.59–4.47) | 0.0002 |
| Multivariate | ||||||
| Positive | 1.00 | 1.00 | 1.00 | |||
| Negative | 1.140 (0.84–1.54) | 0.3935 | 1.798 (0.55–5.84) | 0.3286 | 1.724 (0.89–3.36) | 0.1096 |
|
HER2 | ||||||
| Univariate | ||||||
| Negative | 0.863 (0.56–1.34) | 0.5144 | 0.511 (0.18–1.45) | 0.2060 | 0.601 (0.29–1.23) | 0.1627 |
| Multivariate | ||||||
| Positive | 1.00 | 1.00 | 1.00 | |||
| Negative | 0.685 (0.41–1.13) | 0.141 | 0.369 (0.11–1.19) | 0.0952 | 1.003 (0.47–2.16) | 0.9929 |
|
miR34a | ||||||
| Univariate | ||||||
| High | 0.632 (0.43–0.93) | 0.0199 | 0.393 (0.21–0.74) | 0.0034 | 0.545 (0.30–0.99) | 0.0479 |
| Multivariate | ||||||
| Low | 1.00 | 1.00 | 1.00 | |||
| High | 0.647 (0.43–0.98) | 0.0381 | 0.343 (0.12–0.99) | 0.0468 | 0.727 (0.35–1.5) | 0.4014 |
Abbreviations: CI=confidence interval; HR=hazards ratio.
The P-values are shown for a 20-year follow-up for Cohorts 1 and 2 and 148 months for Cohort 3. Bold entities indicate statistically significant P-values.
The number of patients in each cohort followed by number of patient deaths due to the disease.
A multivariate Cox proportional hazards analysis was used to examine prognostic value of miR34a while controlling for other common prognostic factors as well as potential confounders. miR34a retained its prognostic value such that patients with low expression had a 35% increase in risk of breast cancer death compared with those in the low miR34a group in Cohort 1 (HR=0.647; 95% CI=0.34–0.98; P=0.0381 for Cohort 1, Table 2). Likewise, in Cohort 2 patients with low miR34a expression had high risk of breast cancer death by 65% (HR=0.343; 95% CI=0.12–0.99; P=0.0468, Table 2). However, it lost the prognostic value for the Cohort 3 (P=0.4014, Table 2), which is likely because of fewer years of follow-up as well as fewer number of events (71 out of 795; <10%). In addition to miR34a, nodal status and tumour size were significantly associated with the risk of death from breast cancer in univariate Cox proportional hazard analysis in all three cohorts (Table 2).
Discussion
In this study we evaluated the expression of miR34a in more than 1500 breast cancer patients and correlated it with the disease-free survival outcome. Previously, LNA-modified miR34a probes have been used in FFPE specimens using ISH combined with DAB stain on smaller cohorts with subjective reads (Pena et al, 2009; Nuovo, 2010; Peurala et al, 2011). Our qISH method creates a tumour mask using expression of cytokeratin within a histospot, thereby separating the tumour epithelia from stromal compartment (Giltnane and Rimm, 2004). This continuous scoring of miR34a allows us to identify a quantitative cutpoint in a discovery cohort (Cohort 1), which then applied to two other validation cohorts (Cohorts 2 and 3). Exclusion of stromal compartment is especially important for miR34a as it is expressed in most tissue types including stroma as shown as arrowhead in the Figure 1B. Nonexclusion of stromal compartment could lead to misleading information about its level of expression within tumour cells.
Cell line model systems have been used extensively to establish miR34a as a metastasis, cell proliferation, cell cycle, migration and invasion marker by regulating several key mRNAs in these pathways including Bcl2, Src, CD44, Notch-1, c-Met, E2F1, B-Myb and CDK4/6 (Chen and Hu, 2012). In addition, mouse model systems established using cancer cell lines confirmed the results obtained in in vitro assays and further established the role of miR34a as a tumour suppressor (Pramanik et al, 2011b; Kasinski and Slack, 2012; Kumar et al, 2012; Li et al, 2012). However, less work has been performed to confirm the in vitro data on clinical specimens and to evaluate its potential as a prognostic biomarker in cancer. One recent study used miRNA microarray analysis to identify miR34a as a strong prognostic marker in two small cohorts (48 and 24 patient populations) of pancreatic ductal adenocarcinoma (Jamieson et al, 2012). Loss of miR34a was associated with poor survival and was independent of other clinical variables. In breast cancer a recently published study evaluated the expression of miR34a on 46 individual paired breast cancer tissues and adjacent normal tissues using qPCR with LNA-modified primers (Javeri et al, 2013). This study also evaluated the mutational status of p53 on these patients. They failed to identify a correlation between mutated p53 and miR34a expression that was previously identified using cell line model systems. However, this study was able to identify an association between low expression of miR34a and metastasis. Likewise, in another separate study of more than 1000 breast cancer specimens, Peurala et al (2011) investigated miR34a expression using LNA-modified miR34a probe and DAB-stained ISH method. In this study they found that high expression of miR34a was associated with lower risk for metastasis or death from breast cancer, and identified miR34a as an independent prognostic marker in a multivariate cox proportional hazard analysis in 5-year disease-specific survival (Peurala et al, 2011). This is consistent with our study where we are able to show miR34a as an independent prognostic marker in the two of the three cohorts we examined.
Our qISH method is also able to show that miR34a is a prognostic marker in node-negative patients, independent of clinical biomarkers of ERα, PR and Her2 status. However, this study is limited by a number of issues that will need to be addressed in the future before incorporation into clinical usage. Specifically, this hypothesis generating work is carried out on tissue microarray, and future work would need to be carried out on core biopsies or excision specimens to determine the effect of heterogeneity on evaluation of the marker. Further studies would also need to be carried out to standardise the assay and to test in either a prospective or prospective retrospective setting, as this work was strictly a retrospective collection of cases.
In spite of these limitations, we believe that miR34a has potential future clinical application as it has been historically difficult to identify patients that are node-negative, but are at high risk for developing metastasis. The Oncotype Dx test utilises a 21 gene signature to identify high-risk patients among a subgroup of breast cancer patients who are ERα-positive and node-negative (Kaklamani, 2006; Horne et al, 2014). In future studies, we hope to compare miR34a with Oncotype Dx and to more extensively evaluate the clinical utility of miR34a as a prognostic marker in prospective studies.
Acknowledgments
This work was supported in part by a Susan G Komen for the Cure Investigator Initiated Research Grant to DLR. We thank Pei Chao and Michael Stagner from Information Management Services (Silver Spring, MD) for data management support; the participants, physicians, pathologists, nurses and interviewers from participating centres in Poland for their efforts during field work; and Drs Montserrat Garcia-Closas, Louise Brinton, Jolanta Lissowska, B Peplonska for their contributions to the PBCS study design. We also thank Stephen Hewitt for TMA construction.
Author contributions
SA, JH and DLR participated in the concept, design, coordination of the study and drafting of the manuscript. SA was also involved in the execution, acquisition, analysis and interpretation of all data. DLR, MES and JF participated in providing financial support, patient cohorts, data analysis and interpretation of all data.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Presentation of the manuscript: Portions of this work were presented at the AACR meeting in 2012
Supplementary Material
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116 (2:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17 (15:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8 (11:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10 (21:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- Chen F, Hu SJ. Effect of microRNA-34a in cell cycle, differentiation, and apoptosis: a review. J Biochem Mol Toxicol. 2012;26 (2:79–86. doi: 10.1002/jbt.20412. [DOI] [PubMed] [Google Scholar]
- Cortez MA, Ivan C, Zhou P, Wu X, Ivan M, Calin GA. microRNAs in cancer: from bench to bedside. Adv Cancer Res. 2010;108:113–157. doi: 10.1016/B978-0-12-380888-2.00004-2. [DOI] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10 (10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19 (1:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo E, Navarro A, Vinolas N, Marrades RM, Diaz T, Gel B, Quera A, Bandres E, Garcia-Foncillas J, Ramirez J, Monzo M. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30 (11:1903–1909. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- Genovese G, Ergun A, Shukla SA, Campos B, Hanna J, Ghosh P, Quayle SN, Rai K, Colla S, Ying H, Wu CJ, Sarkar S, Xiao Y, Zhang J, Zhang H, Kwong L, Dunn K, Wiedemeyer WR, Brennan C, Zheng H, Rimm DL, Collins JJ, Chin L. microRNA regulatory network inference identifies miR-34a as a novel regulator of TGF-beta signaling in glioblastoma. Cancer Discov. 2012;2 (8:736–749. doi: 10.1158/2159-8290.CD-12-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltnane JM, Moeder CB, Camp RL, Rimm DL. Quantitative multiplexed analysis of ErbB family coexpression for primary breast cancer prognosis in a large retrospective cohort. Cancer. 2009;115 (11:2400–2409. doi: 10.1002/cncr.24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltnane JM, Rimm DL. Technology insight: identification of biomarkers with tissue microarray technology. Nat Clin Pract. 2004;1 (2:104–111. doi: 10.1038/ncponc0046. [DOI] [PubMed] [Google Scholar]
- Hanna JA, Wimberly H, Kumar S, Slack F, Agarwal S, Rimm DL. Quantitative analysis of microRNAs in tissue microarrays by in situ hybridization. BioTechniques. 2012;52 (4:235–245. doi: 10.2144/000113837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne HN, Sherman ME, Garcia-Closas M, Pharoah PD, Blows FM, Yang XR, Hewitt SM, Conway CM, Lissowska J, Brinton LA, Prokunina-Olsson L, Dawson SJ, Caldas C, Easton DF, Chanock SJ, Figueroa JD. Breast cancer susceptibility risk associations and heterogeneity by E-cadherin tumor tissue expression. Breast Cancer Res Treat. 2014;143 (1:181–187. doi: 10.1007/s10549-013-2771-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson NB, Morran DC, Morton JP, Ali A, Dickson EJ, Carter CR, Sansom OJ, Evans TR, McKay CJ, Oien KA. MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin Cancer Res. 2012;18 (2:534–545. doi: 10.1158/1078-0432.CCR-11-0679. [DOI] [PubMed] [Google Scholar]
- Javeri A, Ghaffarpour M, Taha MF, Houshmand M. Downregulation of miR-34a in breast tumors is not associated with either p53 mutations or promoter hypermethylation while it correlates with metastasis. Med Oncol. 2013;30 (1:413. doi: 10.1007/s12032-012-0413-7. [DOI] [PubMed] [Google Scholar]
- Kaklamani V. A genetic signature can predict prognosis and response to therapy in breast cancer: Oncotype DX. Expert Rev Mol Diagn. 2006;6 (6:803–809. doi: 10.1586/14737159.6.6.803. [DOI] [PubMed] [Google Scholar]
- Kaller M, Liffers ST, Oeljeklaus S, Kuhlmann K, Roh S, Hoffmann R, Warscheid B, Hermeking H. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol Cell Proteomics. 2011;10 (8:M111 010462. doi: 10.1074/mcp.M111.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11 (12:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res. 2012;72 (21:5576–5587. doi: 10.1158/0008-5472.CAN-12-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6 (5:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kumar B, Yadav A, Lang J, Teknos TN, Kumar P. Dysregulation of microRNA-34a expression in head and neck squamous cell carcinoma promotes tumor growth and tumor angiogenesis. PLoS One. 2012;7 (5:e37601. doi: 10.1371/journal.pone.0037601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120 (1:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li L, Xie X, Luo J, Liu M, Xi S, Guo J, Kong Y, Wu M, Gao J, Xie Z, Tang J, Wang X, Wei W, Yang M, Hung MC. Targeted expression of miR-34a using the T-VISA system suppresses breast cancer cell growth and invasion. Mol Ther. 2012;20 (12:2326–2334. doi: 10.1038/mt.2012.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med. 2013;13 (2:109–117. doi: 10.1007/s10238-012-0186-5. [DOI] [PubMed] [Google Scholar]
- Li XJ, Ren ZJ, Tang JH. MicroRNA-34a: a potential therapeutic target in human cancer. Cell Death Dis. 2014;5:e1327. doi: 10.1038/cddis.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, Wiggins JF, Bader AG, Fagin R, Brown D, Tang DG. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17 (2:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Wilson JM, Harvel N, Liu J, Pei L, Huang S, Hawthorn L, Shi H. A systematic evaluation of miRNA:mRNA interactions involved in the migration and invasion of breast cancer cells. J Transl Med. 2013;11:57. doi: 10.1186/1479-5876-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz M, Huppi K, Pitt JJ, Dorsey TH, Ambs S, Caplen NJ. Identification of the receptor tyrosine kinase AXL in breast cancer as a target for the human miR-34a microRNA. Breast Cancer Res Treat. 2011;130 (2:663–679. doi: 10.1007/s10549-011-1690-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeder CB, Giltnane JM, Moulis SP, Rimm DL. Quantitative, fluorescence-based in-situ assessment of protein expression. Methods Mol Biol (Clifton, NJ) 2009;520:163–175. doi: 10.1007/978-1-60327-811-9_12. [DOI] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43 (9:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuovo GJ. In situ detection of microRNAs in paraffin embedded, formalin fixed tissues and the co-localization of their putative targets. Methods (San Diego, Calif) 2010;52 (4:307–315. doi: 10.1016/j.ymeth.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, Tuschl T. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6 (2:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15 (6:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peurala H, Greco D, Heikkinen T, Kaur S, Bartkova J, Jamshidi M, Aittomaki K, Heikkila P, Bartek J, Blomqvist C, Butzow R, Nevanlinna H. MiR-34a expression has an effect for lower risk of metastasis and associates with expression patterns predicting clinical outcome in breast cancer. PLoS One. 2011;6 (11:e26122. doi: 10.1371/journal.pone.0026122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, Mendell JT, Maitra A. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10 (8:1470–1480. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, Mendell JT, Maitra A. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10 (8:1470–1480. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26 (34:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- Welsh AW, Moeder CB, Kumar S, Gershkovich P, Alarid ET, Harigopal M, Haffty BG, Rimm DL. Standardization of estrogen receptor measurement in breast cancer suggests false-negative results are a function of threshold intensity rather than percentage of positive cells. J Clin Oncol. 2011;29 (22:2978–2984. doi: 10.1200/JCO.2010.32.9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Li QJ, Gong ZB, Zhou L, You N, Wang S, Li XL, Li JJ, An JZ, Wang DS, He Y, Dou KF. MicroRNA-34a targets Bcl-2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment. Technol Cancer Res Treat. 2013;13 (1:77–86. doi: 10.7785/tcrt.2012.500364. [DOI] [PubMed] [Google Scholar]
- Yang S, Li Y, Gao J, Zhang T, Li S, Luo A, Chen H, Ding F, Wang X, Liu Z. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2013;32 (36:4294–4303. doi: 10.1038/onc.2012.432. [DOI] [PubMed] [Google Scholar]
- Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni L, Zhang WG, Nan KJ, Song TS, Huang C. MicroRNA profiling of human gastric cancer. Mol Med Rep. 2009;2 (6:963–970. doi: 10.3892/mmr_00000199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.