Abstract
Background:
Treatment planning of localised prostate cancer remains challenging. Besides conventional parameters, a wealth of prognostic biomarkers has been proposed so far. None of which, however, have successfully been implemented in a routine setting so far. The aim of our study was to systematically verify a set of published prognostic markers for prostate cancer.
Methods:
Following an in-depth PubMed search, 28 markers were selected that have been proposed as multivariate prognostic markers for primary prostate cancer. Their prognostic validity was examined in a radical prostatectomy cohort of 238 patients with a median follow-up of 60 months and biochemical progression as endpoint of the analysis. Immunohistochemical evaluation was performed using previously published cut-off values, but allowing for optimisation if necessary. Univariate and multivariate Cox regression were used to determine the prognostic value of biomarkers included in this study.
Results:
Despite the application of various cut-offs in the analysis, only four (14%) markers were verified as independently prognostic (AKT1, stromal AR, EZH2, and PSMA) for PSA relapse following radical prostatectomy.
Conclusions:
Apparently, many immunohistochemistry-based studies on prognostic markers seem to be over-optimistic. Codes of best practice, such as the REMARK guidelines, may facilitate the performance of conclusive and transparent future studies.
Keywords: prostate cancer, prognostic marker, immunohistochemistry, REMARK guidelines
The risk assessment and management of therapeutic strategies for prostate cancer are presently still mainly based on clinical criteria, for example, patient characteristics, clinical stage, serum PSA, and histopathological features, in particular, tumour extent and Gleason score. However, not all low-grade prostate cancers with a Gleason score less than 7 follow an indolent course, and carcinomas with a Gleason score of 7 or higher, on the other hand, may (rarely) present with a rather favourable clinical behaviour. These discrepancies have become increasingly apparent, particularly following the updated recommendations of the 2005 ISUP Gleason System (Harlan et al, 2003), therefore emphasising the pressing need for additional molecular prognostic markers. Interestingly, the lack of reliable molecular biomarkers for the risk assessment of prostate cancer stands in sharp contrast to an ever-emerging number of proposed marker candidates. With nearly 2900 PubMed hits for the search term ‘prostate cancer prognostic marker' (July 2014), no evidently relevant prognostic biomarker has made its way into clinical practise (Kristiansen, 2012). This issue clearly stresses the necessity to reflect on the validity of biomarkers and biomarker development or even some aspects of our scientific publication culture.
Despite the limitation of immunohistochemistry being an only semiquantitative technique, it still represents an essential methodological approach for a large fraction of published biomarker studies. This prompted us to experimentally verify the prognostic value of a wider range of previously published immunohistochemical prognostic markers for prostate cancer in a representative monocentric cohort of radical prostatectomy patients.
Materials and methods
Criteria for identification of prognostic markers
In a first step, prognostic markers for prostate cancer were identified in a PubMed search (‘prostate cancer prognostic/prognosis immunohistochemistry'). Eligibility criteria for study inclusion were: (i) cohort size—a number of patients >50 in studies using conventional slides or >100 for tissue microarray (TMA)-based studies, (ii) multivariate prognostic value demonstrated in Cox analysis (P<0.05), and (iii) conclusive immunohistochemistry protocol and antibody availability. From the pool of resulting candidates, we obtained 28 markers for validation.
Prostate cancer cohort for validation
The cohort consisted of 238 patients having undergone radical prostatectomy for treatment of primary prostate cancer between 1999 and 2006 and for which follow-up data were available. This cohort was compiled as a TMA as described previously, all tumours were reviewed and graded according to the ISUP 2005 recommendations (Gerhardt et al, 2011; Mortezavi et al, 2011; Beer et al, 2012; Gerhardt et al, 2012). Follow-up data were obtained from review of patients' medical records. Median follow-up time for all patients was 60 months (mean 55 months). A biochemical recurrence, defined as a rising PSA level exceeding 0.1 ng ml−1, after having reached a nadir post surgery, occurred in 111 (46.6%) patients. Median time to PSA relapse was 25 months (interquartile range, 12–46 months); among men not experiencing progression, the median follow-up was 72 months (interquartile range, 60–96 months). In total, 13.4% of patients were lost to follow-up in the first 4 years.
The median patient age was 64 years (range 46–75 years). The pT-category was pT2 in 152 patients (63.8%), pT3 in 76 patients (31.9%), and pT4 in 10 patients (4.3%). A total of 151 patients (63.4%) presented with complete surgical resection (R0 resection), whereas 84 patients (35.3%) had undergone a resection with positive margins (R1 resection). The Gleason score was <7 in 45 patients (18.9%), equalled a total score of 7 in 133 patients (55.9%), and was >7 in 60 patients (25.2%). Pre-operative PSA levels ranged from 0.39 ng ml−1 to 357 ng ml−1 (median 10.5 ng ml−1, no data were available for 11 patients). This study was approved by the Cantonal Ethics Committee of Zürich (approval number StV 25-2007).
Immunohistochemistry
Automated immunohistochemical staining was carried out on one of the two technical platforms (Ventana Medical Systems, Tucson, AZ, USA/Leica Microsystems, Melbourne, Australia). If possible, antibodies were selected according to the preceding publications. Staining protocols including antibody dilution, pretreatment, and technical platform are given in Supplementary Table S1.
Evaluation of immunoreactivity
Immunohistochemical slides were evaluated and scored by a single observer (FH) after instruction and supervision of an experienced genito-urinary pathologist (GK). Wherever possible, we adopted the scoring system described in the original publications of the respective markers. Technical details of scoring (localisation, evaluated feature, and scoring scale) are given in Supplementary Table S2.
Statistics
SPSS 20 (IBM SPSS, Version 20.0, Armonk, NY, USA) was used for descriptive statistics. Further data analysis was performed using the R language for statistical computing, version 3.1.0. The statistical significance level was set at 0.05. To verify the prognostic value of the selected markers, univariate and multivariate Cox proportional hazards regression was applied. Multivariate Cox regression was performed with inclusion of the individual selected markers, Gleason score, log10(pre-OP PSA), dichotomous pT status (pT1/2 vs pT3/4), margins (R0 vs R1), and patient age. Additionally, markers were dichotomised using the web-based tool ‘cut off finder' (Budczies et al, 2012) and were analysed using Kaplan–Meier estimates and log rank tests. A post hoc power analysis of the respective models was performed using the PASS 2008 software (NCSS, Kaysville, UT, USA).
Results
Characteristics of selected biomarker studies from the literature
Of the markers matching our inclusion criteria, 27 candidates were selected, 3 of which (androgen receptor, oestrogen receptor-alpha and -beta) each yielded two data points (stromal and epithelial immunoreactivity), resulting in 30 biomarkers for further validation (Table 1). In the originally published studies, cohorts had a median size of 225 patients (range 53–2724), the median follow-up time covered 5.0 years (range 1–12), and the median hazard ratio for disease progression of the reported biomarkers was 2.42 (range 1.1–7.69; values of markers with hazard ratios <1 have been included as their reciprocal value). All selected biomarkers play a role in a broad spectrum of tumour-relevant processes including, for example, proliferation or apoptosis, cell cycle control, cell adhesion, or hormone signalling. Most studies (n=23) were based on radical prostatectomy (RPE) cohorts, four studies described watchful waiting cohorts, and one study described a mixed population (RPE and watchful waiting).
Table 1. Overview of selected prognostic markers for prostate cancer: original studies.
| Marker | Number of patients | Type of cohort | Statistical endpoint | Hazard ratio | P-value (multivariate) | mean+/median* follow-up time (years) | Reference (PMID) |
|---|---|---|---|---|---|---|---|
| ADAM9 | 198 | RPE | BCR | 1.87 | 0.04 | 3.8* | 18061337 |
| AKT1 | 63 | RPE | BCR | 2.12 | 0.02 | 6+ | 16721361 |
| AKT1 | 840 | RPE | BCR, death | 2.86 | 0.027 | 5.1+ | 19417030 |
| ALCAM | 91 | RPE | BCR | 2.22 | 0.057 | 2.5* | 15532095 |
| ALCAM | 2390 | RPE | BCR | 0.71 | 0.02 | 2.9+ | 21683980 |
| AR(nuclear) | 551 | RPE | BCR | 2.42 | 0.004 | 5.5* | 15223964 |
| AR(stromal) | 53 | RPE | DSR | 0.13 | 0.005 | 4.25+ | 15378523 |
| BCL2 | 94 | RPE | BCR | 3.15 | 0.03 | 2.3* | 16080576 |
| BCL2 | 228 | WW | death | 2.14 | 0.004 | 11* | 17850375 |
| CB1R | 372 | WW | death | 2.7 | 0.006 | >12 | 19056257 |
| CD10 | 2724 | RPE | BCR | 1.1 | 0.034 | 2.9+ | 19047112 |
| CD24 | 102 | RPE | BCR | 3.24 | 0.005 | 2.6* | 14716744 |
| CD138 | 551 | RPE | BCR/radiological progression | missing | 0.051 | 5.3* | 12640657 |
| CRGA | 528 | RPE | BCR | 1.66 | 0.049 | 3.8* | 18392627 |
| CRISP3 | 945 | RPE | BCR | 1.59 | 0.007 | 6* | 17634540 |
| E-CADHERIN | 104 | RPE | BCR/clinical progression | 2.5 | 0.019 | 7.9* | 18056176 |
| ER-alpha | 106 | RPE | death | 5.24 | 0.036 | 6.8+ | 17294452 |
| ER-beta | 159 | RPE | BCR | missing | 0.04 | 5.4* | 11454669 |
| EZH2 | 226 | RPE | BCR | 3.14 | 0.05 | 5.5* | 17943722 |
| HDAC2 | 145 | RPE | BCR | 2.36 | 0.019 | 4.2* | 18212746 |
| HSP-27 | 553 | WW | death | 1.5 | 0.03 | 7.9+ | 19707199 |
| Ki-67 | 108 | RPE | BCR/clinical progression | 2.48 | 0.01 | 5.4+ | 8691323 |
| MUC1 | 225 | RPE | BCR/clinical progression | 2.35 | 0.0005 | 8* | 14711987 |
| N-CADHERIN | 104 | RPE | BCR/clinical progression | 5.6 | 0.003 | 7.9* | 18056176 |
| NFkB | 86 | RPE | BCR | 5.0 | 0.002 | 3.6+ | 16278667 |
| p21 | 60 | WW/RPE | death | 6.26 | 0.023 | 3.4+ | 9712417 |
| p27 | 161 | RPE | BCR | 2.44 | 0.014 | 3.75+ | 12629353 |
| p53 | 705 | WW | death | 1.51 | 0.03 | 9.75* | 19239456 |
| p53 | 2514 | RPE | BCR | 1.24 | 0.034 | 2.9+ | 18552821 |
| PSMA | 93 | RPE | BCR | 1.4 | 0.01 | 2.8+ | 17320151 |
| SPINK1 | 817 | RPE | BCR | 2.02 | 0.0004 | missing | 18538735 |
| Vimentin | 287 | RPE | BCR | 2.03 | 0.049 | 7+ | 19447876 |
Abbreviations: BCR=biochemical recurrence; DSR=disease-specific relapse; RPE=radical prostatectomy; WW=watchful waiting/conservative management. +=mean, *=median follow up time.
Immunohistochemistry and expression of selected markers
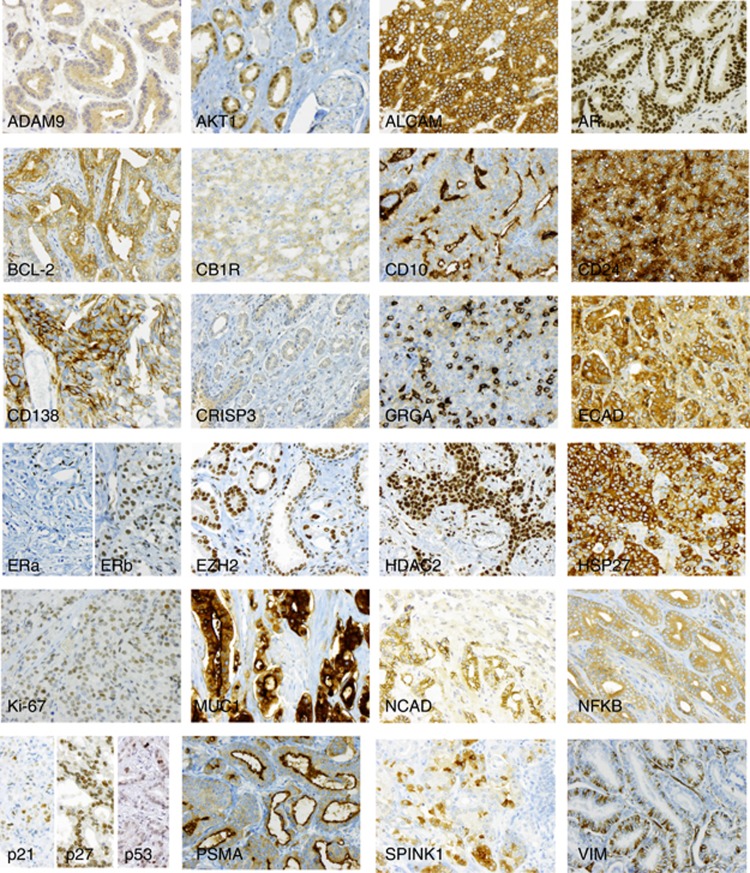
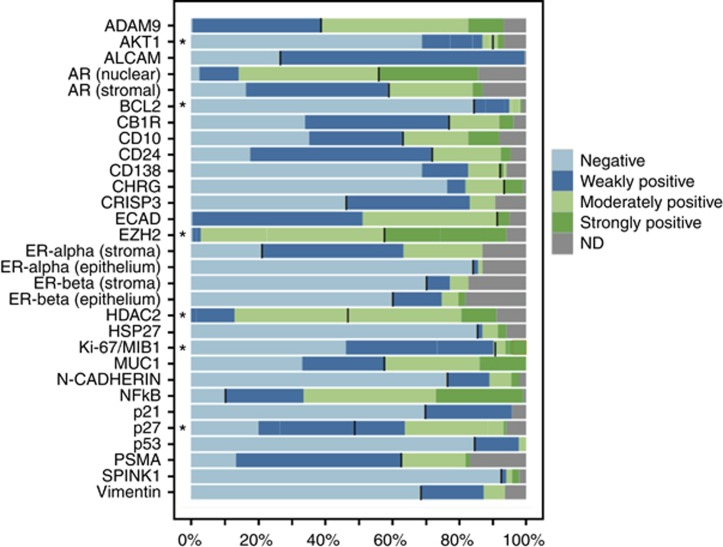
For the markers included in this study, representative examples of positive immunoreactivity in prostate cancer are shown in Figure 1. Expression frequencies and cut-off values are illustrated in Figure 2. We attempted to adopt the cut-off values suggested in the original studies, but allowed for adjustment (optimisation with the cut-off finder tool) in order to ease verification.
Figure 1.
Immunoreactivity of selected prognostic biomarkers. Representative examples of positive immunostaining are shown for each marker, highlighting the typical subcellular localisation (magnification × 200).
Figure 2.
Frequencies of immunohistochemically detected expression of selected prognostic markers in primary prostate carcinomas. Markers labelled with an asterisk were recorded as immunoreactive scores and summarised for easier visualisation (IRS 0: negative, IRS 1–3: weakly positive, IRS 4–8 moderately positive, IRS>8: strongly positive). Black bars indicate the cut-off used for dichotomising the variable for Cox regression analysis. Abbreviation: ND: non determined/missing data.
Associations of biomarkers with clinico-pathological parameters
The association of marker expression in prostate cancer with pT category, Gleason score, and pre-operative serum PSA was analysed in a Spearman rank correlation analysis. As expected for independent prognostic markers, most associations failed significance, even though some dependencies became apparent, particularly with Gleason score and serum PSA, but high correlation coefficients were not noted (Supplementary Table S3).
Prognostic significance of putative prognostic biomarkers
Initially, we conducted a post hoc power analysis and found that the available sample size of 238 analysable patients on the TMA would be sufficient to detect clinically relevant hazard ratios at a significance level of 0.05 and a power of almost 100% (Supplementary Figure S1).
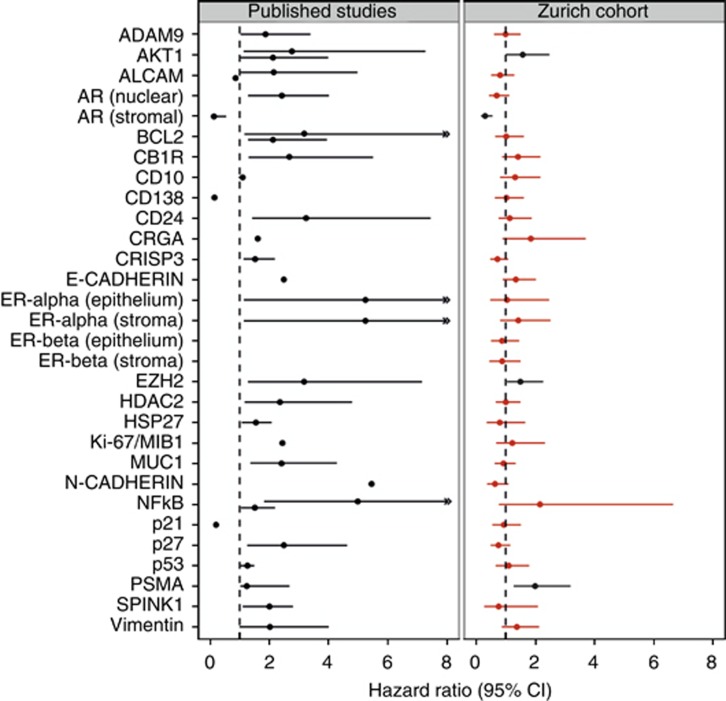
Kaplan–Meier analyses identified 11 markers as being univariate significant factors: AR epithelial/stromal, CB1R, CRGA, E-Cadherin, EZH2, Ki-67, NFkB, p21, p27, and PSMA (Table 2). In univariate Cox analyses, only nine markers were found significant (nuclear and stromal AR, CB1R, CRGA, E-Cadherin, NFkB, p21, p27, and PSMA), CD10 was of borderline significance (P=0.051) and Ki-67 dropped out. Multivariate Cox regression, which was applied to analyse the prognostic value of the markers in combination with other prognostic factors, demonstrated significance for only the four markers AKT1, stromal AR, EZH2, and PSMA (Table 3). A marker-wise comparison of published and verified multivariate prognostic value, illustrated by a forest plot, highlights these differences (Figure 3). In three of the four confirmed prognostic markers, the verified hazard ratio was lower than originally reported; only for one (PSMA), a higher hazard ratio was found.
Table 2. Kaplan–Meier analysis of PSA relapse-free survival for selected prognostic markers following cut-off optimisation.
| Marker | Optimal cut-off | Hazard ratio (95% CI) | P-value |
|---|---|---|---|
| ADAM9 | 2.5 | 0.74 (0.36–1.53) | 0.41 |
| AKT1 | 25% | 1.65 (0.92–2.95) | 0.09 |
| ALCAM | 1.5 | 1.35 (0.89–2.06) | 0.16 |
| AR (epithelial) | 2.5 | 0.60 (0.37––0.95) | 0.028 |
| AR (stromal) | 1.5 | 0.30 (0.17–0.53) | < 0.001 |
| BCL2 | 30% | 1.32 (0.72–2.40) | 0.37 |
| CB1R | 1.5 | 1.70 (1.10–2.62) | 0.017 |
| CD10 | 1.5 | 1.27 (0.86–1.89) | 0.22 |
| CD24 | 1.5 | 0.71 (0.44–1.15) | 0.16 |
| CD138 | 0.5 | 1.67 (0.93–2.99) | 0.08 |
| CRGA | 2.5 | 2.04 (1.03–4.04) | 0.037 |
| CRISP3 | 0.5 | 0.76 (0.51–1.14) | 0.19 |
| E-CADHERIN | 1.5 | 1.67 (1.13–2.45) | 0.009 |
| EZH2 | 8.5 | 1.84 (1.20–2.82) | 0.004 |
| ER-alpha (Epithelium) | n.a. | ||
| ER-alpha (Stroma) | 0.5 | 1.34 (0.81–2.23) | 0.25 |
| ER-beta (Epithelium) | 1.5 | 1.41(0.73–2.72) | 0.3 |
| ER-beta (Stroma) | 1.5 | 0.67 (0.27–1.66) | 0.39 |
| HDAC2 | 8.5 | 1.35 (0.70–2.61) | 0.37 |
| HSP27 | 1.5 | 0.73 (0.32–1.66) | 0.45 |
| Ki-67/MIB1 | 6.5% | 2.05 (0.99–4.21) | 0.047 |
| MUC1 | 0.5 | 0.91 (0.62–1.53) | 0.61 |
| N-CADHERIN | 0.5 | 0.63 (0.37–1.06) | 0.078 |
| NFkB | 0.5 | 2.47 (1.08–5.63) | 0.026 |
| p21 | 2.5% | 2.67 (1.48–4.80) | 0.00067 |
| p27 | 2.5 | 0.58 (0.39–0.86) | 0.0064 |
| p53 | 0.5% | 1.33 (0.81–2.18) | 0.26 |
| PSMA | 1.5 | 2.56 (1.66–3.94) | <0.001 |
| SPINK1 | 0.5 | 0.49 (0.18–1.33) | 0.15 |
| Vimentin | 1.5 | 1.83 (0.95–3.53) | 0.066 |
Abbreviations: CI=confidence interval; n.a.=not applicable; PSA=prostate-specific antigen.
Table 3. Univariate and multivariate Cox analysis of PSA relapse-free survival for verified prognostic markers.
| Marker | Univariate hazard ratio (95% CI) | P-value | Multivariate hazard ratio (95% CI) | P-value | n |
|---|---|---|---|---|---|
| ADAM9 | 1.03 (0.69–1.52) | 0.895 | 1.07 (0.70–1.62) | 0.752 | 208 |
| AKT1 | 1.20 (0.78–1.85) | 0.412 | 1.61 (1.01–2.54) | 0.043 | 208 |
| ALCAM | 0.72 (0.46–1.13) | 0.149 | 0.85 (0.52–1.38) | 0.502 | 223 |
| AR (nuclear) | 0.60 (0.37–0.95) | 0.029 | 0.70 (0.43–1.14) | 0.152 | 193 |
| AR (stromal) | 0.30 (0.17–0.53) | <0.001 | 0.32 (0.17–0.59) | < 0.001 | 196 |
| BCL2 | 1.04 (0.66–1.64) | 0.864 | 0.99 (0.62–1.59) | 0.979 | 220 |
| CB1R | 1.70 (1.10–2.62) | 0.017 | 1.45 (0.91–2.31) | 0.114 | 216 |
| CD10 | 1.55 (1.00–2.40) | 0.051 | 1.27 (0.79–2.05) | 0.319 | 205 |
| CD24 | 0.71 (0.44–1.15) | 0.161 | 1.08 (0.64–1.82) | 0.778 | 214 |
| CD138 | 1.20 (0.77–1.88) | 0.417 | 1.08 (0.67–1.75) | 0.741 | 210 |
| CRGA | 2.04 (1.03–4.04) | 0.042 | 1.85 (0.91–3.76) | 0.089 | 222 |
| CRISP3 | 0.76 (0.51–1.14) | 0.188 | 0.71 (0.47–1.09) | 0.118 | 203 |
| E-CADHERIN | 1.56 (1.06–2.29) | 0.023 | 1.34 (0.89–2.02) | 0.160 | 212 |
| EZH2 | 1.27 (0.86–1.87) | 0.229 | 1.48 (0.98–2.23) | 0.010 | 210 |
| ER-alpha (Epithelium) | 2.00 (0.87–4.60) | 0.100 | 1.03 (0.44–2.41) | 0.953 | 195 |
| ER-alpha (Stroma) | 1.34 (0.81–2.23) | 0.256 | 1.44 (0.81–2.57) | 0.218 | 195 |
| ER-beta (Epithelium) | 1.16 (0.74–1.84) | 0.515 | 0.88 (0.54–1.45) | 0.624 | 184 |
| ER-beta (Stroma) | 1.16 (0.67–2.02) | 0.598 | 0.83 (0.45–1.51) | 0.540 | 186 |
| HDAC2 | 1.18 (0.79–1.75) | 0.415 | 0.99 (0.65–1.51) | 0.961 | 205 |
| HSP27 | 0.81 (0.39–1.67) | 0.568 | 0.77 (0.35–1.70) | 0.520 | 212 |
| Ki-67/MIB1 | 1.43 (0.80–2.56) | 0.226 | 1.21 (0.64–2.28) | 0.552 | 223 |
| MUC1 | 1.05 (0.72–1.53) | 0.814 | 0.93 (0.62–1.40) | 0.722 | 224 |
| N-CADHERIN | 0.63 (0.37–1.06) | 0.080 | 0.66 (0.38–1.16) | 0.149 | 219 |
| NFkB | 2.47 (1.08–5.63) | 0.031 | 2.13 (0.76–5.93) | 0.150 | 222 |
| p21 | 1.72 (1.15–2.56) | 0.008 | 0.87 (0.54–1.40) | 0.556 | 214 |
| p27 | 0.58 (0.39–0.86) | 0.007 | 0.73 (0.48–1.11) | 0.137 | 210 |
| p53 | 1.33 (0.81–2.18) | 0.261 | 1.06 (0.63–1.80) | 0.822 | 224 |
| PSMA | 2.56 (1.66–3.94) | < 0.001 | 1.99 (1.24–3.20) | 0.005 | 186 |
| SPINK1 | 0.73 (0.30–1.80) | 0.501 | 0.80 (0.29–2.20) | 0.661 | 220 |
| Vimentin | 1.34 (0.89–2.03) | 0.163 | 1.32 (0.84–2.07) | 0.232 | 209 |
Abbreviations: CI=confidence interval; n.a.=not applicable; PSA=prostate-specific antigen.
Figure 3.
Forest plots for the prognostic markers predicting biochemical PSA relapse in multivariate analysis. Comparison of hazard ratios and 95% confidence intervals for published studies (left) and the Zurich prostate cancer cohort of 238 patients (right). Statistically significant markers are depicted in black, insignificant in red.
To further analyse the prognostic values in the clinically relevant subgroup of tumours with Gleason scores of 6 or 7, another multivariate model was calculated (Table 4). In this tumour subgroup, again AKT1, stromal AR, and PSMA and additionally CB1R, CD10, E-Cadherin, and N-Cadherin were confirmed.
Table 4. Multivariate Cox analysis of PSA relapse-free survival for verified prognostic markers in the subgroup of tumours with Gleason scores <=7.
| Marker | Multivariate hazard ratio (95% CI) | P-value | n |
|---|---|---|---|
| ADAM9 | 1.27 (0.72–2.24) | 0.402 | 154 |
| AKT1 | 1.94 (1.10–3.42) | 0.023 | 151 |
| ALCAM | 1.08 (0.52–2.24) | 0.834 | 163 |
| AR (nuclear) | 0.94 (0.51–1.73) | 0.840 | 145 |
| AR (stromal) | 0.33 (0.16–0.66) | 0.002 | 148 |
| BCL2 | 1.67 (0.92–3.04) | 0.091 | 161 |
| CB1R | 1.96 (1.09–3.52) | 0.025 | 159 |
| CD10 | 1.86 (1.01–3.39) | 0.045 | 149 |
| CD24 | 0.79 (0.43–1.45) | 0.443 | 156 |
| CD138 | 1.10 (0.59–2.04) | 0.770 | 154 |
| CRGA | 2.13 (0.82–5.54) | 0.121 | 163 |
| CRISP3 | 0.66 (0.38–1.16) | 0.150 | 148 |
| E-CADHERIN | 2.42 (1.41–4.15) | 0.001 | 155 |
| EZH2 | 1.58 (0.92–2.70) | 0.096 | 153 |
| ER-alpha (Epithelium) | 1.43 (0.47–4.30) | 0.528 | 143 |
| ER-alpha (Stroma) | 1.33 (0.61–2.90) | 0.479 | 143 |
| ER-beta (Epithelium) | 1.14 (0.57–2.27) | 0.713 | 134 |
| ER-beta (Stroma) | 1.53 (0.74–3.19) | 0.255 | 136 |
| HDAC2 | 1.27 (0.73–2.21) | 0.395 | 153 |
| HSP27 | 0.65 (0.20–2.17) | 0.488 | 157 |
| Ki-67/MIB1 | 1.84 (0.70–4.84) | 0.219 | 164 |
| MUC1 | 1.11 (0.65–1.90) | 0.701 | 164 |
| N-CADHERIN | 0.35 (0.14–0.89) | 0.027 | 161 |
| NFkB | 7.15 (0.97–52.55) | 0.053 | 162 |
| p21 | 1.70 (0.97–2.97) | 0.062 | 158 |
| p27 | 0.84 (0.49–1.45) | 0.536 | 152 |
| p53 | 1.45 (0.66–3.19) | 0.357 | 164 |
| PSMA | 2.47 (1.27–4.79) | 0.008 | 136 |
| SPINK1 | 0.34 (0.05–2.54) | 0.297 | 162 |
| Vimentin | 1.47 (0.80–2.68) | 0.214 | 151 |
Abbreviations: CI=confidence interval; PSA=prostate-specific antigen.
Discussion
The present study is the first to systematically analyse and verify a larger set of proposed prognostic immunohistochemical markers for prostate cancer that have been demonstrated to be multivariate significant predictors for disease progression in previous studies. Although a series of quality criteria needed to be fulfilled to allow study inclusion, we were unable to confirm the majority of prognostic markers selected. Only 10 out of 28 markers (35%) could be verified in a univariate Cox approach and only 4 (14%) were subsequently confirmed as significant in a multivariate setting. According to these results, the loss of stromal androgen receptor expression was the biomarker with the most significant prognostic impact (hazard ratio 3.33); a finding that doubtlessly deserves further investigation. This rather remarkable discrepancy of published data and this study's results might be explained by either significant shortcomings of this verification study or general deficits in the publication culture concerning prognostic markers that may be either over-optimistic or too permissive.
Concerning the former, great care was taken to minimise potential biases in the conduction of this study. The study cohort, which comprised 238 primary prostate cancer patients, is a well established, contemporary, monocentric prostatectomy cohort which has been extensively used in previous publications (Tischler et al, 2010; Gerhardt et al, 2011; Mortezavi et al, 2011; Beer et al, 2012; Gerhardt et al, 2012). Although the number of patients may appear low at first sight, we want to point out, that it already exceeds the median cohort size of the studies re-analysed. Further, the application of study cases compiled as TMA does not only allow for a convenient high-throughput workflow but also serves as a model of biopsy-detected prostate cancer. In the present setup, each index tumour is represented by a single core. Even though this introduces a potential sampling bias and may compromise the analysis of highly heterogeneously expressed proteins, it also increases the likelihood to identify robust clinical prognostic markers, which ought to be detectable in a single core of a tumour-positive biopsy. Protocols for immunohistochemistry were adapted as closely to the original protocols as possible, and immunohistochemical staining was performed on standardised automated platforms that are also used for routine diagnostics at the Institute of Surgical Pathology of the University Hospital Zurich. To minimise interobserver variability, a single observer evaluated all slides. Our attitude towards statistical marker verification was deliberately benevolent, allowing for optimisation of cut-off values. Even when simplifying the multivariate model, excluding margins and patient age, the number of significantly prognostic biomarkers did not increase (data not shown). However, a caveat to the validity of the current study is that Ki-67, which is assumedly the best verified prognostic marker in prostate cancer, only showed a univariate significance (Kristiansen, 2012; Fisher et al, 2013). Admittedly, the composition of our study cohort may lead to a bias, with cases enriched for high-grade and high-stage tumours, leading to a relapse rate of 46%, which is approximately 15 percentage points above the usual rates of consecutive RPE cohorts. However, this acknowledged bias should be expected to ease the verification of biomarker candidates and is therefore considered useful in this studies' setting.
The most apparent deficit of the original studies lies in their study design, which was generally based on a retrospective analysis of a single cohort. This type of analysis is prone to model overfitting and is nowadays increasingly regarded as only hypothesis-generating because the important validation is missing. A more appropriate approach includes an initial analysis of a training cohort to establish a test hypothesis and a cut-off value, which is then verified in an independent testing cohort. In a way, this study provides the necessary testing cohort for these studies, even though under considerably tightened conditions with probable differences in surgery, in the pre-analytic steps, the immunostaining protocols, and the analytical attitude of the observers in the interpretation of immunostainings. The impact of correct statistical handling in biomarker studies has long been recognised and has led to several recommendations (Harris, 2005; McShane et al, 2005b), most importantly, the REMARK (‘REporting recommendations for tumor MARKer prognostic studies') guidelines (Simon and Altman, 1994; Hayes et al, 1996; Altman, 1998). These provide a comprehensive description of quality criteria for prognostic biomarker studies but are still not widely accepted, as Mallett et al (2010) recently demonstrated in a meta-analysis; an observation we can only confirm.
An additional potential bias lies in the composition of the respective cohort under analysis, which may also influence biomarker performance (Braun et al, 2011). It remains unclear whether a biomarker that performs well in clinically detected populations shows similar performance in a contemporary PSA screen-detected cohort. Also, a possible influence of patient ethnicity cannot be excluded. Last but not least, the evolution of Gleason grading in the last decade (ISUP 2005) may introduce a significant bias and it is unclear which type of Gleason grading the individual studies were based on.
A general deficit of most TMA-based studies is the missing demonstration of relevant construction bias. Ideally, a new TMA should be verified with a panel of established markers prior to the analysis of new candidates, and this confirmative data should be included in the Supplementary Material of the first publication. We further suggest publicising the immunohistochemistry raw data sets in order to allow re-analyses and insightful meta-analytic studies. This would allow for proper post-publication data review and would prospectively increase the quality of published papers. Even though high-quality journals demand the deposition of genomic data in centralised repositories, there is no widely acknowledged infrastructure for immunohistochemistry data. This may be partly due to the widespread notion of immunohistochemistry being a somewhat arbitrary technique yielding semiquantitative results. However, the growing number of immunohistochemistry-based biomarker studies indicates the necessity for better tools to increase their quality and to allow for a better comparison of the published data.
The main restriction of immunohistochemistry as a technique is missing normalisation. Many factors influence the final signal intensity, considering differences in pre-analytic steps, section thickness, antigen retrieval techniques, antibody quality and concentration, and detection systems. To our knowledge, the approach of normalising an immunohistochemical staining signal by simultaneous measurement of a ‘housekeeping protein' has not been pursued widely and is a field that needs to be developed in the future. Certainly, this would necessitate computer-aided image analysis, which, taken by itself, has not been a major breakthrough so far (Rizzardi et al, 2012).
The search for molecular prognostic markers for malignant tumours has been a central aim of biomedical researchers in the last two decades. A more concise diagnosis provided by a molecular marker may facilitate individualised patient treatment. Additionally, prognostic markers might help to unravel the molecular background of tumour progression and even represent an attractive new therapeutic target. Ideally, a prognostic factor only has a few degrees of freedom and allows for a dichotomous or trichotomous readout (e.g., negative/positive or nil/low/high) to ensure a high degree of reproducibility. Unfortunately, this does not exactly hold true for the majority of biomarkers measured on expression level, which necessitate cut-off values to delineate meaningful prognostic subgroups. As these cut-off values are mainly arbitrarily chosen, and the most popular platform to determine protein expression in tumour tissues is immunohistochemistry, it does not surprise that the reproducibility of these prognostic markers is limited (Altman et al, 1994). Promising future exceptions could be genetic events of prognostic value, which may be detected with mutation-specific antibodies. Although it is difficult to overcome the inherent obstacles of contemporary immunohistochemistry, other biological platforms may offer a more robust alternative. As an example, several mRNA expression signatures have been proposed to predict disease progression and are currently undergoing intense verification (Cuzick et al, 2011; Karnes et al, 2013; Wu et al, 2013).
Although most authors of prognostic biomarker studies in prostate cancer investigate markers for therapy planning at the biopsy stage, these studies, as exemplified in our selection, mainly use RPE specimens (and not preoperatively sampled biopsy material) and analyse disease progression following radical prostatectomy. The inherent but to our knowledge unproven assumption is that markers of disease progression following surgery can also be used upfront to estimate the tumours' endogenous aggressiveness; or, in other words, that predictive markers are also prognostic markers. The distinction of prognostic markers that estimate the natural course of disease and predictive markers that estimate the response to therapy is still often ignored. This aspect should be considered carefully, for surgery might indeed heal and not only ameliorate the disease. As the TMPRSS2-ERG translocation illustrates, a biomarker may well allow prognostication in untreated patients (Demichelis et al, 2007; Attard et al, 2008) but can still fail to predict progression following therapy (Minner et al, 2011; Pettersson et al, 2012). Other markers may work in both settings, as demonstrated by the cell cycle proliferation signature, proposed by Cuzick et al (2011).
We are convinced that the final and most crucial step in verification of a biomarker for therapy planning at the initial biopsy stage would be a prospective trial in an active surveillance cohort. A reliable biomarker should then be able to identify insignificant tumours that can safely be kept under surveillance for a longer time and do not necessitate active treatment because of the criteria of tumour progression. Even then, long-term follow-up data with either cancer-specific death or onset of castration refractory disease as an endpoint would be highly desirable. Another open point of discussion is, if the commonly used surrogate marker of disease progression, PSA relapse, is delineating a clinically meaningful endpoint, because many patients with a PSA progression will die of other, non-cancer related causes (Attard and de Bono, 2009). To complicate the matter, the definition of PSA progression also varies between studies (Nielsen and Partin, 2007). It also has to be kept in mind, that it is highly unlikely that molecular prognostic biomarkers solely can supersede the clinico-pathological parameters that build the basis of commonly used nomograms, but are only able to add prognostic information to these. This is in principle good news for histopathologists who should strengthen their efforts to provide even more standardised reports in the future, irrespective of molecular developments.
In summary, this study sheds some very critical light on contemporary immunohistochemistry studies that aim to identify prognostic biomarkers for prostate cancer. Acknowledging the inherent limitations of this comprehensive meta-analysis and verification study, the majority of published biomarkers could not be confirmed. This is disappointing but in excellent concordance with the skeptical view of biostatisticians and may also be true for other tumour entities (Ioannidis, 2005, 2013). We feel that in addition to the suggestions made above, the REMARK guidelines, which summarise important cornerstones of biomarker investigations, clearly deserve a wider reception, better acknowledgment, and stricter adherence in order to increase the quality of published data in the future (McShane et al, 2005a).
Acknowledgments
We thank Silvia Behnke (immunohistochemistry) and Martina Storz (TMA facility) for truly excellent technical support.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Altman DG. Suboptimal analysis using 'optimal' cutpoints. Br J Cancer. 1998;78 (4:556–557. doi: 10.1038/bjc.1998.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman DG, Lausen B, Sauerbrei W, Schumacher M. Dangers of using "optimal" cutpoints in the evaluation of prognostic factors. J Natl Cancer Inst. 1994;86 (11:829–835. doi: 10.1093/jnci/86.11.829. [DOI] [PubMed] [Google Scholar]
- Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, Berney D, Foster CS, Fletcher A, Gerald WL, Moller H, Reuter V, De Bono JS, Scardino P, Cuzick J, Cooper CS. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27 (3:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attard G, de Bono JS. Prostate cancer: PSA as an intermediate end point in clinical trials. Nat Rev Urol. 2009;6 (9:473–475. doi: 10.1038/nrurol.2009.130. [DOI] [PubMed] [Google Scholar]
- Beer M, Montani M, Gerhardt J, Wild PJ, Hany TF, Hermanns T, Muntener M, Kristiansen G. Profiling gastrin-releasing peptide receptor in prostate tissues: clinical implications and molecular correlates. Prostate. 2012;72 (3:318–325. doi: 10.1002/pros.21434. [DOI] [PubMed] [Google Scholar]
- Braun M, Scheble VJ, Menon R, Scharf G, Wilbertz T, Petersen K, Beschorner C, Reischl M, Kuefer R, Schilling D, Stenzl A, Kristiansen G, Rubin MA, Fend F, Perner S. Relevance of cohort design for studying the frequency of the ERG rearrangement in prostate cancer. Histopathology. 2011;58 (7:1028–1036. doi: 10.1111/j.1365-2559.2011.03862.x. [DOI] [PubMed] [Google Scholar]
- Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S, Denkert C. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7 (12:e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, Mesher D, Speights VO, Stankiewicz E, Foster CS, Moller H, Scardino P, Warren JD, Park J, Younus A, Flake DD, 2nd, Wagner S, Gutin A, Lanchbury JS, Stone S. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12 (3:245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, Hoshida Y, Mosquera JM, Pawitan Y, Lee C, Adami HO, Mucci LA, Kantoff PW, Andersson SO, Chinnaiyan AM, Johansson JE, Rubin MA. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26 (31:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- Fisher G, Yang ZH, Kudahetti S, Moller H, Scardino P, Cuzick J, Berney DM. Prognostic value of Ki-67 for prostate cancer death in a conservatively managed cohort. Br J Cancer. 2013;108 (2:271–277. doi: 10.1038/bjc.2012.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt J, Montani M, Wild P, Beer M, Huber F, Hermanns T, Muntener M, Kristiansen G. FOXA1 promotes tumor progression in prostate cancer and represents a novel hallmark of castration-resistant prostate cancer. Am J Pathol. 2012;180 (2:848–861. doi: 10.1016/j.ajpath.2011.10.021. [DOI] [PubMed] [Google Scholar]
- Gerhardt J, Steinbrech C, Buchi O, Behnke S, Bohnert A, Fritzsche F, Liewen H, Stenner F, Wild P, Hermanns T, Muntener M, Dietel M, Jung K, Stephan C, Kristiansen G. The androgen-regulated Calcium-Activated Nucleotidase 1 (CANT1) is commonly overexpressed in prostate cancer and is tumor-biologically relevant in vitro. Am J Pathol. 2011;178 (4:1847–1860. doi: 10.1016/j.ajpath.2010.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan SR, Cooperberg MR, Elkin E, Lubeck DP, Meng M, Mehta SS, Carroll PR. Time trends and characteristics of men choosing watchful waiting for initial treatment of localized prostate cancer: results from CaPSURE. J Urol. 2003;170 (5:1804–1807. doi: 10.1097/01.ju.0000091641.34674.11. [DOI] [PubMed] [Google Scholar]
- Harris AL. REporting recommendations for tumour MARKer prognostic studies (REMARK) Br J Cancer. 2005;93 (4:385–386. doi: 10.1038/sj.bjc.6602730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes DF, Bast RC, Desch CE, Fritsche H, Jr., Kemeny NE, Jessup JM, Locker GY, Macdonald JS, Mennel RG, Norton L, Ravdin P, Taube S, Winn RJ. Tumor marker utility grading system: a framework to evaluate clinical utility of tumor markers. J Natl Cancer Inst. 1996;88 (20:1456–1466. doi: 10.1093/jnci/88.20.1456. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2 (8:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP. Biomarker failures. Clin Chem. 2013;59 (1:202–204. doi: 10.1373/clinchem.2012.185801. [DOI] [PubMed] [Google Scholar]
- Karnes RJ, Bergstralh EJ, Davicioni E, Ghadessi M, Buerki C, Mitra AP, Crisan A, Erho N, Vergara IA, Lam LL, Carlson R, Thompson DJ, Haddad Z, Zimmermann B, Sierocinski T, Triche TJ, Kollmeyer T, Ballman KV, Black PC, Klee GG, Jenkins RB. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190 (6:2047–2053. doi: 10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen G. Diagnostic and prognostic molecular biomarkers for prostate cancer. Histopathology. 2012;60 (1:125–141. doi: 10.1111/j.1365-2559.2011.04083.x. [DOI] [PubMed] [Google Scholar]
- Mallett S, Timmer A, Sauerbrei W, Altman DG. Reporting of prognostic studies of tumour markers: a review of published articles in relation to REMARK guidelines. Br J Cancer. 2010;102 (1:173–180. doi: 10.1038/sj.bjc.6605462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumor MARKer prognostic studies (REMARK) Nat Clin Pract Urol. 2005;2 (8:416–422. [PubMed] [Google Scholar]
- McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97 (16:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- Minner S, Enodien M, Sirma H, Luebke AM, Krohn A, Mayer PS, Simon R, Tennstedt P, Muller J, Scholz L, Brase JC, Liu AY, Schluter H, Pantel K, Schumacher U, Bokemeyer C, Steuber T, Graefen M, Sauter G, Schlomm T. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin Cancer Res. 2011;17 (18:5878–5888. doi: 10.1158/1078-0432.CCR-11-1251. [DOI] [PubMed] [Google Scholar]
- Mortezavi A, Hermanns T, Seifert HH, Baumgartner MK, Provenzano M, Sulser T, Burger M, Montani M, Ikenberg K, Hofstadter F, Hartmann A, Jaggi R, Moch H, Kristiansen G, Wild PJ. KPNA2 expression is an independent adverse predictor of biochemical recurrence after radical prostatectomy. Clin Cancer Res. 2011;17 (5:1111–1121. doi: 10.1158/1078-0432.CCR-10-0081. [DOI] [PubMed] [Google Scholar]
- Nielsen ME, Partin AW. The impact of definitions of failure on the interpretation of biochemical recurrence following treatment of clinically localized prostate cancer. Rev Urol. 2007;9 (2:57–62. [PMC free article] [PubMed] [Google Scholar]
- Pettersson A, Graff RE, Bauer SR, Pitt MJ, Lis RT, Stack EC, Martin NE, Kunz L, Penney KL, Ligon AH, Suppan C, Flavin R, Sesso HD, Rider JR, Sweeney C, Stampfer MJ, Fiorentino M, Kantoff PW, Sanda MG, Giovannucci EL, Ding EL, Loda M, Mucci LA. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21 (9:1497–1509. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzardi AE, Johnson AT, Vogel RI, Pambuccian SE, Henriksen J, Skubitz AP, Metzger GJ, Schmechel SC. Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagnostic pathology. 2012;7:42. doi: 10.1186/1746-1596-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Altman DG. Statistical aspects of prognostic factor studies in oncology. Br J Cancer. 1994;69 (6:979–985. doi: 10.1038/bjc.1994.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler V, Fritzsche FR, Wild PJ, Stephan C, Seifert HH, Riener MO, Hermanns T, Mortezavi A, Gerhardt J, Schraml P, Jung K, Moch H, Soltermann A, Kristiansen G. Periostin is up-regulated in high grade and high stage prostate cancer. BMC Cancer. 2010;10:273. doi: 10.1186/1471-2407-10-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CL, Schroeder BE, Ma XJ, Cutie CJ, Wu S, Salunga R, Zhang Y, Kattan MW, Schnabel CA, Erlander MG, McDougal WS. Development and validation of a 32-gene prognostic index for prostate cancer progression. Proc Natl Acad Sci USA. 2013;110 (15:6121–6126. doi: 10.1073/pnas.1215870110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.