Abstract
Background:
Invasion of the surrounding tissue is part of the metastatic cascade. Here, we examined the invasion of pancreatic ductal adenocarcinoma (PDAC) cells into the mesothelial barrier and identified the related microRNA (miRNA) expression profiles.
Methods:
The interactions between PDAC cells and mesothelial monolayers were characterised and quantified using a specific time-lapse videomicroscopy assay. Pancreatic ductal adenocarcinoma cells were further evaluated using the adhesion assay, and miRNA, mRNA and protein expressions were determined using microarray, q-RT–PCR and western blots, respectively. These data were correlated with in vivo dissemination scores.
Results:
Two groups of PDAC cell lines were distinguished by their integration capacity into the mesothelial monolayer using mean elongation factors (MEFs). Adhesion assays showed a concordant relation between adhesive properties and integration capacity. The distant metastases scores were reverse correlated with MEFs. Microarray analysis of these groups revealed that miR-23a and/or miR-24 target for FZD5, HNF1B and/or TMEM92, respectively, and that they are significantly deregulated.
Conclusions:
MiR-23a and/or miR-24 overexpression leads to gene silencing of FZD5, TMEM92 and/or HNF1B. Their downregulation induces deregulated expression and degradation of E-cadherin and β-catenin causing destabilisation of the cadherin/catenin complex, and altered the expression of Wnt-related genes. We propose a molecular (epi)genetic mechanism by which increased EMT-like cell shape transformation and integration into mesothelial monolayers of PDAC cells can be observed.
Keywords: PDAC, peritoneal dissemination, EMT, miR-23a, miR-24, FZD5, TMEM92, HNF1B
Approximately 3% of all new cancer cases in the United States of America are caused by pancreatic malignancies (Siegel et al, 2012). The median survival time of all patients accounts for 6 months (Sohn et al, 2000; Jang et al, 2013).
In spite of intensive efforts in basic research and advances in clinical management, the high mortality rate has not been improved significantly in recent years (Jemal et al, 2009). A main cause of persisting mortality is the cancer's early dissemination, which is often in the form of peritoneal carcinomatosis. Further investigation of the mechanisms underlying pancreatic ductal adenocarcinoma (PDAC) cell dissemination may identify novel therapeutic targets, and provide a basis for increased survival of treatment strategies.
Expression of the adhesion molecule E-cadherin is a key factor for cell dissemination in many settings, ranging from embryogenesis to metastasis. Gain or loss of E-cadherin expression and the ensuing abrogation or maintenance of intercellular adherens junctions leads to EMT (epithelial–mesenchymal transition) or its reverse mechanism MET (mesenchymal–epithelial transition). Loss of E-cadherin results in EMT and allows cells to detach from cell agglomerations and to migrate. The morphological correlate manifests as loss of cell polarity towards spindle-shaped transformation (reviewed in Thiery, 2002; Thiery et al, 2009; Lim and Thiery, 2012).
In the past two decades, epigenetic alterations have been recognised as crucial elements of the genesis of many carcinomas, including PDAC cells (Suter et al, 2004; McCleary-Wheeler et al, 2013). Gene silencing by microRNAs (miRNAs) is an epigenetic mechanism (Bushati and Cohen, 2007) that has an important role in the development of pancreatic malignancies (Bloomston et al, 2007; Lee et al, 2007; Szafranska et al, 2007).
Phenotype investigation, that is, microscopic investigation, of the development of peritoneal carcinomatosis in combination with correlated miRNA expression profiles could lead to a better understanding of the dissemination process.
We hypothesised that the ability to metastasise relies on successful invasion of the surrounding tissue, and that this process is bound to miRNA deregulation. To identify relevant miRNAs, we investigated the dissemination of PDAC cells via migration and adhesion assays and correlated these findings with miRNA expression profiles.
Materials and methods
Cell culture: PDAC cell lines
Fifteen established human PDAC cell lines (Table 1) were investigated and maintained as described in Mees et al (2010).
Table 1. Initial EF (EF-0).
| Cell line | EF-0 |
|---|---|
| A818 | 1.18±0.22 |
| ASPC1 | 1.16±0.10 |
| BxPc3 | 1.11±0.06 |
| Capan1 | 1.08±0.03 |
| Capan2 | 1.14±0.09 |
| HPaf | 1.21±0.20 |
| MiaPaCa2 | 1.13±0.12 |
| MPanc96 | 1.11±0.04 |
| Panc1 | 1.09±0.05 |
| Patu8902 | 1.12±0.04 |
| Patu8988S | 1.11±0.04 |
| Patu8988T | 1.11±0.06 |
| Su8686 | 1.11±0.12 |
| Suit0028 | 1.11±0.11 |
| Suit007 | 1.15±0.09 |
| Average | 1.13±0.03 |
Abbreviations: EF=elongation factors; HMC=human mesothelial cell; PDAC=pancreatic ductal adenocarcinoma.
This table shows all PDAC cell lines initially chosen for our investigations.
EF-0s, corresponding standard deviations and averaged value of all examined PDAC cell lines are listed in alphabetical order. The factored-0s range from EF-0Capan1 1.08±0.03 to EF-0HPaf 1.21±0.2, and average EF-0all cell lines 1.13±0.03. All PDAC cell lines show round shapes at the beginning of their cell–cell interactions with HMCs.
Cell culture: human mesothelial cells
After obtaining written consent, specimens of the greater omentum were obtained from patients undergoing abdominal surgery for reasons other than malignancy or acute/chronic inflammatory diseases. Specimens (1 mm3) were incubated for 30 min at 37 °C in 1% collagenase II solution (Worthington Biochemical Corporation, Lakewood Township, NJ, USA).
The resuspended human mesothelial cells (HMCs) were cultivated in Medium 199 (PAA Laboratory GmbH, Pasching, Austria) in fibronectin- (Sigma-Aldrich, Taufkirchen, Germany) coated tissue dishes. Monolayers were confluent after ∼7 days of growth and then introduced into assays.
Migration assay
We added suspended and stained (Cell-Tracker-Red; Invitrogen, Darmstadt, Germany) PDAC cells on a confluent HMC monolayer, which is stained with the fluorescence dye Cell-Tracker-Green (Invitrogen).
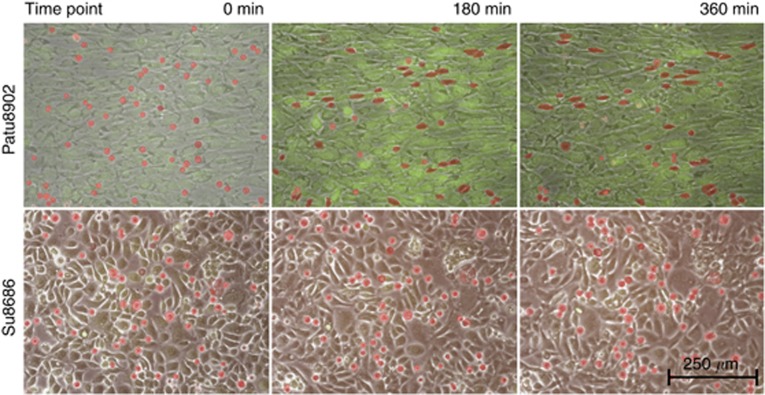
Over a 6-h time period, pictures were taken every 4 min to produce a time-lapse video clip using the Eclipse TE2000U inverted microscope (Nikon, Düsseldorf, Germany) at × 100 magnification (Figure 1 and Supplementary Data).
Figure 1.
Sequences of different integration patterns. Suspended PDAC cells were stained with Cell-Tracker-Red (Invitrogen) and added to a confluent HMC monolayer, which is stained with Cell-Tracker-Green (Invitrogen). Fluorescence pictures using the appropriate wavelength and control pictures in low light intensity were obtained. The three pictures of the two sequences represent 0, 180 and 360 min examination time. The carcinoma cells are highlighted in red owing to the difficulty to identify them in motionless pictures when integrated into the HMC layer. These highlights were used as the basis for the partly automatic, partly manual morphometrical analyses (see Figure 2A). The upper sequence shows an example of the observed migration pattern of cell lines with PDAC cell elongation (Patu8902). Most carcinoma cells show round or nearly round shapes at the beginning (0 min). The following pictures of the series (180 and 360 min) illustrate the changes of most carcinoma cells toward an elongated shape and their integration into the mesothelial monolayer. The lower sequence illustrates the migration pattern of Su8686 exemplarily for cell lines without tendencies to elongate. Only marginal changing of the initial round cell shapes is observed over the entire examination time.
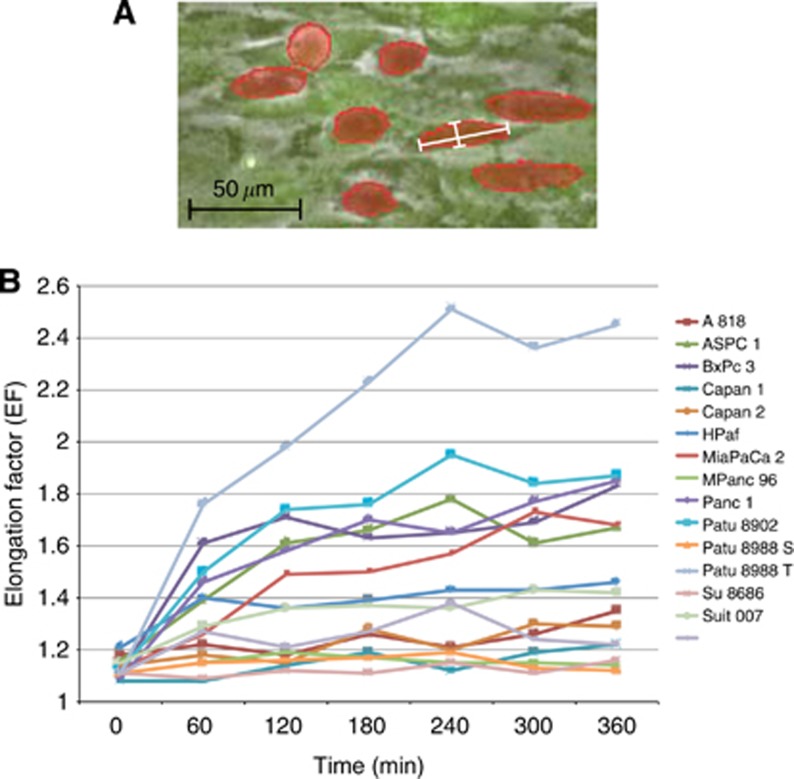
In areas with <30 evaluable PDAC cells, abnormal cell accumulation or inconsistent HMC monolayers were excluded from examination. The peripheral cells were also disregarded and excluded from further examination. The Nikon Imaging software Advanced Research 3.1 (Nikon, Düsseldorf, Germany; NIS AR Version 3.1) was used for recording and quantitative examination. Cell shape transformation was quantified by calculating the extent of elongation using the following formula:
Elongation factor (EF)=maxFeret/minFeret (NIS AR 3.1) (Figure 2A)
Figure 2.
(A) Illustration of EF measurement. The details of the microscopy picture illustrate exemplarily how the EF was calculated. The PDAC cells were marked red along their cell margins. (B) EF development. Development of the EF of PDAC cell lines plotted against time. This diagram excludes standard deviations for better readability. Once elongated, the cells showed no tendency to relinquish this development and to revert to their initial round shapes.
where Feret's diameter is the distance between two parallel lines restricting the object perpendicular to its specified direction and max/minFeret is the maximal/minimal Feret's diameter of an object. EF=1 is equivalent to a circle; the higher the EF value, the more elongated the investigated shape.
Of each time-lapse video clips, seven frames at intervals of 60 min were analysed, starting with frame number one at 0 min (Figure 2B).
Statistics
The cell lines were divided into three groups according to their mean elongation capacity (mean EFs as MEF classification) (Table 2). Cell lines falling into the intermediate group were disregarded and excluded from further investigations.
Table 2. MEF classification.
| Group | Cell line | MEF |
|---|---|---|
| Group A (high) | Patu8988T | 2.05±1.15 |
| Patu8902 | 1.67±0.62 | |
| BxPc3 | 1.60±0.63 | |
| Panc1 | 1.59±0.71 | |
| ASPC1 | 1.54±0.83 | |
| MiaPaCa2 | 1.49±0.54 | |
| Intermediate | HPaf | 1.38±0.45 |
| Suit007 | 1.34±0.44 | |
| Group B (low) | Suit0028 | 1.25±0.70 |
| A818 | 1.23±0.26 | |
| Capan2 | 1.22±0.45 | |
| Capan1 | 1.15±0.30 | |
| MPanc96 | 1.15±0.14 | |
| Patu8988S | 1.15±0.12 | |
| Su8686 | 1.12±0.11 |
Abbreviation: MEF=mean elongation factor.
Classification according to the MEFs distinguishes cell lines of group A with major elongation capacity from those of group B with minor elongation capacity. Cell lines excluded by modified median split (HPaf and Suit007) are listed to maintain completeness.
Adhesion assay
Human mesothelial cells were plated on fibronectin- (Sigma-Aldrich, Saint Louis, MO, USA) coated microtiter plates (96 wells; Greiner BioOne, Frickenhausen, Germany) and cultivated till confluent monolayers were achieved. Adhesion properties of AM-Calcein (Invitrogen)-labelled PDAC cells were determined every 10 min for 90 min by measuring the fluorescence intensity. Background autofluorescence induced by the genuine PDAC cells and HMCs served as negative controls. Negative and positive controls (total load) were conducted routinely. Measurements were conducted in triplicates and repeated at least two times in independent experiments.
Statistics
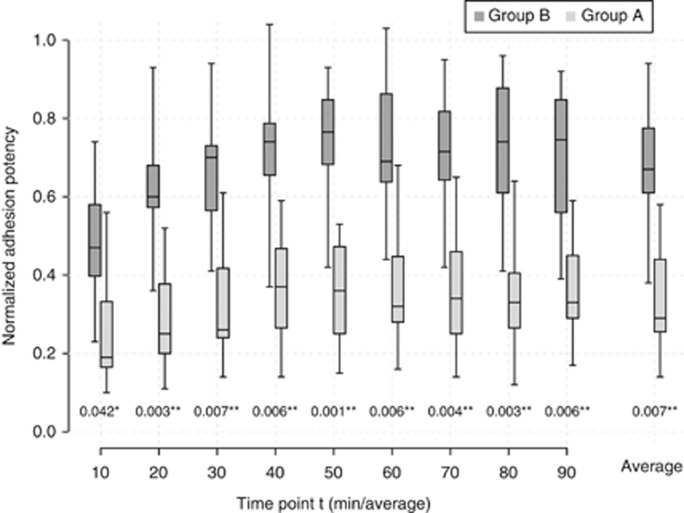
For statistical analysis, background autofluorescence was subtracted from the recorded values and the relative adhesion properties were calculated by the ratio related to the positive control. The normalised relative adhesion potentials of the two groups were compared using the unpaired t-test for each time point (Figure 3). P<0.05 was considered significant.
Figure 3.
Adhesion assay. The PDAC cell lines were classified by MEF into group A (dark grey) and group B (light grey). Boxplots illustrate the mean normalised adhesion potencies of the two groups plotted against time at every time point of measurement and in toto. Significantly increased adhesion potency for group A (*P<0.05; **P<0.01) was observed for each time point.
Correlation of the MEF classification and dissemination scores
Our group has previously published dissemination scores for the cell lines used in the orthotopic mouse models (Mees et al, 2010). These scores were then used for the present study.
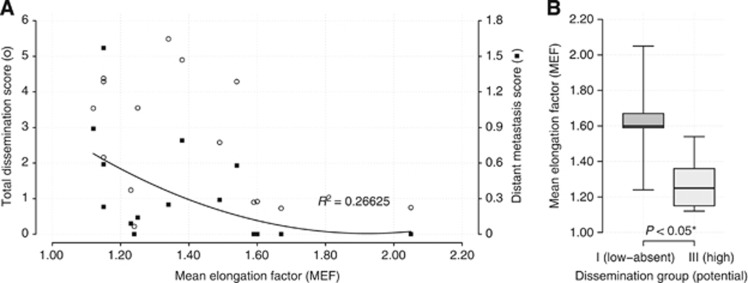
Mean EFs for each cell line were plotted against the score of the total dissemination score and the distant metastases score, respectively (Figures 4A and B).
Figure 4.
(A) Correlation of cell elongation to different aspects of metastasis formation. Total dissemination scores (○) were not related to MEF, but cellular elongation showed moderate relationship with liver-dominated distant metastasis scores (▪). Trendline with coefficient of determination is provided for distant metastasis scores only. (B) Correlation boxplots. Significant differences of MEFs between cell lines with high and low metastatic potential (P<0.05).
Microarray analysis
RNA isolation was performed according to the manufacturer's instructions using the RNeasy Mini-Kit (Qiagen, Hilden, Germany) for mRNA and miRNeasy Mini-Kit for miRNA (Qiagen). The prepared RNA was preserved at −80 °C. Only samples with RNA integrity number >8.8 for miRNA and >8.7 for mRNA were accepted for further processing.
Array analyses were performed using RNA 6000 Nano LabChipVR reagent set (Agilent Technologies, Boeblingen, Germany), Low RNA Input Linear Amplification Kit PLUS, One-Color (Agilent Technologies), Gene Expression Hybridisation Kit (Agilent) and miRNA Labelling Reagent and Hybridisation Kit (Agilent Technologies).
The Microarray Scanner G2505B (Agilent Technologies) was used for washing and scanning of miRNA and mRNA arrays.
Statistics
Statistical and bioinformatical analysis of microarray data from mRNA and miRNA microarrays were performed using the GeneSpring GX Software Version 12.0 (Agilent Technologies).
Non-biological influences and systemic variation were corrected using a normalisation algorithm. First, false-positive values due to background signals were excluded by setting a threshold of raw signals to 1.0 and by transforming them to the logarithm base two (log2(1)=0). Second, to avoid systemic biases of the individual array chips, a percentile shift was implemented to adjust all spot intensities. The 75th percentile was used for mRNA microarray and the 90th percentile was used for miRNA microarray. Additionally, the normalised mRNA data was subjected to baseline transformation (baseline corresponds to the median of all samples) to identify similar expression profiles more easily.
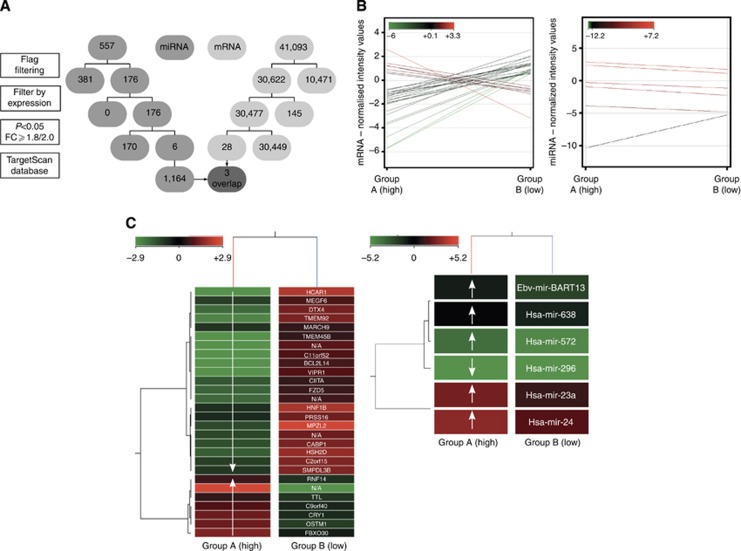
Cell lines were separated into two groups according to the MEF classification, which were evolved from the migration assay results. (HPaf and Suit007 had already been excluded after the migration assay. Capan-2 had to be excluded from further microarray analysis because of insufficient reproducibility.) Normalised data was averaged into groups. RNAs with a sufficient fluorescence signal in at least one of the analysed cell lines were displayed in a profile plot corresponding to the two provided groups and were analysed further. All miRNAs/mRNAs passed two steps of filtering (Figure 5A). The first filter was based on an acceptable fluorescence signal of all values in one of the two compared groups (flag filtering). The second filter excluded non-changing genes with a normalised signal intensity value in a range from 0.5 to −0.5 of all values in both groups (filter by expression). A moderated t-test corrected by Westfall–Young permutation and the fold change analysis were used for statistical analysis. Entities with satisfying P-values (cutoff P<0.05) as well as fold changes (cutoff FC ⩾1.8 for miRNA and FC ⩾2.0 for mRNA, respectively) identified significantly deregulated miRNAs/mRNAs. These differently regulated miRNAs/mRNAs were clustered hierarchically in a two-dimensional dendrogram corresponding to their distance metric values (uncentred Pearson's correlation) generated by the average-linked correlation (Figure 5B and C). MiRNA microarray analysis was completed by searching the TargetScan database for mRNAs, which were targeted by the differently regulated miRNAs.
Figure 5.
(A) Flowchart of microarrays. The flowchart displays the four steps of the microarray analysis for miR (middle grey) and for mRNA (light grey), and the subsequent reduction of the initially detected mRNA/miR. The final overlap is highlighted as dark grey and arrows. (B) Profile plots of microarrays. Profile plots show the mRNA/miR expression profiles for entities with a fold change cutoff of ⩾2.0/⩾1.8. The normalisation algorithms, baseline transformation in case of mRNA microarray, flag filtering, subtraction of non-changing entities (range +0.5 to −0.5) and performed moderated t-test (P-value cutoff <0.05) were conducted previously. The normalised intensity values are displayed according to an analogically coloured scale with a 100% range and an asymmetric centre. A total of 28 mRNAs were differentially regulated in the two examined groups. Seven mRNAs were significantly upregulated and 21 downregulated in group A. In all, 41 065 initially detected mRNAs were excluded by filtering. Five miRs were significantly upregulated and one downregulated in group A. Five hundred and fifty-one miRs were previously eliminated. (C) Heat maps of microarrays. Heat maps demonstrate two-dimensional dendrograms of the differentially expressed entities related to the provided groups. The hierarchical structure of the mRNA/miRs correlates with the averaged metric distances of the entities to each other. The mRNA/miR normalised intensity values are displayed according to an analogically coloured scale of reduced ranges (intensity value range: mRNA, ±2.9; miR, ±5.2) with a symmetric centre. In addition, the names of the identified mRNA/miRs are listed. Exposed genes and miRs – resulting from the performed overlap – are marked in bold type.
Finally, an overlap of the potential miRNA targets and the significantly deregulated mRNA entities was executed (Table 3).
Table 3. Deregulation of genes and miRNA.
| Gene/miRNA | Expression in group B (low) (vs group A (high)) | Log2 (fold change) | Fold change |
|---|---|---|---|
| FZD5 | Upregulation | 2.5188 | 5.7312 |
| TMEM92 | Upregulation | 3.9888 | 15.8762 |
| HNF1B | Upregulation | 1.2885 | 2.4428 |
| Hsa-mir-23a | Downregulation | 2.4 | 5.278 |
| Hsa-mir-24 | Downregulation | 2.2 | 4.5947 |
Abbreviations: miRNA=microRNA.
FZD5 is 2.5-fold, TMEM92 is 4.0-fold and HNF1B is 1.3-fold upregulated in group B. MiR-24 and miR-23a are 2.4- and 2.2-fold, respectively, downregulated in group B cell lines. (The deregulation is calculated as the fold change of logarithm to the base 2 (log2(FC)). Hsa-miR-24 targets for all three genes, TMEM92, HNF1B and FZD5. In addition to hsa-miR-24, FZD5 is also targeted by hsa-miR-23a.
Quantitative real-time PCR validation
All steps of miRNA and mRNA amplification and evaluation by quantitative RT–PCR were carried out according to the manufacturer's specifications (Qiagen).
MiRNA normalisation was performed using small nuclear RNU 6-2, SNORD 68_1 and SNORD 96A_1 (Qiagen), and mRNA normalisation was performed using PPIB1 and HPRT1 (Qiagen). MiRNA and mRNA quantitative RT–PCR, including no-template controls and real-time minus controls, were performed in triplicate and run in a CFX 384 Touch Real-Time PCR Detection System (Bio-Rad, Munich, Germany).
Statistics
The extracted data was statistically analysed with the Bio-Rad CFX Manager 2.1 software (Bio-Rad), and the relative expression was calculated using the 2−ΔΔCT method.
For comparison of both experimental groups, t-tests for dependent or independent groups were applied as appropriate. Array and RT–PCR data were statistically analysed as described above.
Western blot
Western blots were performed as previously described according to the manufacturer's instructions (Mees et al, 2010).
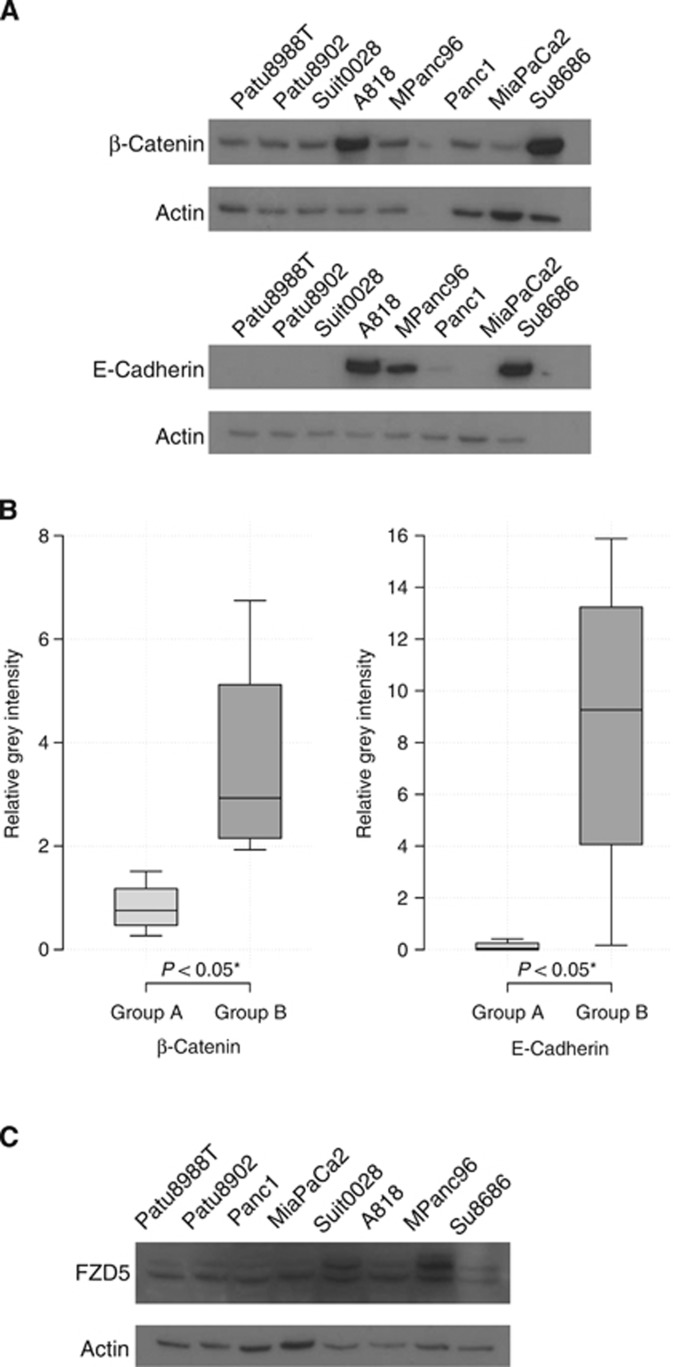
FZD5, E-cadherin and β-catenin were detected with rabbit antibodies (Cell Signaling Technology, Danvers, MA, USA). HNF1B was detected with a mouse antibody (Santa Cruz Biotechnology, Dallas, TX, USA) and TMEM92 was detected with a goat antibody (Santa Cruz Biotechnology). Four exemplary cell lines were used for each group with actin as the reference.
Statistics
Quantitative densitometric analysis was performed using the ImageJ densitometry software (Bio-Rad) (version 1.48; NIH, Bethesda, MD, USA) and selected bands were quantified based on their relative intensities. For comparison of both experimental groups, t-tests for dependent or independent groups were applied as appropriate (Figure 6).
Figure 6.
(A) Western blot. Western blots of β-catenin and E-cadherin were performed for four representative cell lines in each group. Actin was used as the reference. (B) Western boxplots. Expression level is displayed as relative grey intensity. E-cadherin was significantly more expressed in group B cell lines (8.65±6.55) vs group A cell lines (0.13±0.20) (*P<0.05). Likewise, the expression of β-catenin was significantly lower in group A (P<0.05) than in group B (group A, 0.82±0.52; group B, 3.63±2.18). (C) Western blot. Western blots of FZD5 were performed for four representative cell lines in each group. Actin was used as the reference. FZD5 showed a significant electrophoretic shift towards slower mobility in group B vs group A (*P<0.05).
Results
Time-lapse analysis
Three specific motility characteristics were observed:
Pancreatic ductal adenocarcinoma cells were almost exclusively located at cell–cell contacts of HMCs.
Pancreatic ductal adenocarcinoma cell lines differed with regard to cellular elongation during contact with HMC layers.
Some PDAC cell lines integrated into the HMC layer, but did not show transmesothelial migration.
Initially, all PDAC cells showed nearly round shapes with an average EF of 1.13±0.03 (Table 1). During examination, about half of the cell lines elongated, whereas the other half kept their primary shapes (Figure 1 and Supplementary Data).
The cell elongation was quantified (Figure 2A), and the MEF observed in every cell line was determined using seven standardised time-lapse frames (Figure 2B). The MEFs ranged from MEFSu8686 1.12±0.11 to MEFPatu8988T 2.05±1.15 and were subsequently used to classify the PDAC cell lines into two groups (Table 2). According to this MEF classification, group A cell lines showed a major elongation capacity of PDAC cells with an average MEF of 1.66±0.2, which is significantly increased (P<0.05) compared with the initial MEFs and the MEFs of PDAC cells in group B (1.18±0.05). The overall average MEF for all cell lines for all time points was 1.36±0.03. Pancreatic ductal adenocarcinoma cells of two cell lines (HPaf MEF 1.34±0.44 and Suit 007 MEF 1.38±0.45) showed an intermediate elongation capacity using a modified median split and were excluded from further investigations.
Adhesion assay
With regard to the adhesive capacity of PDAC cell lines, we compared group A (major elongation capacity) results with group B cell lines (minor elongation capacity) (Figure 3). Using the unpaired t-test for statistical analysis, group A showed significantly increased adhesion (P<0.01–0.05) at every single time point, as well as on an average throughout the observation period. Adhesion potency of group A ranged between 0.48±0.08 and 0.73±0.09 with an average of 0.68±0.10. The adhesion potency of group B was less (0.36±0.07) and ranged from 0.28±0.05 to 0.39+0.07.
Correlation of the MEF classification and dissemination scores
For assessment and comparison of cell elongation with dissemination properties, we used dissemination scores previously published by our group (Mees et al, 2010). The score of total metastatic formation as well as its subgroup of distant metastatic formation were used. Total metastatic formation includes lymph nodes, whereas distant metastatic formation is mainly determined by liver metastases. Cellular elongation showed moderate relationship to distant metastasis scores, whereas correlation between the score of total metastatic formation and MEF values was not observed (Figure 4A). Comparing the MEFs of both groups with their potential for distant metastasis formation, group A cell lines (high MEFs) were found in low metastatic cell lines, whereas high metastatic cell lines showed significantly lower MEF values (P<0.05) (Figure 4B).
Microarray analysis
mRNA and miRNA expression profiles of the above-defined groups (MEF classification) were determined using microarray analysis to detect gene and miRNA expression differences as a potential rationale for the observed differing cell behavior (Figure 5A). Further, investigated miRNAs and mRNAs were validated by quantitative rRT–PCR and the corresponding protein expression was determined by western blot.
Five miRNAs (hsa-miR-23a, hsa-miR-24, hsa-miR-638, hsa-miR-572 and ebv-miR-BART13) were upregulated and one miRNA (hsa-miR-296) was downregulated in group A (Figure 5B and C, right). The following analysis of the TargetScan database identified 1146 gene targets influenced by the expression of these six miRNAs.
Seven mRNAs were significantly upregulated and 21 mRNAs were downregulated in group A (Figure 5B and C, left).
One thousand one hundred and sixty-four mRNAs were identified as potential targets for the six deregulated miRNAs. Three mRNAs overlap between the potential miRNA targets and the observed deregulated mRNAs: FZD5, TMEM92 and HNF1B. Two of the six deregulated miRNAs (hsa-miR-23a and hsa-miR-24) are reported to target the three overlapping mRNAs (Table 3).
Western blot
E-cadherin and β-catenin protein expression in both groups was compared by western blot. Group A cell lines displayed a significantly reduced protein expression of E-cadherin and β-catenin compared with group B cell lines (P<0.05). For E-cadherin, densitometric quantification showed relative grey intensities of 0.1269±0.1973 for group A and of 8.6483±6.5454 group B. For β-catenin, densitometric quantification showed relative grey intensities of 0.8234±0.5187 for group A and of 3.6340±2.1762 group B (Figures 6A and B). Western blot of TMEM92 and HNF1B did not show different protein amounts in both groups (not shown), but for FZD5 a significant electrophoretic shift towards slower mobility was observed in group B vs group A (Figure 6C).
Discussion
We implemented a novel migration assay for PDAC cells at HMC monolayers to analyse the peritoneal metastasis formation. Instead of the expected general transmigration, all PDAC cells exhibited a specific migration pattern and were almost exclusively found at cell–cell boundaries of the epithelial monolayers. During this process, several cell lines underwent EMT typical morphological changes, such as the development of an elongated shape from an initial round cell shape. Cell elongation appeared to be a significant process during PDAC peritoneal metastasis, and we thus classified the investigated cell lines according to their exhibited elongation (MEF classification).
Besides quantitative analysis via MEF classification, the qualitative observation of the elongation suggested that PDAC cells were influenced by de novo intercellular adhesion with the HMCs.
To discriminate between EMT- and adhesion-associated effects, the adhesion potency and velocity of the PDAC cell lines were compared: the significantly more elongated cell lines of group A showed significantly higher adhesion properties than the more round group B cell lines (P<0.05). This result underlines the relevant influence of HMC shape adaption by adhesion and appears to be contradictory. It is possible that the observed cell adhesion characteristics reflect beginning of the reversal of EMT, that is, MET induced by HMC contacts. However, the migrating PDAC cells remained spindle shaped even within an extended observation time (Figure 2B) challenging this assumption.
Thus, it appears that the integrating PDAC cells could have been influenced by de novo intercellular adhesion with the HMCs, independent of E-cadherin (Thiery, 2002), or, more likely, during interactions with HMCs, the integrating PDAC cells can modulate the availability of E-cadherin using the identified targets FZD5, HNF1B and TMEM via miR23a/miR24 deregulation.
In summary, the present results indicate that the development of PDAC peritoneal carcinomatosis is characterised by integration into HMC monolayers rather than by transmigration. Different PDAC cell lines exhibited varying ability to integrate into the monolayer of the mesothelium. The disposition to integrate could be advantageous for cell survival and for the development of peritoneal dissemination and consequently could lead to more metastatic phenotypes.
Surprisingly, integration properties of PDAC cells into HMC monolayers inversely correlated with the previously reported extent of distant metastasis formation, mainly determined by liver colonisation (Mees et al, 2010). The contradicting correlations between liver colonisation and peritoneal metastasis model data suggest organ-specific involvement of epigenetic regulations in cancer progression. This is likely determined by microenvironmental factors such as HMCs vs liver sinusoidal cells.
We hypothesised that epigenetic mechanisms have a role not only for the pancreatic cancer development (Bloomston et al, 2007) but also for its organ-specific dissemination process. Cell lines with major elongation capacity (group A, MEF 1.66±0.2) showed higher expression levels of miR-23a (2.4-fold) and miR-24 (2.2-fold) than cell lines with minor integration capacity (group B, MEF 1.18±0.05). Additionally, group A cell lines exhibited lower expression of their target proteins FZD5, TMEM92 and HNF1B.
The expression of Wnt-related genes leads to cell shape changes toward elongation (Brown et al, 1986), and the canonical Wnt/β-catenin signalling cascade has been revealed to be deregulated in PDAC cells (Zeng et al, 2006; Bu et al, 2008). β-Catenin, the central molecule of the Wnt/β-catenin pathway, connects it to the main intercellular adhesion mechanism, the cadherin/catenin complex. Deregulations of this complex are also known to occur in PDAC cells (Perl et al, 1998; Pryczynicz et al, 2010).
Group B cell lines showed increased expression of E-cadherin and β-catenin in western blot analysis (P<0.05). These results support our theory of a misbalance of cadherin and catenin in group A cell lines, resulting in the loss of the stable cadherin/catenin complex and resulting in EMT-like cell lengthening. Howard et al (2011) suggested that permanent interactions of intracellular β-catenin rather than its absolute amount seem to facilitate EMT, which could serve as an explanation for the limited absolute difference in our analysis.
Similarly, our microarray results showed an overall decreased expression of E-cadherin in group A cell lines, which however remained below the threshold for significance (5.08-fold increased expression in group B after altered algorithm (without the correction by Westfall–Young permutation)) (Supplementary Data).
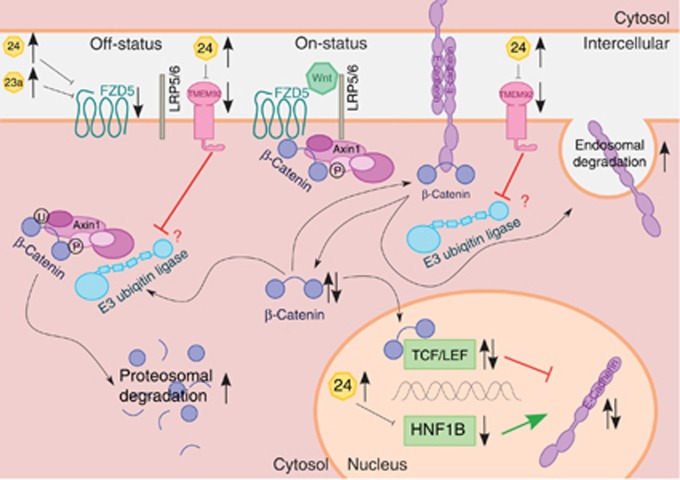
The present data suggest that each of the three deregulated targets (FZD5, TMEM92 and HNF1B) – independently or in combination – could influence the two highly connected EMT-related systems: the canonical Wnt/β-catenin pathway and the intercellular adhesion process by the cadherin/catenin complex (Figure 7).
Figure 7.
Molecular genetic model (adapted from van Roy and Berx, 2008; Clevers and Nusse, 2012; Pei and Grishin, 2012; Van den Bossche et al, 2012). This in silico model is highly simplified and points out the crucial essentials of our results. (Top, left; canonical Wnt/β-catenin pathway) The destruction complex Axin1 leads to the phosphorylation of β-catenin. Phosphorylated β-catenin is ubiquitinated by the E3 ubiquitin ligase and then degraded proteasomally (off-status). The activation of the two receptors FZD and LRP5 by Wnt leads to membrane recruitment of the destruction complex. The E3 ubiquitin ligase is no longer able to ubiquitinate β-catenin of membrane-associated Axin1 (on-status). Consequently, the cytoplasmatic amount of β-catenin increases. MiR-23a and miR-24 lead to the downregulation of FZD5. Subsequently, Wnt cannot adhere to its receptors, the signal transduction is precluded and the Wnt/β-catenin signalling pathway remains in its off-status. In this case, the β-catenin phosphorylation, ubiquitination and degradation take place without any regulatory inhibition. The cytoplasmatic β-catenin amount decreases. Consequently, two inverse effects seem to occur. First, the lack of cytoplasmatic β-catenin could result in the destabilisation of catenation between the cadherin–catenin complex and the actin cytoskeleton. This could be responsible for the observed loss of cell polarity in the cell lines of group A. Second, an increase of cytoplasmatic β-catenin leads to an increase of nuclear β-catenin as well. βeta-catenin bound to TCF/LEF acts as a negative transcription factor (Jamora et al, 2003). It suppresses the expression of E-cadherin. Subsequently, the decrease of β-catenin leads to an increased expression of E-cadherin. This function of β-catenin seems to be a compensatory long-term feedback mechanism to maximise the small amount of cytoplasmatic β-catenin trying to restabilise the connection between the CCC and the cytoskeleton. (Bottom, right) Further, this attempt of compensatory feedback mechanism could be abrogated by the simultaneous downregulation of HNF1B. The expression of E-cadherin is enhanced by HNF1B serving as a positive transcription factor (Goomer et al, 1994). Thus, our results emphasise that the upregulation of miR-24 leads to the downregulation of HNF1B followed by decreased expression of E-sadherin with the loss of cell polarity. (Top, right; CCC) The cadherin–catenin complex (CCC) forms homologous cell–cell adhesion. The CCC includes the stable membrane-associated β-catenin pool. The E3 ubiquitin ligase leads to the ubiquitination of E-cadherin. Previous to the following endosomal degradation of E-cadherin, β-catenin dissociates from E-cadherin. Subsequently, the cytoplasmatic pool of β-catenin increases. We suppose that, most likely, TMEM92 interacts competitively with the E3 ubiquitin ligase and stabilises the amount of the CCC. MiR-24 leads to the downregulation of TMEM92. Downregulated TMEM92 fails to block the E3 ubiquitin ligase. Consequently, E-cadherin degradation is increased. The amount of E-cadherin decreases, cytoplasmatic β-catenin increases after dissociation, and the CCC and intercellular adhesion are destabilised. Possibly, the increase of dissociated β-catenin is immediately intercepted, because the same mechanism leads to a decreased pool of cytoplasmatic β-catenin. By competitive binding, TMEM92 inhibits the E3 ubiquitin ligase and consequently blocks β-catenin degradation. Thus, the gene silencing of TMEM92 by the upregulation of miR-24 appears to increase the degradation of β-catenin as well as E-cadherin. This mechanism also leads to the destabilisation of the CCC.
In the Frizzled family, FZD5 is one of eleven known receptor proteins (FZD1-10, SMO). These seven-pass transmembrane receptors (Vinson et al, 1989) are indispensable to the activation of the canonical Wnt/β-catenin signalling pathway (Bhanot et al, 1996). For the FZD5 electrophoretic mobility shift, likely due to serine phosphorylation, regulatory consequences are uncertain.
Although correlations between HNF1B and the Wnt/β-catenin pathway were not observed, HNF1B serves as a positive transcription factor for E-cadherin (Goomer et al, 1994).
TMEM92 could act as an adaptor for E3 ubiquitin ligases and could support the degradation process of β-catenin and E-cadherin. In contrast, TMEM92 could also act as a competitive binding partner of the E3 ubiquitin ligase and also inhibit the ubiquitination and the following degradation of both β-catenin and E-cadherin, which appears to be more likely (Konstas et al, 2002). Further, TMEM92 could act variably depending on its substrates. Thus, the quality of this interaction remains unclear and the consequences of TMEM92 deregulation are currently unknown, but gene silencing of TMEM92 by the upregulation of miR-24 appears to increase the degradation of β-catenin as well as that of E-cadherin.
Our explanatory model presents complex, fragile and instable molecular mechanisms with an epigenetic background followed by EMT-like cell shape transformation as observed in the time-lapse microscopy. We theorise that the disequilibrium of miR-23a and/or miR-24 is at its basis. In silico pathway analysis supported by mRNA and protein expression data delivers an explanation for how the miRNA-mediated downregulation of FZD5, TMEM92 and HNF1B could influence the canonical Wnt/β-catenin pathway and the intercellular adhesion mediated by the cadherin/catenin Complex. This influence could be exerted by the deregulated expression and/or degradation of the two key components β-catenin and E-cadherin.
To the best of our knowledge, this is the first report correlating the Wnt/β-catenin pathway or the cadherin/catenin complex with the peritoneal dissemination of PDAC. At the same time, peritoneal dissemination seems to be an organ-specific form of metastasis formation that differs from other host organ sites.
Taken together, the unexpected PDAC cell elongation during mesothelial invasion seems to be the product of coincidental loss and acquisition of intercellular adhesion between PDAC cells and the HMCs. This coincidence is likely caused by deregulation of the sensitive interaction between the cadherin/catenin Complex and the Wnt/β-catenin pathway. The immediate microenvironment of the PDAC cells produced by the HMCs may trigger this deregulation.
Acknowledgments
We thank F Spiecker, D Pütz (Nikon, Duesseldorf, Germany), A Raem (arrows biomedical Deutschland GmbH, Muenster, Germany), J Schnekenburger (Biomedical Technology Center Technologiezentrum, Westfalian Wilhelm-University Muenster, Germany) and B Löffler (Institute of Medical Microbiology, University Hospital of Muenster, Muenster, Germany). This project was partially funded by an unrestricted grant of the Foerderverein Peter Geiger, Beilstein, Germany.
SW is employed at arrows biomedical Deutschland GmbH, a company that distributes molecular biology services. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- Brown AM, Wildin RS, Prendergast TJ, Varmus HE. A retrovirus vector expressing the putative mammary oncogene int-1 causes partial transformation of a mammary epithelial cell line. Cell. 1986;46:1001–1009. doi: 10.1016/0092-8674(86)90699-9. [DOI] [PubMed] [Google Scholar]
- Bu XM, Zhao CH, Zhang N, Gao F, Lin S, Dai XW. Hypermethylation and aberrant expression of secreted frizzled-related protein genes in pancreatic cancer. World J Gastroenterol. 2008;14:3421–3424. doi: 10.3748/wjg.14.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. MicroRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Goomer RS, Holst BD, Wood IC, Jones FS, Edelman GM. Regulation in vitro of an L-CAM enhancer by homeobox genes HoxD9 and HNF-1. Proc Natl Acad Sci USA. 1994;91:7985–7989. doi: 10.1073/pnas.91.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard S, Deroo T, Fujita Y, Itasaki N. A positive role of cadherin in Wnt/β-catenin signalling during epithelial–mesenchymal transition. PLoS One. 2011;6:e23899. doi: 10.1371/journal.pone.0023899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamora C, DasGupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 2003;422:317–322. doi: 10.1038/nature01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JY, Kang MJ, Heo JS, Choi SH, Choi DW, Park SJ, Han SS, Yoon DS, Yu HC, Kang KJ, Kim SG, Kim SW. A prospective randomized controlled study comparing outcomes of standard resection and extended resection, including dissection of the nerve plexus and various lymph nodes, in patients with pancreatic head cancer. Ann Surg. 2013;259 (4:656–664. doi: 10.1097/SLA.0000000000000384. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Konstas AA, Shearwin-Whyatt LM, Fotia AB, Degger B, Riccardi D, Cook DI, Korbmacher C, Kumar S. Regulation of the epithelial sodium channel by N4WBP5A, a novel Nedd4/Nedd4-2-interacting protein. J Biol Chem. 2002;277:29406–29416. doi: 10.1074/jbc.M203018200. [DOI] [PubMed] [Google Scholar]
- Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Thiery JP. Epithelial–mesenchymal transitions: insights from development. Development. 2012;139:3471–3486. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- McCleary-Wheeler AL, Lomberk GA, Weiss FU, Schneider G, Fabbri M, Poshusta TL, Dusetti NJ, Baumgart S, Iovanna JL, Ellenrieder V, Urrutia R, Fernandez-Zapico ME. Insights into the epigenetic mechanisms controlling pancreatic carcinogenesis. Cancer Lett. 2013;328:212–221. doi: 10.1016/j.canlet.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mees ST, Mardin WA, Wendel C, Baeumer N, Willscher E, Senninger N, Schleicher C, Colombo-Benkmann M, Haier J. EP300 – a miRNA-regulated metastasis suppressor gene in ductal adenocarcinomas of the pancreas. Int J Cancer. 2010;126:114–124. doi: 10.1002/ijc.24695. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. Unexpected diversity in Shisa-like proteins suggests the importance of their roles as transmembrane adaptors. Cell Signal. 2012;24:758–769. doi: 10.1016/j.cellsig.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- Pryczynicz A, Guzińska-Ustymowicz K, Kemona A, Czyzewska J. Expression of the E-cadherin–catenin complex in patients with pancreatic ductal adenocarcinoma. Folia Histochem Cytobiol. 2010;48:128–133. doi: 10.2478/v10042-008-0089-1. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Sohn TA, Yeo CJ, Cameron JL, Koniaris L, Kaushal S, Abrams RA, Sauter PK, Coleman J, Hruban RH, Lillemoe KD. Resected adenocarcinoma of the pancreas-616 patients: results, outcomes, and prognostic indicators. J Gastrointest Surg. 2000;4:567–579. doi: 10.1016/s1091-255x(00)80105-5. [DOI] [PubMed] [Google Scholar]
- Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- Thiery JP. Epithelial–mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial–mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Van den Bossche J, Malissen B, Mantovani A, De Baetselier P, Van Ginderachter JA. Regulation and function of the E-cadherin/catenin complex in cells of the monocyte–macrophage lineage and DCs. Blood. 2012;119:1623–1633. doi: 10.1182/blood-2011-10-384289. [DOI] [PubMed] [Google Scholar]
- van Roy F, Berx G. The cell–cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65:3756–3788. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson CR, Conover S, Adler PN. A Drosophila tissue polarity locus encodes a protein containing seven potential transmembrane domains. Nature. 1989;338:263–264. doi: 10.1038/338263a0. [DOI] [PubMed] [Google Scholar]
- Zeng G, Germinaro M, Micsenyi A, Monga NK, Bell A, Sood A, Malhotra V, Sood N, Midda V, Monga DK, Kokkinakis DM, Monga SP. Aberrant Wnt/beta-catenin signaling in pancreatic adenocarcinoma. Neoplasia. 2006;8:279–289. doi: 10.1593/neo.05607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.