Abstract
Transcriptional deregulation plays a key role in a large array of cancers, and successful targeting of oncogenic transcription factors that sustain diseases has been a holy grail in the field. Acute promyelocytic leukaemia (APL) driven by chimeric transcription factors encoding retinoic acid receptor alpha fusions is the paradigm of targeted cancer therapy, in which the application of all-trans retinoic acid (ATRA) treatments have markedly transformed this highly fatal cancer to a highly manageable disease. The extremely high complete remission rate resulted from targeted therapies using ATRA in combination with arsenic trioxide will likely be able to minimise or even totally eliminate the use of highly toxic chemotherapeutic agents in APL. In this article, we will review the molecular basis and the upcoming challenges of these targeted therapies in APL, and discuss the recent advance in our understanding of epigenetics underlying ATRA response and their potential use to further improve treatment response and overcome resistance.
Keywords: epigenetics, acute promyelocytic leukaemia pathogenesis, histone methylation, PHF8, histone demethylase, ATRA, target therapy
From the initial discovery of the recurring chromosomal translocation encoding chimaeric PML–retinoic acid receptor alpha (RARalpha) fusion to the successful application of all-trans retinoic acid (ATRA) treatment for induction of complete remission, acute promyelocytic leukaemia (APL), which accounts for ∼10% of all cases of acute myeloid leukaemias (AMLs), has become the paradigm of differentiation therapy and one of the most successfully targeted cancers. ATRA plus anthracycline-based chemotherapy that has been the standard regimen for APL therapy in the last decades can achieve long-term remissions close to 80% (Sanz and Lo-Coco, 2011). The revolutionary development of ATRA was closely followed by another major discovery of arsenic trioxide (ATO) treatment that has markedly improved the management of relapsed and refractory APL patients, and now rapidly enters therapy regimens for newly diagnosed low-to-intermediate-risk APL patients in combination with ATRA (Breccia and Lo-Coco, 2012; Lo-Coco et al, 2013). ATRA/ATO with reduced haematological toxicity compared with anthracycline-based regimens is likely to further improve long-term outcomes for APL patients (Shen et al, 2004; Wang and Chen, 2008). Thus, hope is arising that chemotherapy in APL may become largely dispensable and be solely replaced by targeted therapeutic approaches. In spite of these successes, the role of ATRA/ATO for high-risk patients has not yet been clarified. Also relapsed and refractory APL patients, due to ATRA and/or ATO resistance, still occur among all prognostic subgroups of APL patients, and remain a clinically significant problem in the field (Goto et al, 2011; Fung and So, 2013; Tomita et al, 2013). An improved understanding of the mechanisms underlying the oncogenic transformation and treatment response in APL is urgently needed for development of better therapeutic strategies. In this article, we will review the molecular basis, in particular, the transcriptional and epigenetic machineries that are critically involved in APL development, as well as their roles in mediating response and resistance to ATRA treatments. Although the epigenetic regulation has a key role in mediating ATRA response/resistance, it is unlikely the only answer to all APL therapies such as ATO treatment. Since the role of ATRA/ATO-mediated onco-fusion degradation in disease remission has been extensively reviewed elsewhere (Ablain and de The, 2011; Lallemand-Breitenbach et al, 2012), the current review will mainly focus on the epigenetic aspects, and its potential future development in APL treatment.
Epigenetic in APL
Although transcriptional deregulation has a central role in a large array of cancers including acute leukaemia, it is evident that epigenetic machineries including DNA methylation and post-translational histone modifications constitute integral functions of the oncogenic transcriptional complexes in mediating the aberrant transcriptional programmes (Cheung and So, 2011). Consistently, we and others have revealed that RARalpha fusions form high-order homotetramers (Lin and Evans, 2000; Minucci et al, 2000; Kwok et al, 2006; Sternsdorf et al, 2006) that aberrantly recruit the DNA-binding cofactor, RXRalpha (Zeisig et al, 2007; Zhu et al, 2007), as well as epigenetic-modifying enzymes such as histone deacetylases (HDACs) (Grignani et al, 1998; Lin et al, 1998), DNA methyltransferases (DNMTs) (Di Croce et al, 2002), SUV39H1 (Carbone et al, 2006), and polycomb repressive complexes (PRCs) 1 and 2 (Villa et al, 2007; Boukarabila et al, 2009; Smith et al, 2011) to suppress expression of downstream targets critical for differentiation and tumour suppression (Figure 1A).
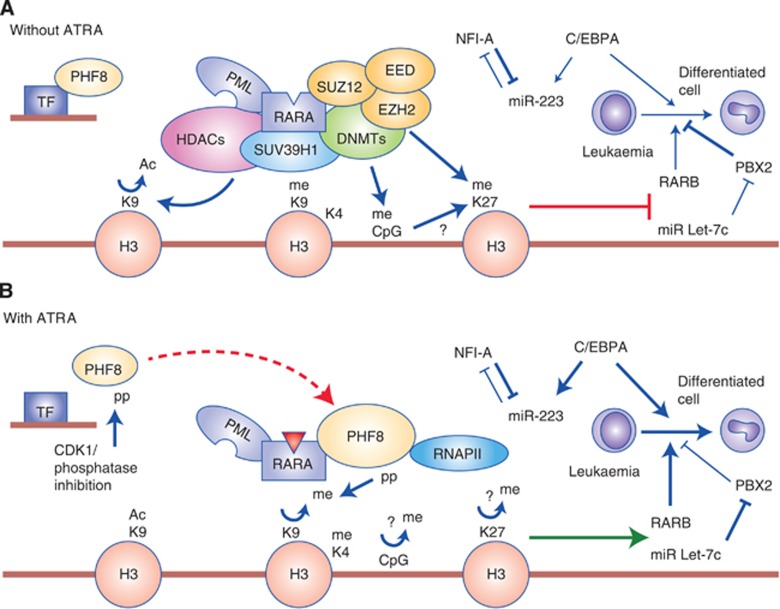
Figure 1.
Epigenetic functions of PML–RARalpha in APL pathogenesis and treatment response. (A) In APL cells, homotetrameric PML–RARalpha (for simplicity, the homotetramer is not illustrated in the figure) recruits multiple repressive epigenetic modifiers including DNMTs (green), PRC complexes (yellow), lysine methyltransferases (blue), and the histone lysine deactylase complex (pink). In the absence of ATRA, PHF8 is hypo-phosphorylated and associates with transcription factors (TFs) to binds to different chromatin regions (naive promoters). (B) Upon ATRA treatment, ATRA (red triangle) binds to RARalpha moiety and induces conformational changes that allow dissociation of the co-repressor complex. On the other hand, PHF8 can be phosphorylated by CDK1. It dissociates from its naive chromatin-binding sites and is recruited by RARalpha fusions (illustrated by a big dashed red arrow). PHF8 removes the repressive H3K9me2 mark, but promotes active histone marks (H3K4 hypermethylation and K9 hyperacetylation) and recruitment of RNA polymerase II to turn on gene expression. Genes/proteins to be activated are bolded. Normal arrow indicates activation; blunted arrow indicates suppression; dashed arrow represents translocation of PHF8 protein; thickened arrows represent enhanced processes. ‘me' is hypermethylation; ‘Ac' is hyperacetylation; ‘p' is phosphorylation and ‘?' indicates the mechanism remains unclear.
On the other hand, epigenetic mechanisms driving APL leukaemogenesis also include the emerging new epigenetic factor, miRNAs (micro RNAs) such as Let-7c (Saumet et al, 2009), which is upregulated to promote granulocytic differentiation of APL cells in part by suppressing PBX2 upon ATRA treatment (Pelosi et al, 2012; Figure 1B). miRNA-223 regulated by C/EBPalpha and NFI-A is also activated upon ATRA for granulocytic differentiation of APL cells (Fazi et al, 2005; Figure 1B). In addition, upregulation of miRNA-125b has been shown to contribute to paediatric APL and is associated with increased drug resistance (Zhang et al, 2011). Although abnormal expression of miRNAs in APL may provide extended diagnostic options and potential targets for molecular therapy, development of the pharmacological mean for targeting specific miRNAs is still in an early experimental stage. At the same time, the highly tractable epigenetic-modifying enzymes that are critical for pathogenesis and treatment response will likely present potential targets for the next wave of targeted therapies.
DNA methylation in APL
DNA methylation is a well-characterised epigenetic process regulated by DNMTs that transfer methyl groups to cytosine in cytosine–guanine dinucleotide (CpG) islands commonly found in promoter regions. DNA methylation stably regulates cellular gene expression and mostly associates with gene suppression. Given its critical functions in gene regulation, it is not surprising that DNA methylation has a crucial role in the development of various cancers including AML (Baylin and Jones, 2011; Schoofs and Muller-Tidow, 2011). The first hint of DNA methylation involvement in APL came from the studies by Di Croce et al (2002), who demonstrated that PML–RARalpha directly interacted with DNMTs (e.g., DNMT1 and DNMT3a) leading to hypermethylation and subsequent silencing of downstream targets, such as RARbeta crucial for haematopoietic differentiation (Figure 1A). Consistently, APL patients are characterised by a specific DNA methylation pattern that is distinctive from other AML subtypes (Figueroa et al, 2010). More importantly, overexpression of the DNMT3a collaborates with PML–RARalpha to promote APL leukaemogenesis in vivo (Subramanyam et al, 2010), and aberrant DNA methylation in p15 and p16 genes has negative prognostic impact in APL patients (Chim et al, 2001; Teofili et al, 2003), suggesting a critical role of aberrant DNA methylation in APL pathogenesis.
However, recent global epigenetic analyses revealed that ATRA treatment induced major changes in post-translational modifications such as histone acetylation, but not DNA methylation in APL cells (Martens et al, 2010; Mikesch et al, 2010). Although DNA hypermethylation in APL cells occurred frequently at genomic regions regulated by PRC2, such as SUZ12- and REST-binding sites in embryonic stem cells (Schoofs et al, 2013), no major difference in the DNA methylation signature at PML–RARalpha DNA-binding sites or its vicinities was found in comparison with controls (Martens et al, 2010; Wang et al, 2010; Schoofs et al, 2013). These studies suggest that the change of DNA methylation patterns may be relatively late events in APL leukaemogenesis, and contribute to APL maintenance rather than to leukaemia initiation (Schoofs et al, 2013). Nevertheless, the DNA demethylating agent decitabine can induce apoptosis of APL cells in vitro via activation of the TRAIL pathway (Soncini et al, 2013). Thus, although it may mainly target the late collaborative events, reversal of the aberrant DNA methylation status appears to be an attractive therapeutic approach, in particular, when combined with ATRA/ATO for APL treatment.
A key role of histone acetylation in APL pathogenesis and ATRA response
Compared with DNA methylation, aberrant DNA binding and histone modifications including acetylation and methylation may have had even more important roles in APL pathogenesis and treatment response. Global epigenetic studies have shown that, although wild-type RARalpha binding is mostly restricted to canonical retinoic acid response element (RARE)-binding sites, around 30% of the PML–RARalpha-binding sites are atypical RARE motifs (Martens et al, 2010; Wang et al, 2010), which are probably attributed by the homo/hetero-oligomeric nature of the fusions (Zeisig et al, 2007; Zhu et al, 2007). More importantly, these studies also revealed that the vast majority of PML–RARalpha-binding sites, associated with distinct histone modifications characterised by low H3-acetylation (H3ac), reduced H3K27 trimethylation (H3K27me3) and increased H3K9me3 (Hoemme et al, 2008; Martens et al, 2010). Strikingly, ATRA treatment induced a significant increase of H3ac in about 80% of the PML–RARalpha-binding sites, including genes that are crucial for haematopoietic differentiation (Figure 1B). On contrast, H3K27me3 and H3K9me3 levels remained largely unchanged (Martens et al, 2010). CpG DNA methylation, which has been shown to crosstalk with H3K27me3, also did not show significant changes upon ATRA treatment either. Although these studies suggest histone acetylation rather than H3K27me3 to be critical for ATRA response in APL, it is noted that a small number of PML–RARalpha-binding sites do correlate with the changes of H3K27me3 and DNA methylation upon ATRA treatment (Martens et al, 2010). Future functional validation is needed to further investigate the importance of these changes and to define the major epigenetic determinants for the ATRA treatment response (Mikesch et al, 2010).
The role of histone modification in APL pathogenesis is also highlighted by the study of a variant APL fusion, PLZF–RARalpha, which has a higher binding ability to SMRT/NcoR/HDAC (Grignani et al, 1998; Lin et al, 1998), and is in general more resistant to ATRA treatment (Zelent et al, 2001). Interestingly, although recent genome-wide studies revealed overlapping targets by PLZF–RARalpha and PML–RARalpha (Rice et al, 2009; Spicuglia et al, 2011), PLZF–RARalpha significantly increased H3K27me3 and concomitantly decreased H3K9K14ac at targeted sites (Spicuglia et al, 2011). Thus, different histone codes may account for the differences in ATRA response, and can potentially be exploited to improve treatment response.
Histone demethylase PHF8 as a crucial co-activator governing ATRA response
Although the aberrant recruitment of transcriptional co-repressor complexes by RARalpha is critical for APL pathogenesis, it becomes evident that the recruitment of opposing activator complexes upon ATRA treatment has a central role in mediating treatment response (Mikesch et al, 2010). Identification of these missing ATRA-responsive co-activators may hold a key to improve not only our understanding of the biology of the disease, but also development of more effective therapeutic strategies. Very recently, a histone demethylase and a member of the plant homeodomain finger (PHF) family, PHF8 has been identified as a key co-activator specially recruited by RARalpha fusions to mediate ATRA response in APL (Arteaga et al, 2013). PHF8 contains a N-terminal plant homeodomain as well as an active JmjC domain, which is able to recognise H3K4me3 mark and mediates lysine demethylation, respectively (Feng et al, 2010; Fortschegger et al, 2010; Kleine-Kohlbrecher et al, 2010; Liu et al, 2010; Loenarz et al, 2010; Qi et al, 2010). In the presence of ATRA, PHF8 is specifically recruited by RARalpha fusions to activate expression of their downstream target genes (Arteaga et al, 2013). Recruitment of PHF8 leads to a reduction of the H3K9me2-repressive mark and an increase in the H3K4me3 and H3K9Ac activation marks for active gene expression (Figure 1B). Consistently, PHF8 expression is downregulated in ATRA-resistant human APL cells. Forced expression of PHF8 resurrects ATRA sensitivity, whereas its suppression leads to resistance (Arteaga et al, 2013). PHF8 binds to promoter regions of genes involved in cell cycle progression and dissociates from these promoters upon phosphorylation of S33/S84 residues by CDK1 (Liu et al, 2010; Figure 1A). ATRA can induce nuclear translocation of CDK1 that promotes PHF8 phosphorylation and binding to the promoter regions of RARalpha-fusion target genes for epigenetic reprogramming (Arteaga et al, 2013). Consistently, expression of hyperphosphorylated PHF8 alone can activate expression of PML–RARalpha downstream targets and suppress transformation of ATRA-resistant APL cells (Figure 1B). Together, these studies reveal PHF8 as a key molecular sensor that governs ATRA response in APL. More importantly, these functions of PHF8 critically depend on both its enzymatic activity and phosphorylation status, which can potentially be therapeutically exploited (Arteaga et al, 2013).
Targeting epigenetic machinery to overcome ATRA resistance in APL
Resistance to ATRA still occurs in a proportion of APL patients and remains a clinically significant problem in APL therapy (Fung and So, 2013) Thus, uncovering the underlying mechanisms and designing specific therapeutic strategies to overcome ATRA resistance is of a high priority in the field. Point mutations within the PML–RARalpha ligand-binding domain (LBD) have been found and account for ∼40% of ATRA-resistant APL (Gallagher et al, 2012). Given the central role of epigenetic reprogramming underlying ATRA response, another major resistant mechanism, which may also apply to the variant PLZF–RARalpha fusion, is due to the formation of aberrant repression complexes that cannot be easily dissociated by ATRA treatment (McNamara et al, 2008).
To improve the potency of ATRA response, a significant amount of effort has been made to develop synthetic retinoids with a higher RARalpha-binding affinity (Figure 2). However, most of the derivatives were not able to definitively overcome ATRA resistance or were associated with significant toxicity to the patients (Petrie et al, 2007). As recruitment of HDACs by RARalpha fusions has a major role in oncogenic transformation in APL, the idea of using HDAC inhibitors (HDACi) to overcome ATRA resistance has gained significant attention in the past few years (Figure 2). In fact, re-sensitization of ATRA-resistant APL cells can be achieved by HDACi (He et al, 2001; Petrie et al, 2007; Botrugno et al, 2009). Valproic acid (VPA) alone has been shown to induce differentiation of APL blasts and transient remission in mouse models (Leiva et al, 2012). In spite of these, HDACi seem to be effective only for a small subset of ATRA-resistant PML–RARalpha mutants carrying mutations affecting helix 12, but not helix 5/6 of the LBD in the fusion (Cote et al, 2002), and may upregulate multidrug resistance 1 (MDR1) gene expression resulting in drug resistance (Tabe et al, 2006). Moreover, HDACs can have a dual tumour suppressor and oncogenic role, depending on the stage of the disease. Although suppression of HDAC1 prolonged disease latency in APL mouse models, inhibition of HDAC1 (and HDAC2) would accelerate APL pathogenesis in the early transformation stage owing to its effect on the expansion of a subset of APL cells with leukaemia-initiating features (Santoro et al, 2013). In fact, VPA treatment in an APL mouse model failed to target leukaemia-initiating cells and only resulted in transient responses (Leiva et al, 2012). Thus, overcoming these limitations is required to fully exploit the therapeutic potentials of HDACi in APL therapy in the future.
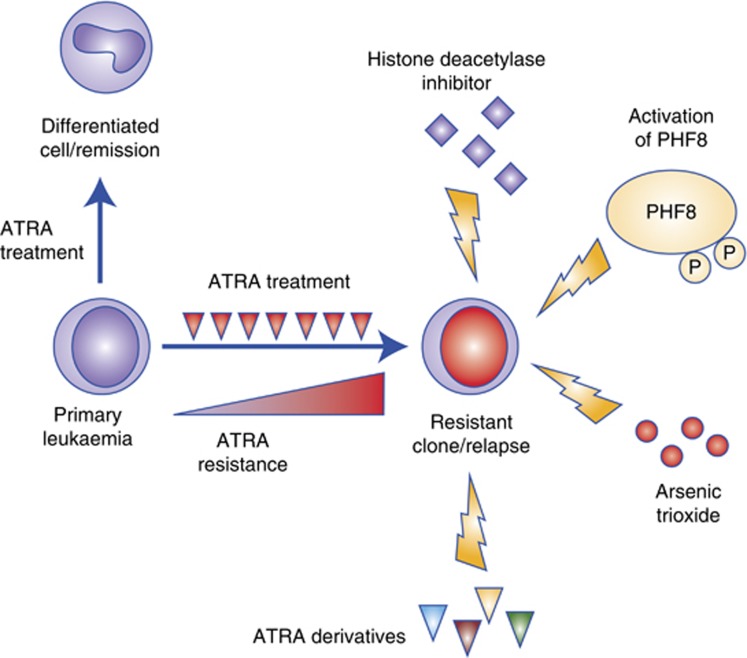
Figure 2.
Targeting ATRA-resistant APL. In most of the cases, ATRA treatment induces degradation of RARalpha fusion and APL cell differentiation, resulting in complete remission. However, some APL patients are refractory to ATRA, or in some cases ATRA treatment may select/evolve drug-resistant clones that are no longer responsive to ATRA. Several approaches have been proposed to target ATRA-resistant APL. These include (1) retinoid derivatives with a higher affinity to the fusions; (2) ATO that binds to the PML moiety of the fusion and subsequently induces degradation of the onco-fusion; (3) HDACi that facilitate histone acetylation; and (4) overexpression or hyperphosphorylation of PHF8 that removes the repressive H3K9me2 mark to turn on the differentiation transcriptional programme.
The recent discovery of PHF8 as a molecular sensor that regulates ATRA response in APL has suggested a novel avenue for overcoming ATRA resistance (Figure 2). PHF8 is capable of resurrecting ATRA sensitivity in resistant APL cells. The critical regulation of PHF8 activity by specific phosphorylation has provided an alternative pharmacological approach for overcoming ATRA resistance in APL cells. Indeed, inhibition of PHF8 dephosphorylation by the phosphatase inhibitor Okadaic acid has been shown to sensitise ATRA-resistant human APL cells to the treatment in vitro and in vivo, resulting in a significant extension of disease latency in xenograft models (Arteaga et al, 2013). Moreover, PHF8 exhibits a broad range of in vitro and in vivo activities against ATRA resistance due to LBD mutations or formation of aberrant transcriptional repression complexes. In contrast to ATRA resistance, ATO resistance associates with mutations affecting the PML moiety that disrupt ATO-mediated degradation of the onco-fusion (Goto et al, 2011). Given the nature of PHF8 binding to the RARalpha moiety of the fusions, it is tempting to speculate its potential usefulness in overcoming ATO resistance (Fung and So, 2013). Thus, future identification of specific phosphatases inhibitors or the pharmacological activator of PHF8 may represent an alternative avenue for targeting treatment resistance in APL.
Outlook
ATRA- and ATO-containing regimens have significantly improved the overall and disease-free survival of de novo and relapsed APL patients (Figure 2). The recent results of a phase III clinical trial using ATRA plus ATO as the first-line therapy for low-to-intermediate-risk APL patients show improved 2 years disease-free and overall survival with a significant reduction of haematologic toxicity and lower rates of infections, revealing the promise of curing APL patients without administration of chemotherapy (Chen and Chen, 2013; Lo-Coco et al, 2013). However, their role in high-risk patients has yet to be determined. Other challenging issues, such as the reduction of early haemorrhagic death rate in APL therapy still remains. Also, some patients, especially those >60 years of age are not eligible for aggressive therapy regimens, and for younger patients, late toxicity of chemotherapy can induce serious complications. Finally, relapsed and refractory APL patients due to ATRA and/or ATO resistance can be a major challenge for these targeted APL therapies (Fung and So, 2013). While it becomes clear that the largely reversible nature of epigenetic modifications provides an unprecedented opportunity to target oncogenic transcription factors frequently mutated in acute leukaemia (Cheung and So, 2011; Zeisig et al, 2012), ongoing and future studies in dissecting and targeting tractable epigenetic modifying enzymes critical for pathogenesis will likely represent the next wave of successful targeted cancer therapies.
Acknowledgments
We apologize that not all publications related to the field could be cited in this mini-review article. The work in our lab is supported by Leukaemia and Lymphoma Research, Cancer Research UK, the Association for International Cancer Research, Medical Research Council, and Kay Kendall Leukaemia Fund.
References
- Ablain J, de The H. Revisiting the differentiation paradigm in acute promyelocytic leukemia. Blood. 2011;117 (22:5795–5802. doi: 10.1182/blood-2011-02-329367. [DOI] [PubMed] [Google Scholar]
- Arteaga MF, Mikesch JH, Qiu J, Christensen J, Helin K, Kogan SC, Dong S, So CW. The histone demethylase PHF8 governs retinoic acid response in acute promyelocytic leukemia. Cancer Cell. 2013;23 (3:376–389. doi: 10.1016/j.ccr.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 2011;11 (10:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botrugno OA, Santoro F, Minucci S. Histone deacetylase inhibitors as a new weapon in the arsenal of differentiation therapies of cancer. Cancer Lett. 2009;280 (2:134–144. doi: 10.1016/j.canlet.2009.02.027. [DOI] [PubMed] [Google Scholar]
- Boukarabila H, Saurin AJ, Batsche E, Mossadegh N, van Lohuizen M, Otte AP, Pradel J, Muchardt C, Sieweke M, Duprez E. The PRC1 polycomb group complex interacts with PLZF/RARA to mediate leukemic transformation. Genes Dev. 2009;23 (10:1195–1206. doi: 10.1101/gad.512009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breccia M, Lo-Coco F. Arsenic trioxide for management of acute promyelocytic leukemia: current evidence on its role in front-line therapy and recurrent disease. Expert Opin Pharmacother. 2012;13 (7:1031–1043. doi: 10.1517/14656566.2012.677436. [DOI] [PubMed] [Google Scholar]
- Carbone R, Botrugno OA, Ronzoni S, Insinga A, Di Croce L, Pelicci PG, Minucci S. Recruitment of the histone methyltransferase SUV39H1 and its role in the oncogenic properties of the leukemia-associated PML-retinoic acid receptor fusion protein. Mol Cell Biol. 2006;26 (4:1288–1296. doi: 10.1128/MCB.26.4.1288-1296.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SJ, Chen Z. Targeting agents alone to cure acute promyelocytic leukemia. N Engl J Med. 2013;369 (2:186–187. doi: 10.1056/NEJMe1304762. [DOI] [PubMed] [Google Scholar]
- Cheung N, So CW. Transcriptional and epigenetic networks in haematological malignancy. FEBS Lett. 2011;585 (13:2100–2111. doi: 10.1016/j.febslet.2011.03.068. [DOI] [PubMed] [Google Scholar]
- Chim CS, Liang R, Tam CY, Kwong YL. Methylation of p15 and p16 genes in acute promyelocytic leukemia: potential diagnostic and prognostic significance. J Clin Oncol. 2001;19 (7:2033–2040. doi: 10.1200/JCO.2001.19.7.2033. [DOI] [PubMed] [Google Scholar]
- Cote S, Rosenauer A, Bianchini A, Seiter K, Vandewiele J, Nervi C, Miller WH., Jr Response to histone deacetylase inhibition of novel PML/RARalpha mutants detected in retinoic acid-resistant APL cells. Blood. 2002;100 (7:2586–2596. doi: 10.1182/blood-2002-02-0614. [DOI] [PubMed] [Google Scholar]
- Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, Fuks F, Lo Coco F, Kouzarides T, Nervi C, Minucci S, Pelicci PG. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295 (5557:1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123 (5:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Feng W, Yonezawa M, Ye J, Jenuwein T, Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol. 2010;17 (4:445–450. doi: 10.1038/nsmb.1778. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, Schifano E, Booth J, van Putten W, Skrabanek L, Campagne F, Mazumdar M, Greally JM, Valk PJ, Lowenberg B, Delwel R, Melnick A. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17 (1:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortschegger K, de Graaf P, Outchkourov NS, van Schaik FM, Timmers HT, Shiekhattar R. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol Cell Biol. 2010;30 (13:3286–3298. doi: 10.1128/MCB.01520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TK, So CW. Overcoming treatment resistance in acute promyelocytic leukemia and beyond. Oncotarget. 2013;4 (8:1128–1129. doi: 10.18632/oncotarget.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher RE, Moser BK, Racevskis J, Poire X, Bloomfield CD, Carroll AJ, Ketterling RP, Roulston D, Schachter-Tokarz E, Zhou DC, Chen IM, Harvey R, Koval G, Sher DA, Feusner JH, Tallman MS, Larson RA, Powell BL, Appelbaum FR, Paietta E, Willman CL, Stock W. Treatment-influenced associations of PML-RARalpha mutations, FLT3 mutations, and additional chromosome abnormalities in relapsed acute promyelocytic leukemia. Blood. 2012;120 (10:2098–2108. doi: 10.1182/blood-2012-01-407601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto E, Tomita A, Hayakawa F, Atsumi A, Kiyoi H, Naoe T. Missense mutations in PML-RARA are critical for the lack of responsiveness to arsenic trioxide treatment. Blood. 2011;118 (6:1600–1609. doi: 10.1182/blood-2011-01-329433. [DOI] [PubMed] [Google Scholar]
- Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, Zamir I, Seiser C, Lazar MA, Minucci S, Pelicci PG. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391 (6669:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- He LZ, Tolentino T, Grayson P, Zhong S, Warrell RP, Jr, Rifkind RA, Marks PA, Richon VM, Pandolfi PP. Histone deacetylase inhibitors induce remission in transgenic models of therapy-resistant acute promyelocytic leukemia. J Clin Invest. 2001;108 (9:1321–1330. doi: 10.1172/JCI11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoemme C, Peerzada A, Behre G, Wang Y, McClelland M, Nieselt K, Zschunke M, Disselhoff C, Agrawal S, Isken F, Tidow N, Berdel WE, Serve H, Muller-Tidow C. Chromatin modifications induced by PML-RARalpha repress critical targets in leukemogenesis as analyzed by ChIP-Chip. Blood. 2008;111 (5:2887–2895. doi: 10.1182/blood-2007-03-079921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N, Shi X, Gozani O, Rappsilber J, Salcini AE, Helin K. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 2010;38 (2:165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok C, Zeisig BB, Dong S, So CW. Forced homo-oligomerization of RARalpha leads to transformation of primary hematopoietic cells. Cancer Cell. 2006;9 (2:95–108. doi: 10.1016/j.ccr.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Zhu J, Chen Z, de The H. Curing APL through PML/RARA degradation by As2O3. Trends Mol Med. 2012;18 (1:36–42. doi: 10.1016/j.molmed.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Leiva M, Moretti S, Soilihi H, Pallavicini I, Peres L, Mercurio C, Dal Zuffo R, Minucci S, de The H. Valproic acid induces differentiation and transient tumor regression, but spares leukemia-initiating activity in mouse models of APL. Leukemia. 2012;26 (7:1630–1637. doi: 10.1038/leu.2012.39. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Evans RM. Acquisition of oncogenic potential by RAR chimeras in acute promyelocytic leukemia through formation of homodimers. Mol Cell. 2000;5 (5:821–830. doi: 10.1016/s1097-2765(00)80322-6. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391 (6669:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, Desai A, Dorrestein PC, Glass CK, Rosenfeld MG. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466 (7305:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo-Coco F, Avvisati G, Vignetti M, Thiede C, Orlando SM, Iacobelli S, Ferrara F, Fazi P, Cicconi L, Di Bona E, Specchia G, Sica S, Divona M, Levis A, Fiedler W, Cerqui E, Breccia M, Fioritoni G, Salih HR, Cazzola M, Melillo L, Carella AM, Brandts CH, Morra E, von Lilienfeld-Toal M, Hertenstein B, Wattad M, Lubbert M, Hanel M, Schmitz N, Link H, Kropp MG, Rambaldi A, La Nasa G, Luppi M, Ciceri F, Finizio O, Venditti A, Fabbiano F, Dohner K, Sauer M, Ganser A, Amadori S, Mandelli F, Dohner H, Ehninger G, Schlenk RF, Platzbecker U, Gruppo Italiano Malattie Ematologiche dell'Adulto, the German–Austrian Acute Myeloid Leukemia Study Group, Study Alliance Leukemia Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N Engl J Med. 2013;369 (2:111–121. doi: 10.1056/NEJMoa1300874. [DOI] [PubMed] [Google Scholar]
- Loenarz C, Ge W, Coleman ML, Rose NR, Cooper CD, Klose RJ, Ratcliffe PJ, Schofield CJ. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nepsilon-dimethyl lysine demethylase. Hum Mol Genet. 2010;19 (2:217–222. doi: 10.1093/hmg/ddp480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JH, Brinkman AB, Simmer F, Francoijs KJ, Nebbioso A, Ferrara F, Altucci L, Stunnenberg HG. PML-RARalpha/RXR alters the epigenetic landscape in acute promyelocytic leukemia. Cancer Cell. 2010;17 (2:173–185. doi: 10.1016/j.ccr.2009.12.042. [DOI] [PubMed] [Google Scholar]
- McNamara S, Wang H, Hanna N, Miller WH., Jr Topoisomerase IIbeta negatively modulates retinoic acid receptor alpha function: a novel mechanism of retinoic acid resistance. Mol Cell Biol. 2008;28 (6:2066–2077. doi: 10.1128/MCB.01576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikesch JH, Gronemeyer H, So CW. Discovery of novel transcriptional and epigenetic targets in APL by global ChIP analyses: emerging opportunity and challenge. Cancer Cell. 2010;17 (2:112–114. doi: 10.1016/j.ccr.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Minucci S, Maccarana M, Cioce M, De Luca P, Gelmetti V, Segalla S, Di Croce L, Giavara S, Matteucci C, Gobbi A, Bianchini A, Colombo E, Schiavoni I, Badaracco G, Hu X, Lazar MA, Landsberger N, Nervi C, Pelicci PG. Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol Cell. 2000;5 (5:811–820. doi: 10.1016/s1097-2765(00)80321-4. [DOI] [PubMed] [Google Scholar]
- Pelosi A, Careccia S, Lulli V, Romania P, Marziali G, Testa U, Lavorgna S, Lo-Coco F, Petti MC, Calabretta B, Levrero M, Piaggio G, Rizzo MG. miRNA let-7c promotes granulocytic differentiation in acute myeloid leukemia. Oncogene. 2012;32 (31:3648–3654. doi: 10.1038/onc.2012.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie K, Prodromou N, Zelent A. Histone deacetylase inhibitors in APL and beyond. Curr Top Microbiol Immunol. 2007;313:157–203. doi: 10.1007/978-3-540-34594-7_10. [DOI] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, Yaghi NK, Lim H, Garcia BA, Brizuela L, Zhao K, Roberts TM, Shi Y. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466 (7305:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice KL, Hormaeche I, Doulatov S, Flatow JM, Grimwade D, Mills KI, Leiva M, Ablain J, Ambardekar C, McConnell MJ, Dick JE, Licht JD. Comprehensive genomic screens identify a role for PLZF-RARalpha as a positive regulator of cell proliferation via direct regulation of c-MYC. Blood. 2009;114 (27:5499–5511. doi: 10.1182/blood-2009-03-206524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro F, Botrugno OA, Dal Zuffo R, Pallavicini I, Matthews GM, Cluse L, Barozzi I, Senese S, Fornasari L, Moretti S, Altucci L, Pelicci PG, Chiocca S, Johnstone RW, Minucci S. A dual role for Hdac1: oncosuppressor in tumorigenesis, oncogene in tumor maintenance. Blood. 2013;121 (17:3459–3468. doi: 10.1182/blood-2012-10-461988. [DOI] [PubMed] [Google Scholar]
- Sanz MA, Lo-Coco F. Modern approaches to treating acute promyelocytic leukemia. J Clin Oncol. 2011;29 (5:495–503. doi: 10.1200/JCO.2010.32.1067. [DOI] [PubMed] [Google Scholar]
- Saumet A, Vetter G, Bouttier M, Portales-Casamar E, Wasserman WW, Maurin T, Mari B, Barbry P, Vallar L, Friederich E, Arar K, Cassinat B, Chomienne C, Lecellier CH. Transcriptional repression of microRNA genes by PML-RARA increases expression of key cancer proteins in acute promyelocytic leukemia. Blood. 2009;113 (2:412–421. doi: 10.1182/blood-2008-05-158139. [DOI] [PubMed] [Google Scholar]
- Schoofs T, Muller-Tidow C. DNA methylation as a pathogenic event and as a therapeutic target in AML. Cancer Treat Rev. 2011;37 (Suppl 1:S13–S18. doi: 10.1016/j.ctrv.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Schoofs T, Rohde C, Hebestreit K, Klein HU, Gollner S, Schulze I, Lerdrup M, Dietrich N, Agrawal-Singh S, Witten A, Stoll M, Lengfelder E, Hofmann WK, Schlenke P, Buchner T, Hansen K, Berdel WE, Rosenbauer F, Dugas M, Muller-Tidow C. DNA methylation changes are a late event in acute promyelocytic leukemia and coincide with loss of transcription factor binding. Blood. 2013;121 (1:178–187. doi: 10.1182/blood-2012-08-448860. [DOI] [PubMed] [Google Scholar]
- Shen ZX, Shi ZZ, Fang J, Gu BW, Li JM, Zhu YM, Shi JY, Zheng PZ, Yan H, Liu YF, Chen Y, Shen Y, Wu W, Tang W, Waxman S, De The H, Wang ZY, Chen SJ, Chen Z. All-trans retinoic acid/As2O3 combination yields a high quality remission and survival in newly diagnosed acute promyelocytic leukemia. Proc Natl Acad Sci USA. 2004;101 (15:5328–5335. doi: 10.1073/pnas.0400053101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LL, Yeung J, Zeisig BB, Popov N, Huijbers I, Barnes J, Wilson AJ, Taskesen E, Delwel R, Gil J, Van Lohuizen M, So CW. Functional crosstalk between Bmi1 and MLL/Hoxa9 axis in establishment of normal hematopoietic and leukemic stem cells. Cell Stem Cell. 2011;8 (6:649–662. doi: 10.1016/j.stem.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Soncini M, Santoro F, Gutierrez A, Frige G, Romanenghi M, Botrugno OA, Pallavicini I, Pelicci P, Di Croce L, Minucci S. The DNA demethylating agent decitabine activates the TRAIL pathway and induces apoptosis in acute myeloid leukemia. Biochim Biophys Acta. 2013;1832 (1:114–120. doi: 10.1016/j.bbadis.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Spicuglia S, Vincent-Fabert C, Benoukraf T, Tiberi G, Saurin AJ, Zacarias-Cabeza J, Grimwade D, Mills K, Calmels B, Bertucci F, Sieweke M, Ferrier P, Duprez E. Characterisation of genome-wide PLZF/RARA target genes. PLoS One. 2011;6 (9:e24176. doi: 10.1371/journal.pone.0024176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternsdorf T, Phan VT, Maunakea ML, Ocampo CB, Sohal J, Silletto A, Galimi F, Le Beau MM, Evans RM, Kogan SC. Forced retinoic acid receptor alpha homodimers prime mice for APL-like leukemia. Cancer Cell. 2006;9 (2:81–94. doi: 10.1016/j.ccr.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Subramanyam D, Belair CD, Barry-Holson KQ, Lin H, Kogan SC, Passegue E, Blelloch R. PML-RAR{alpha} and Dnmt3a1 cooperate in vivo to promote acute promyelocytic leukemia. Cancer Res. 2010;70 (21:8792–8801. doi: 10.1158/0008-5472.CAN-08-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabe Y, Konopleva M, Contractor R, Munsell M, Schober WD, Jin L, Tsutsumi-Ishii Y, Nagaoka I, Igari J, Andreeff M. Up-regulation of MDR1 and induction of doxorubicin resistance by histone deacetylase inhibitor depsipeptide (FK228) and ATRA in acute promyelocytic leukemia cells. Blood. 2006;107 (4:1546–1554. doi: 10.1182/blood-2004-10-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teofili L, Martini M, Luongo M, Diverio D, Capelli G, Breccia M, Lo Coco F, Leone G, Larocca LM. Hypermethylation of GpG islands in the promoter region of p15(INK4b) in acute promyelocytic leukemia represses p15(INK4b) expression and correlates with poor prognosis. Leukemia. 2003;17 (5:919–924. doi: 10.1038/sj.leu.2402907. [DOI] [PubMed] [Google Scholar]
- Tomita A, Kiyoi H, Naoe T. Mechanisms of action and resistance to all-trans retinoic acid (ATRA) and arsenic trioxide (As2O 3) in acute promyelocytic leukemia. Int J Hematol. 2013;97 (6:717–725. doi: 10.1007/s12185-013-1354-4. [DOI] [PubMed] [Google Scholar]
- Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, Vire E, Nomdedeu JF, Jenuwein T, Pelicci PG, Minucci S, Fuks F, Helin K, Di Croce L. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11 (6:513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Wang K, Wang P, Shi J, Zhu X, He M, Jia X, Yang X, Qiu F, Jin W, Qian M, Fang H, Mi J, Xiao H, Minden M, Du Y, Chen Z, Zhang J. PML/RARalpha targets promoter regions containing PU.1 consensus and RARE half sites in acute promyelocytic leukemia. Cancer Cell. 2010;17 (2:186–197. doi: 10.1016/j.ccr.2009.12.045. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Chen Z. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood. 2008;111 (5:2505–2515. doi: 10.1182/blood-2007-07-102798. [DOI] [PubMed] [Google Scholar]
- Zeisig BB, Kulasekararaj AG, Mufti GJ, So CW. SnapShot: acute myeloid leukemia. Cancer Cell. 2012;22 (5:698–698 e1. doi: 10.1016/j.ccr.2012.10.017. [DOI] [PubMed] [Google Scholar]
- Zeisig BB, Kwok C, Zelent A, Shankaranarayanan P, Gronemeyer H, Dong S, So CW. Recruitment of RXR by homotetrameric RARalpha fusion proteins is essential for transformation. Cancer Cell. 2007;12 (1:36–51. doi: 10.1016/j.ccr.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Zelent A, Guidez F, Melnick A, Waxman S, Licht JD. Translocations of the RARalpha gene in acute promyelocytic leukemia. Oncogene. 2001;20 (49:7186–7203. doi: 10.1038/sj.onc.1204766. [DOI] [PubMed] [Google Scholar]
- Zhang H, Luo XQ, Feng DD, Zhang XJ, Wu J, Zheng YS, Chen X, Xu L, Chen YQ. Upregulation of microRNA-125b contributes to leukemogenesis and increases drug resistance in pediatric acute promyelocytic leukemia. Mol Cancer. 2011;10:108. doi: 10.1186/1476-4598-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Nasr R, Peres L, Riaucoux-Lormiere F, Honore N, Berthier C, Kamashev D, Zhou J, Vitoux D, Lavau C, de The H. RXR is an essential component of the oncogenic PML/RARA complex in vivo. Cancer Cell. 2007;12 (1:23–35. doi: 10.1016/j.ccr.2007.06.004. [DOI] [PubMed] [Google Scholar]