Abstract
Background:
Lung cancer remains the leading cause of cancer-related death, largely owing to the lack of effective treatments. A tumour vascular targeting strategy presents an attractive alternative; however, the molecular signature of the vasculature in lung cancer is poorly explored. This work aimed to identify novel tumour vascular targets in lung cancer.
Methods:
Enzymatic digestion of fresh tissue followed by endothelial capture with Ulex lectin-coated magnetic beads was used to isolate the endothelium from fresh tumour specimens of lung cancer patients. Endothelial isolates from the healthy and tumour lung tissue were subjected to whole human genome expression profiling using microarray technology.
Results:
Bioinformatics analysis identified tumour endothelial expression of angiogenic factors, matrix metalloproteases and cell-surface transmembrane proteins. Predicted novel tumour vascular targets were verified by RNA-seq, quantitative real-time PCR analysis and immunohistochemistry. Further detailed expression profiling of STEAP1 on 82 lung cancer patients confirmed STEAP1 as a novel target in the tumour vasculature. Functional analysis of STEAP1 using siRNA silencing implicates a role in endothelial cell migration and tube formation.
Conclusions:
The identification of cell-surface tumour endothelial markers in lung is of interest in therapeutic antibody and vaccine development.
Keywords: microarray analysis, RNA-seq, tumor endothelial marker
Lung cancer is now the leading cause of cancer mortality in the UK, accounting for 24% of cancer deaths in men and 21% in women (Office for National Statistics, 2011). Worldwide rates vary markedly; overall lung cancer accounted for 18% of all cancer deaths in 2008 (Ferlay et al, 2010). Rates have been impacted little by the advances in diagnosis and treatment to date, and in some groups, such as British women (Office for National Statistics, 2011), they continue to rise. Lung cancer is divided into two broad categories – non-small cell (NSCLC) and small cell. More than 80% of lung cancers are NSCLC, which is comprised mostly of squamous cancers and adenocarcinomas. Key changes over the past 10 years in NSCLC include the reclassification of tumour types (Nair et al, 2011), use of biological markers to guide certain therapies, use of adjuvant therapy after selected complete resections and advances in selection and planning for surgery and radiotherapy – concepts that have been reviewed comprehensively elsewhere (Goldstraw et al, 2011). The most promise has been shown by CT-based screening, though concerns have been raised about the potential for multiple invasive tests and cost-effectiveness of the strategy (Bach et al, 2012). Since many patients present with advanced disease surgery is often impossible, hence there is a great need for novel therapeutic agents. This is particularly important as current first line regimes add little >2 months to the average survival (Goldstraw et al, 2011). In addition, many therapies are less beneficial (or indeed more harmful) in squamous tumours, such as the antifolate agent pemetrexed (Scagliotti et al, 2011), anti-vascular endothelial growth factor (VEGF) antibody bevacuzimab (Johnson et al, 2004) and tyrosine kinase inhibitor (TKI) gefitinib (Wang et al, 2012). Selection for the latter drug usually also involves testing for EGFR mutation (Wang et al, 2012), meaning that the pool of patients in whom each drug works is increasingly small. Therefore, the identification of novel biomarkers or therapeutic targets is a priority for lung cancer.
A functional vasculature contributes to tumour progression and malignant cell metastasis. Endothelial cells lining the tumour vasculature are exposed to molecular factors and mechanical forces that are absent in healthy tissue. For example, the vasculature in solid tumours is often in a hypoxic environment (Dachs and Chaplin, 1998) and is exposed to elevated the levels of hypoxically induced angiogenic factors such as VEGF (Relf et al, 1997). Tumour vessels may also be leaky, tortuous, sometimes blind ended and have poor vascular smooth muscle and pericyte coverage (Baluk et al, 2005). As a result, the tumour endothelial transcriptome is markedly different from that in healthy tissue and provides a unique source for cancer target identification. In the last decade, attempts to identify tumour endothelial markers (TEMs) have included construction of SAGE libraries from freshly isolated endothelium (ST Croix et al, 2000), use of microarray platforms (Ho et al, 2003), proteomic analysis of freshly isolated endothelial cell membranes (Ho et al, 2003; Oh et al, 2004) as well as bioinformatics data mining (Huminiecki and Bicknell, 2000; Herbert et al, 2008). These efforts identified several targets including the EDB domain of fibronectin, a series of numbered TEMs, annexin A and recently CLEC14A reviewed in Meyer, 2010.
Recent studies have shown that TEMs are often tissue dependent, and that endothelial transcriptomes have been documented for colon (Van Beijnum et al, 2006), breast (Bhati et al, 2008; Jones et al, 2012) and ovarian cancer (Sasaroli et al, 2011). Known TEMs are often weakly expressed in lung tumours (Mura et al, 2012) and this prompted us to investigate TEMs in the lung. For successful expression profiling it is essential to obtain a pure endothelial population. We demonstrate in this study that rapid tissue digestion followed by magnetic bead isolation yields pure endothelial isolates. Following expression profiling of endothelial isolates from NSCLC patient samples, novel cell-surface targets of lung tumour vasculature were identified, presenting attractive potential for developing targeted therapies for lung cancer.
Materials and methods
Ulex-bead isolation
Healthy and tumour lung tissue was obtained immediately following surgery with full patient consent and ethics approval (Heartlands Hospital, REC. 07/MRE08/42). Minced tissue was digested in DMEM containing 2 mg ml−1 collagenase type V (Sigma, Gillingham, UK), 7.4 mg ml−1 of actinomycin (Sigma) and 30 kU ml−1 of DNAse I (Qiagen, Crawley, UK) at 37oC. Endothelial cells were isolated from the digested cell suspension by positive selection using Ulex europaeus lectin-coated magnetic beads (Invitrogen, Paisley, UK).
Microarray
RNA extracted from Ulex-bead isolated samples was converted to cRNA, then subjected to amplification and labelling. Labelled cRNA samples were then hybridised to an Agilent 4 x 44k whole human gene expression microarray (Agilent, Wokingham, UK). The Bioconductor packages preprocess Core and Limma were used to subtract background and quantile-normalise probe signal intensities prior to performing differential gene expression analyses. Principle component analysis (PCA) was performed in R.
RNA-seq
Seventy-nine and 84 million paired end reads (50 bp+35 bp) were sequenced on the SOLiD4 2nd generation sequencer (Applied Biosystems, Foster City, CA, USA) for endothelium from fresh tumour and healthy tissue, respectively. Reads were mapped to the Human genome (University California Santa Cruz, version hg19) with Tophat 1.3.3 (Trapnell et al, 2009). Default parameters for colour space mapping were used with the exception of the following: 1 g/—max-multihits was set to 1 to identify the single best mapped read; 2 library-type was set to fr-secondstrand to reflect the sequencing library preparation; 3 G provided Tophat with a model set of gene annotation genome positions from the Refseq hg19 transcriptome. The Tophat output bam files were sorted using samtools (Version: 0.1.8, (Li et al, 2009)), and 'HTSeq-count' version 0.4.7p4 (Anders, 2010) was used, in conjunction with the Human transcriptome GTF Refseq version 19, to assign gene counts and produce a tab delimited file of transcript/gene counts. Differential gene expression analysis and P-value generation on the count data was carried out using the R Bioconductor package DESeq v1.5 (Anders and Huber, 2010).
Quantitative real-time PCR
RNA extraction, complementary DNA preparation and quantitative real-time PCR (qPCR) were performed using LightCycler real-time quantitative PCR (Roche, Burgess Hill, UK) by following previously described methods (Armstrong et al, 2008). The Primer sequences are provided in Supplementary Table 1. The double delta Ct method was used to compare the expression levels in tumour relative to healthy endothelial isolates.
Immunohistochemistry
Immunohistochemistry of placental tissue or lung tumour sections were immunostained with antisera to the targets (all antisera from Abcam, Cambridge, UK). The sections were then visualised using ImmPRESS universal antibody kit and NovaRed chromagen (Vector labs, Burlingame, CA, USA). Finally, the sections were counterstained with Mayers hematoxylin, dehydrated and mounted in distyrene–plasticizer–xylene resin.
Functional assay with siRNA knockdown
Transfection with siRNA and functional assays were performed as previously described (Armstrong et al, 2008). STEAP1 siRNA duplexes were:
D1: sense: 5′-CUAUAUUCAGAGCAAGCUATT-3′ anti-sense: 5′-UAGCUUGCUCUGAAUAUAGTG-3′
D2: sense: 5′-GAAUAAGUGGAUAGAUAUATT-3′ anti-sense: 5′-UAUAUCUAUCCACUUAUUCCA-3′ (Ambion, Chipping Norton, UK). The open area of the wound was quantified using a cell intelligence quotient analyzer or Image J software (Image J website, rsbweb.nih.gov). The effect of STEAP1 knockdown on Matrigel assays was analysed by Angiogenesis Analyzer for ImageJ. All images were acquired using a Leica DM IL microscope (Leica, Milton Keynes, UK) and USB 2.0 2M Xli camera (XL Imaging LLC, Carrollton, TX, USA).
Results
Isolation of lung endothelium from fresh tissue
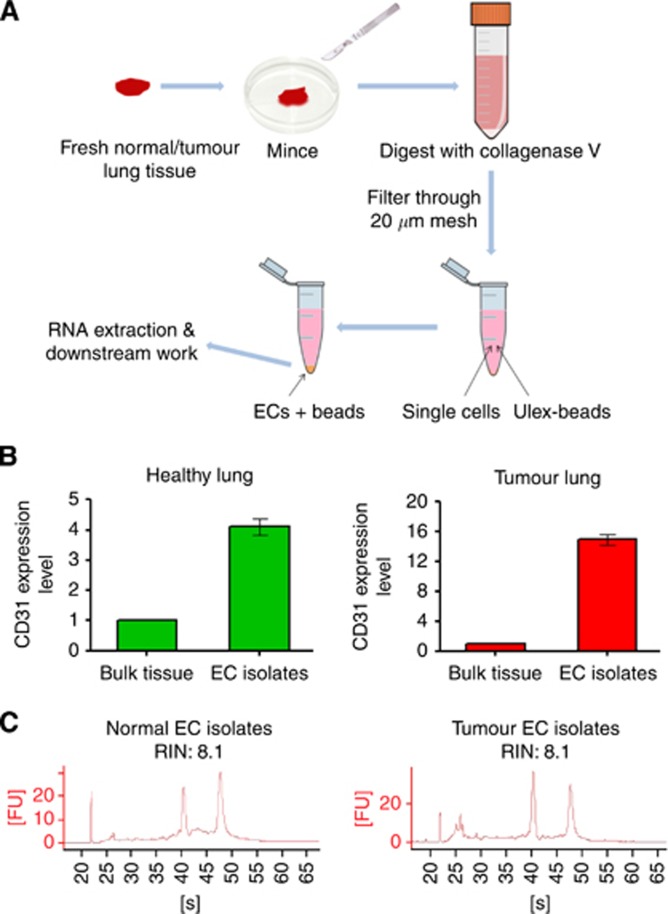
Previous studies have shown that a high purity of endothelial isolates can be achieved using Ulex-conjugated beads (Jackson et al, 1990) but has not yet been applied to human lung tissue. Ulex agglutinin I is a lectin that specifically binds to L-fucose residues present in glycoproteins on the human endothelial surface (Holthofer et al, 1982). Here we examine this approach for the isolation of endothelium from fresh lung specimens. Fresh healthy or tumour lung tissue samples (1–3 g) were processed within 3 h post surgery. The tumour tissue was resected from the viable region of the tumour core and the patient-matched healthy tissue was resected>10 cm away from the tumour core. Endothelial cells were positively isolated using magnetic beads coupled to Ulex lectin (workflow illustrated in Figure 1A). To verify endothelial enrichment, expression of the universal endothelial marker CD31 was examined by qPCR in the endothelial isolates and compared with that in the bulk tissue. A 15-fold enrichment of endothelium was achieved in the bead isolated samples when compared with whole tumour extracts. Afour-fold enrichment was seen in endothelial cells isolated from healthy lung (Figure 1B). The differing fold increase in CD31 seen in healthy and tumour samples is likely owing to the proportion of endothelial cells being higher in healthy lung (30%) than in tumours (3%–5%). RNA integrity analysis of a typical RNA isolate is shown in Figure 1C. The data confirm that the Ulex-bead isolation approach can effectively isolate the endothelial population from lung.
Figure 1.
A Ulex-coated magnetic bead isolation achieved substantial endothelial enrichment and gave good quality RNA. (A) The workflow of the main steps in Ulex-bead isolation of endothelial cells from healthy/tumor lung tissue. (B) Confirmation of endothelial enrichment using the Ulex-beads approach. Real-time PCR using a primer set for the endothelial marker CD31 was performed on the bead isolated endothelial cells and bulk tissue. Expression of CD31 in the bead isolated sample was normalised to that in the bulk tissue (n=3). (C) Good quality RNA (RIN>7) was obtained from Ulex-bead isolated endothelial cells from healthy and tumour lung tissue. ECs=Endothelial cells.
Microarray of endothelial isolates from lung cancer patients
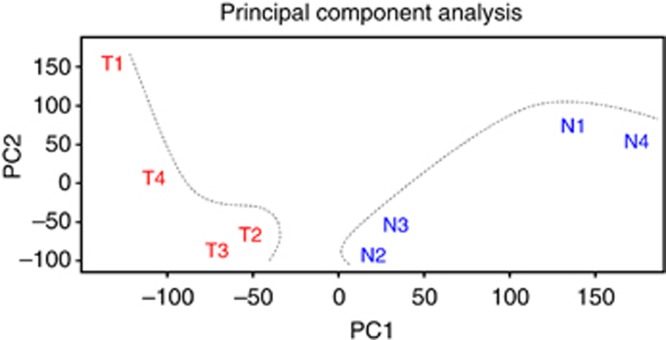
For expression profiling, a microarray analysis was performed on four pairs of NSCLC patient-matched healthy and tumour lung endothelial isolates. Clinical and pathologic data was obtained from Birmingham Heartlands Hospital (Table 1, patients 1–4). A PCA plot shows variation in both tumour and healthy lung samples and between the samples of each group. This was to be expected as samples were collected and extracted from different patients and statistically significant genes are those that are consistent across replicate samples. Despite this the tumour and healthy isolates fall into two discrete groups (Figure 2).
Table 1. Clinical-pathological data of lung cancer patients used in the genomic analysis.
| ID | Age | Gender | Pack years smoked | Histology of tumour | Tumour stage | Application |
|---|---|---|---|---|---|---|
| 1 | 65 | M | 15 | Squamous | T1N0M0 | Microarray |
| 2 | 71 | M | 40 | Squamous | T1N0M0 | Microarray |
| 3 | 63 | F | 17 | Squamous | T1N1M0 | Microarray |
| 4 | 67 | M | 50 | Squamous | T3N0M0 | Microarray |
| 5 | 73 | F | 25 | Adeno | T2N0M0 | RNA-Seq |
| 6 | 83 | M | 35 | Adeno | T2N1M0 | RNA-Seq |
| 7 | 52 | M | 25 | Adeno | T2N1M0 | RNA-Seq |
Figure 2.
Principle component analysis plot of microarray of the four pairs of healthy and tumour lung endothelial isolates. A PCA plot showing that the endothelial transcriptomes of healthy and tumour lung show a clear difference. The separation between healthy and tumour lung endothelium was highlighted by dotted lines.
To better understand the role of known angiogenesis-associated genes in NSCLC, a differential expression analysis was performed using the programme Limma. The analysis revealed a panel of known angiogenesis-associated genes including COL1A1, VEGF-A, TEM7, TNC, EPHB2, IL8, FGF1, ANGPTL2 and TEM8 to be elevated in lung tumour endothelium (Table 2). As tumour angiogenesis proceeds by proteolysis of the extracellular matrix (Sottile, 2004), elevated matrix metalloprotease (MMP) activity is associated with active angiogenesis and tumour progression. The analysis also identified a number of MMPs that are upregulated in lung tumour compared with healthy lung endothelium (Table 3).
Table 2. Upregulated angiogenesis-associated genes in lung cancer.
| Gene ID | Gene symbol | GenBank accession no. | LogFC | P-value |
|---|---|---|---|---|
| Collagen, type I, alpha 1 | COL1A1 | NM_000088 | 5.11 | 0.00 |
| Vascular endothelial growth factor A | VEGF-A | NM_001025366 | 2.59 | 0.00 |
| Plexin domain containing 1 | PLXDC1(TEM7) | NM_020405 | 2.04 | 0.01 |
| Tenascin C | TNC | NM_002160 | 1.95 | 0.01 |
| Eph receptor B2 | EPHB2 | NM_004442 | 1.76 | 0.00 |
| Interleukin 8 | IL8 | ENST00000401931 | 1.34 | 0.16 |
| Fibroblast growth factor 1 (Acidic) | FGF1 | NM_000800 | 0.82 | 0.08 |
| Angiopoietin-like 2 | ANGPTL2 | NM_012098 | 0.69 | 0.40 |
| Anthrax toxin receptor 1 | ANTXR1(TEM8) | NM_032208 | 0.65 | 0.15 |
Abbreviation: LogFC=log2 fold change; TEM=tumour endothelial marker. Differential expression analysis of microarray data for the identification of elevated known angiogenesis-associated genes. Listed genes were ranked by LogFC in descending order.
Table 3. Upregulated matrix metallopeptidases in lung cancer.
| Gene ID | Gene Symbol | GenBank accession no. | LogFC | P-value |
|---|---|---|---|---|
| Matrix metallopeptidase 11 | MMP11 | NM_005940 | 4.10 | 0.00 |
| Matrix metallopeptidase 9 | MMP9 | NM_004994 | 4.00 | 0.00 |
| Matrix metallopeptidase 12 | MMP12 | NM_002426 | 3.80 | 0.00 |
| Matrix metallopeptidase 7 | MMP7 | NM_002423 | 3.29 | 0.17 |
| Matrix metallopeptidase 1 | MMP1 | NM_002421 | 2.49 | 0.05 |
| Matrix metallopeptidase 3 | MMP3 | NM_002422 | 1.81 | 0.02 |
| Matrix metallopeptidase 10 | MMP10 | NM_002425 | 1.74 | 0.12 |
| Matrix metallopeptidase 14 | MMP14 | NM_004995 | 1.55 | 0.00 |
| Matrix metallopeptidase 13 | MMP13 | NM_002427 | 1.21 | 0.04 |
| Matrix metallopeptidase 2 | MMP2 | NM_004530 | 1.03 | 0.12 |
Abbreviations: Log FC=log fold change; MMP=matrix metalloprotease. Differential expression analysis of microarray data for the identification of elevated matrix metallopeptidases. Listed genes were ranked by logFC in descending order.
Identification and validation of putative tumour vascular targets in NSCLC
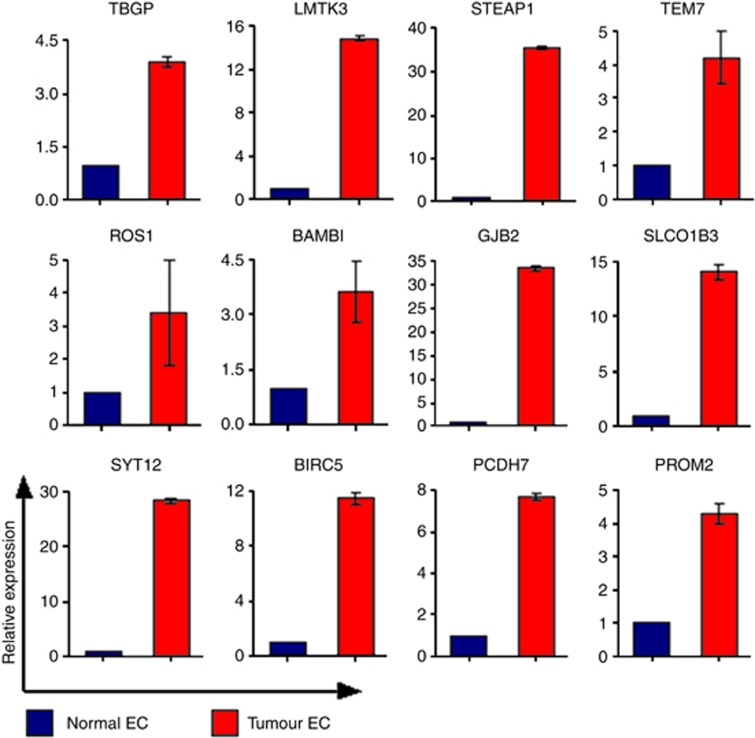
For target identification, differentially expressed genes from the microarray data were filtered through several selection criteria: Log2 fold change magnitude>1, a P-value<0.5 and containing a transmembrane or signal peptide domain, which generated a list comprised of 584 genes. Twelve target candidates were chosen for further validation based on additional criteria including the level of association with endothelial cells, previously published work, sites of expression and relation to known genes with interesting functional properties (Table 4). To validate putative targets, a qPCR was performed on the four pairs of endothelial isolates used in the microarray. Figure 3 shows that all candidates had elevated expression in tumour compared with that in healthy endothelium ranging from a 3-to 35-fold increase in expression.
Table 4. Putative vascular targets in lung cancer.
| Gene ID | Gene Symbol | GenBank accession no. | LogFC | P-value | TM |
|---|---|---|---|---|---|
| Six transmembrane epithelial antigen of the prostate 1 | STEAP1 | NM_012449 | 4.19 | 0.00 | 6 |
| Synaptotagmin Xii | SYT12 | NM_177963 | 4.16 | 0.00 | 1 |
| Gap junction protein, β 2, 26Kda | GJB2 | NM_004004 | 4.13 | 0.00 | 4 |
| Solute Carrier organic anion transporter family, member 1B3 | SLCO1B3 | NM_019844 | 3.65 | 0.00 | 11 |
| Baculoviral Iap Repeat containing 5 | BIRC5 | NM_001012271 | 3.45 | 0.00 | 0 |
| Protocadherin 7 | PCDH7 | NM_002589 | 2.29 | 0.00 | 1 |
| Prominin 2 | PROM2 | NM_001165978 | 2.05 | 0.00 | 6 |
| Plexin Domain Containing 1 | PLXDC1(TEM7) | NM_020405 | 2.04 | 0.01 | 1 |
| Bmp and activin membrane-bound inhibitor homologue | BAMBI | NM_012342 | 1.99 | 0.00 | 1 |
| Lemur tyrosine kinase 3 | LMTK3 | NM_001080434 | 1.94 | 0.07 | 2 |
| Trophoblast glycoprotein | TPBG | NM_006670 | 1.48 | 0.03 | 1 |
| C-Ros oncogene 1 , receptor tyrosine kinase | ROS1 | ENST00000403284 | 1.39 | 0.22 | 1 |
Abbreviations: Log FC, log fold change; TEM, tumour endothelial marker. Differential expression analysis of microarray data for the identification of putative vascular targets for lung cancer. Listed genes were ranked by logFC in descending order.
Figure 3.
Validation of putative lung vascular targets by qPCR. Quantitative real-time PCR validation of tumour vascular target candidates in the endothelial cells isolated from the healthy and tumour lung tissue. Flotillin 2 was used as the house keeping gene to which the data was normalised. The double delta Ct method was used to compare the expression levels in tumour relative to healthy endothelial isolates.
Expression profiling of lung endothelial isolates by RNA-seq
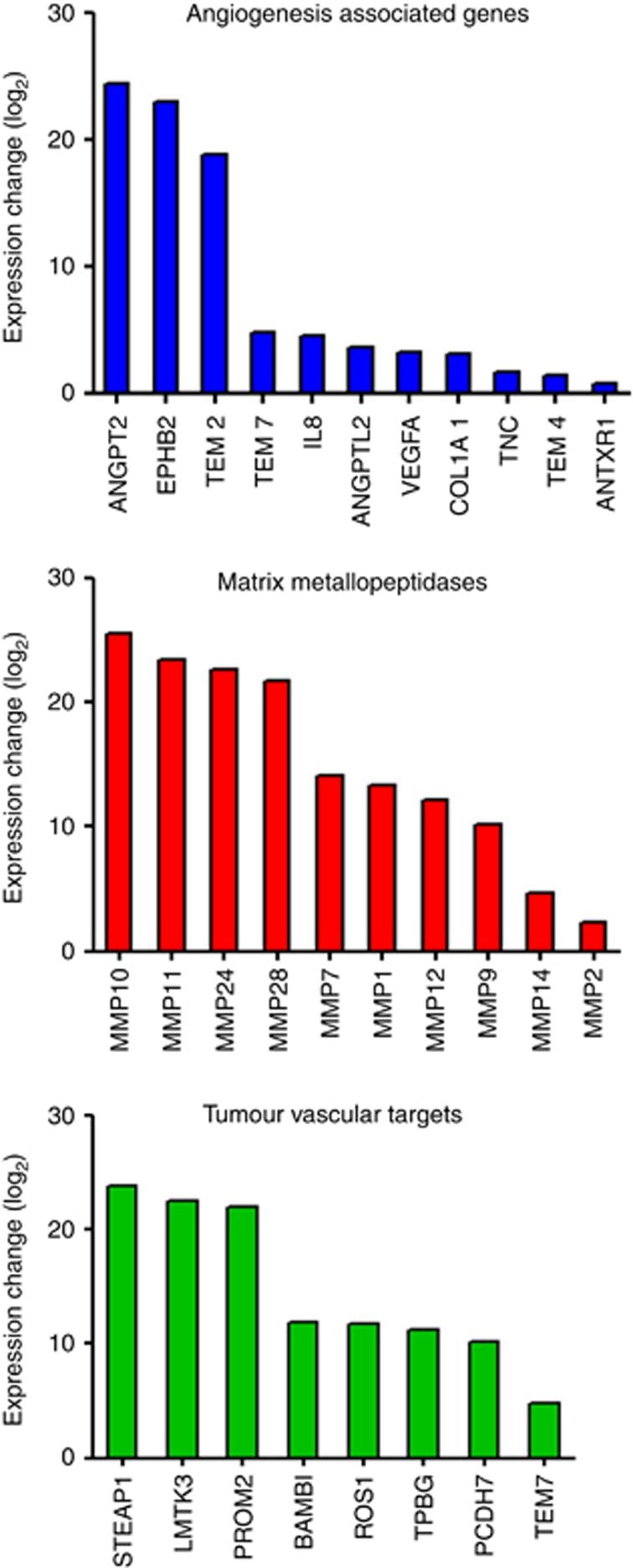
RNA-seq using deep sequencing technology provides an in-depth resolution of RNA snapshots by generating millions of reads that can be assembled and mapped to a known transcriptome, allowing the measurement of differential gene expression. RNA-seq has the advantage of querying novel transcripts and does not rely on prior knowledge and annotation. Here we used RNA-seq to verify the genes that had been identified through the microarray analysis. We note that a lower yield of RNA was obtained from healthy lung tissue compared with that from tumour. This was possibly owing to the endothelial cells in healthy lung tissue being in a quiescent state compared with the active endothelium in tumours. For this reason, endothelial RNA isolated from three healthy lung samples (pooled; Table 1, patients 5–7) and one tumour lung tissue (Table 1, patient 6) were sequenced as one healthy and one tumour sample on a SOLiD4 sequencer. The differential gene expression analysis of the RNA-seq data was performed using the DESeq v1.5 package (Anders and Huber, 2010). The analysis confirmed most of the unregulated angiogenesis-associated genes, MMPs and putative targets identified through the microarray analysis (Figure 4). Analysis of the RNA-seq data alone generated a list of 477 genes with the same criterion used in the microarray analysis for target identification. The intersection of the microarray and RNA-seq gene pools comprises a list of 122 genes, which provides a rich source for target identification (Supplementary Table 2). The discrepancy between the two analyses is likely owing to the cancer type (squamous vs. adeno) and the individual patient variability.
Figure 4.
Confirmation of upregulated angiogenesis-associated genes, MMP and putative vascular targets in lung cancer by RNA-seq. Differential gene expression analysis of RNA-seq data confirmed a panel of elevated angiogenesis-associated genes, MMP genes and lung cancer vascular target candidates identified through microarray analysis.
Expression of TEM candidates in angiogenic tissue and lung cancer
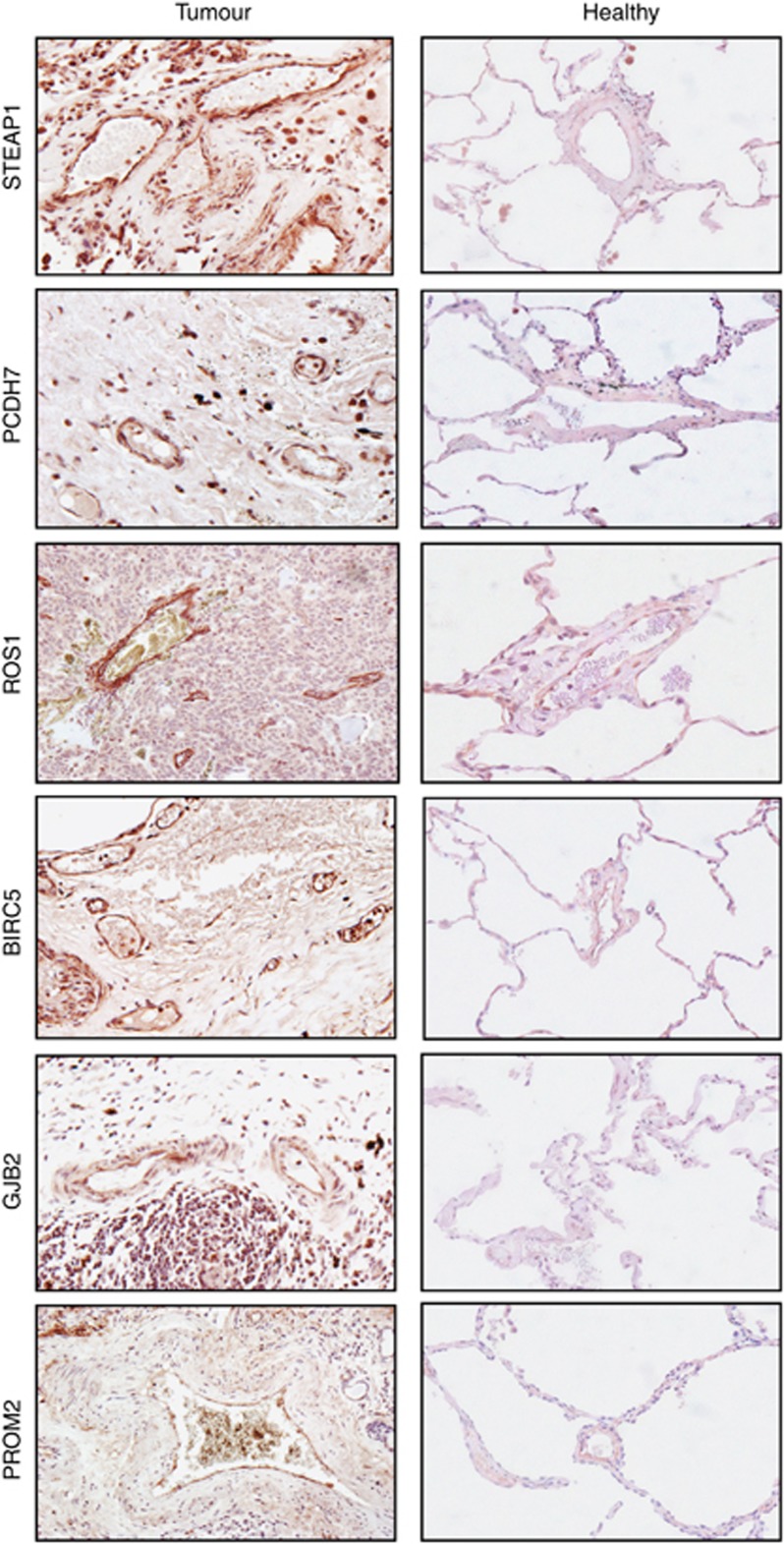
To further validate the candidate targets, we investigated protein expression by immunohistochemistry. We have previously shown that placental vasculature is a rich source of endothelial gene expression. Thus, immunohistochemical staining was performed first on the human placental tissue using antibodies to the lung TEM targets. Amongst the twelve targets, six genes: ROS1, PCDH7, BIRC5, STEAP1, GJB2 and PROM2 showed expression in human placental vessels (Supplementary Figure 1). The tumour and healthy lung tissue was then immunostained and these six candidates are indeed overexpressed in the lung tumour vessels, whereas absent or at a low level in the healthy lung tissue (Figure 5). Some of the targets were not restricted to the tumour endothelium; for example, ROS1 and STEAP1 also showed positive expression on some tumour cells or macrophages and this may be beneficial for developing drugs targeting the tumour and its vasculature simultaneously. It is also worth mentioning that other target candidates should not be completely eliminated for further investigation simply owing to the lack of antibody reactivity in immunochemistry.
Figure 5.
Validation of putative lung vascular targets by immunohistochemistry. Identified putative lung TEMs were validated by immunohistochemistry. Representative immunohistochemistry for lung vascular target candidates on the healthy and tumour lung tissue.
Expression of STEAP1 in lung cancer
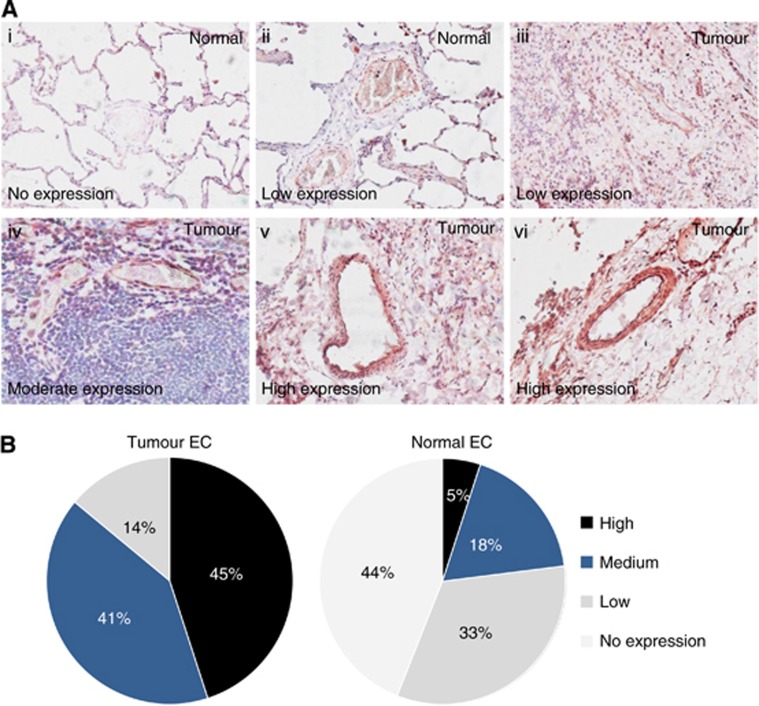
We then focused on the top-ranked target STEAP1. To confirm whether STEAP1 is differentially expressed in the endothelium within the healthy and tumour lung tissue, an expression profiling was carried out on human lung cancer tissues by immunohistochemistry. Eighty-two patients were examined (Table 5). The intensity of the signal was classified as absent, low, medium or high. Representative images of STEAP1 staining in lung cancer are shown in Figure 6A. From the 82 cases examined, a clear overexpression of STEAP1 in tumour vessels was observed; for example 45% of the vessels highly expressed STEAP1 in lung cancer vs only 5% in matching healthy lung. The proportion of vessels that are ‘low' and ‘no expression' of STEAP1 was 77% in healthy lung, but only 14% in lung tumours (Figure 6B). These data confirm that STEAP1 is differentially expressed between the tumour and healthy lung vasculature and presents a possible vascular target for lung cancers.
Table 5. Clinical characteristics of lung cancer patients for STEAP1 profiling.
| Patients (n) | % | |
|---|---|---|
| Age | 63.8±9.5 | |
|
Sex | ||
| Male | 64 | 78% |
| Female | 18 | 22% |
|
Histology | ||
| Squamous cell carcinoma | 47 | 57.3% |
| Adenocarcinomas | 19 | 23.2% |
| Large cell carcinoma | 5 | 6.1% |
| Bronchioloalveolar carcinoma | 7 | 8.5% |
| Carcinosarcoma | 2 | 2.4% |
|
Clinical stage | ||
| I | 34 | 41.5% |
| II | 35 | 42.7% |
| III | 13 | 15.8% |
Figure 6.
Expression profiling of STEAP1 expression in clinical lung cancer samples. (A) Representative images of STEAP1 in healthy (i–ii) and tumour (iii–vi) lung tissue; expression level classified as no expression (i), low (ii–iii), medium (iv) and high (v–vi). Images were acquired using an optical microscope at a magnification of × 20. (B) Expression profiling of STEAP1 in clinical samples by immunohistochemistry (n=82).
Function of STEAP1 in endothelial cells
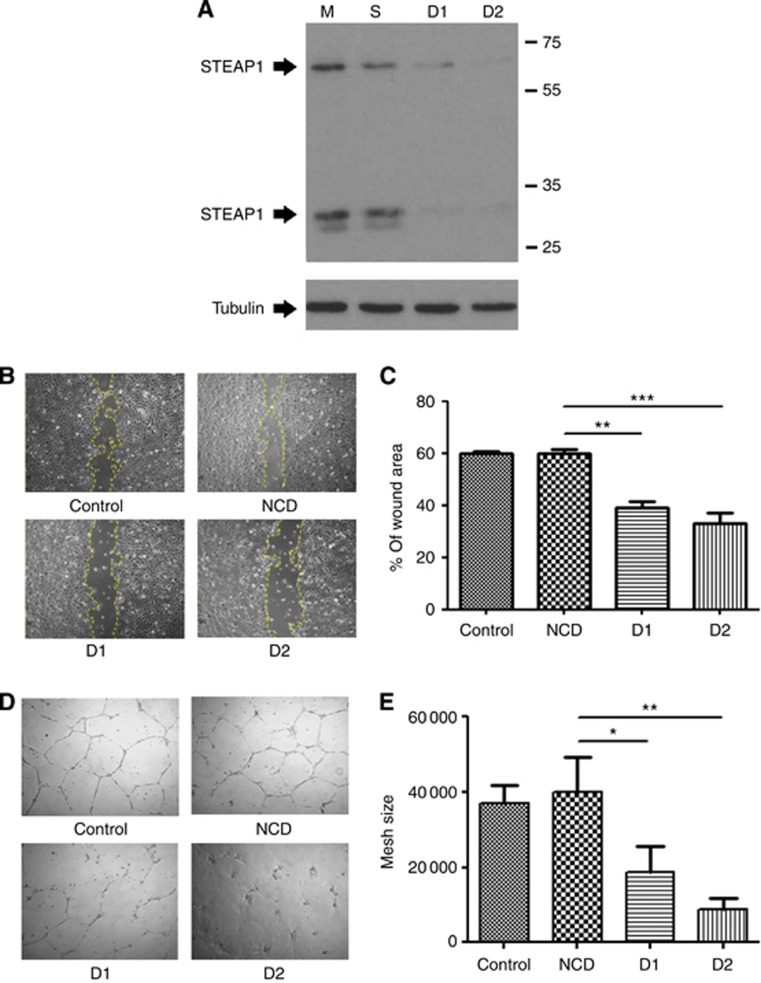
We next used siRNA knockdown to seek a function of STEAP1 in the endothelial cells. STEAP1 protein expression was efficiently knocked down by two independent siRNA duplexes (Figure 7A). Migration of HUVEC after STEAP1 knockdown was compared with that of mock and negative siRNA-transfected controls in a scratch-wound assay. At 24 h, control wounds showed 60% closure, whereas in STEAP1 knockdown cells the wound had only closed by 35%–40% (Figure 7B and C). STEAP1 knockdown also compromised tube formation on Matrigel. Tubes showed a significant decrease in mesh size compared with controls (Figure 7D and E).
Figure 7.
Functional analysis of STEAP1 in endothelial cells. (A) Western blot showing that two independent siRNA duplexes efficiently knock down the STEAP1 protein in HUVEC. Tubulin was used as a loading control. STEAP1 appears as two bands, the 65-kDa glycosylated mature protein and the 30-kDa unglycosylated precursor. (B–C) Scratch-wound assay and (D–E) Matrigel tube forming assay in HUVEC transfected with mock, negative control or STEAP1 siRNA duplexes (± s.e.m.).
Discussion
The ineffectiveness of current treatments for NSCLC prompted the search for alternatives. Although TKIs of EGFR initially showed promising outcomes in several trials, resistance developed in all the patients (Lovly and Carbone, 2011). Antiangiogenic/anti-tumour vascular therapy remains a viable alternative (Vasudev and Reynolds, 2014). VEGF (receptor) blockers have been used to treat NSCLC patients in early clinical trials; however, concerns have arisen from the limited efficacy in achieving tumour regression and the tendency to develop resistance (Pallis and Syrigos, 2013). In addition, the lack of effective biomarkers for patient pre-treated selection, emphasises the need for novel targets and biomarkers. Our study describes the first molecular profiling of endothelium from NSCLC patient tissue.
A major obstacle that hinders the expression profiling of in vivo endothelium is the challenge to obtain pure endothelial isolates. Favre et al described attempts for mouse lung endothelial isolates, using the unpurified sample as the control, aimed at identifying endothelial specific genes (Favre et al, 2003). An effort to isolate mouse endothelium from a Lewis lung carcinoma tumour model was reported by Allport and Weissleder, (2003); however, the isolated endothelial cells were subjected to in vitro culture and only the characterised angiogenesis genes were investigated (Allport and Weissleder, 2003). In this study, we demonstrate that the Ulex-bead isolation approach has proven to be an effective approach to obtain a pure endothelial population from the human lung. The purity of the endothelial isolates allows for the first time to precisely document the transcriptome of the lung vasculature using microarray analysis and deep sequencing platforms.
Our microarray analysis of endothelial isolates from cancer patients identified a panel of angiogenesis-associated genes and MMPs elevated in tumour endothelium, which have potential to be biomarkers for NSCLC. The elevated MMP2 expression is in agreement with a reported increased MMP2 level in serum that was shown to be a predictor of metastasis in NSCLC patients (Guo et al, 2007). MMP2 and MMP9 have also been reported to associate with tumour grade in various solid tumours (John and Tuszynski, 2001). The expression of MMP7 and MMP9 were previously found to be significantly upregulated in NSCLC compared with that of healthy lung and benign lung tumours (Safranek et al, 2009). Although clinical trials investigating inhibitors targeting multiple MMPs yielded limited efficacy (Heath et al, 2001; Goffin et al, 2005), possibilities remain to design drugs that are more specific to those highly expressed MMPs in NSCLC.
RNA-seq using deep sequencing technology has been intensively applied to molecular profiling of many tissue types and cell populations from different species (Guduric-Fuchs et al, 2012; Voellenkle et al, 2012). This work is the first description of RNA-seq for profiling the endothelium from human lung cancer. Further, cross referencing the microarray data identified 122 genes as lung vascular target candidates. Immunohistochemical analysis confirmed PCHD7, STEAP1, ROS1, BIRC5, GJB2 and RPOM2 to be novel tumour vascular targets in NSCLC.
STEAP1 was the first member of a family of metalloreductases described as a cell-surface antigen in prostate tissue (Hubert et al, 1999; Yang et al, 2001). High STEAP1 expression has been found in prostate, breast, bladder, colon and ovarian carcinomas and in Ewing's sarcoma, (Gomes et al, 2012) whereas low or absent in healthy human tissues, suggesting that STEAP1 may be a wide ranging tumour antigen (Hubert et al, 1999). Alves et al (2006) showed that STEAP1 peptides can be used to stimulate CD8+ T cells, suggesting that STEAP1 may be a useful target for cancer immunotherapy. Indeed, Maria de la Luz and colleagues also showed the efficacy of a vaccine against STEAP1 in prophylactic and therapeutic tumour models (Garcia-Hernandez Mde et al, 2007). To date, expression of STEAP1 in endothelium has not been described. Our data have shown for the first time that STEAP1 expression is upregulated in endothelial cells in the vessels of human lung tumours.
PCDH7 belongs to the protocadherin gene family and encodes a single-pass transmembrane protein. First reported in 1998, study of PCDH7 has concentrated on neuronal function (Yoshida et al, 1998; Kim et al, 2007; Blevins et al, 2011), and a role in endothelium or lung cancer has not been explored. An exception was a genome-wide genotyping of the frozen tumour tissue from NSCLC patients that identified five nucleotide polymorphisms (SNPs) located in PCDH7 that are prognostic for the overall survival in early-stage NSCLC (Huang et al, 2009). ROS1 is a proto-oncogene and highly expressed in a variety of tumour cell lines (Birchmeier et al, 1987; Sharma et al, 1989). ROS1 belongs to the sevenless subfamily of tyrosine kinase receptors and remains an orphan receptor. Recently, chromosomal rearrangement of ROS1 was detected in a subpopulation of NSCLC patients (Bergethon et al, 2012; Janne and Meyerson, 2012). Despite intensive study of ROS1 in lung cancer, there has been no previous report of its expression in the tumour endothelium. BIRC5 or survivin, belongs to a family of inhibitors of apoptosis. BIRC5 inhibits the caspase activation regulating apoptosis. Disruption of the BIRC5 signalling pathway leads to tumour cell apoptosis and growth delay. BIRC5 protein is often present in tumour cells and fetal tissues but has rarely been described in the healthy tissue (Sah et al, 2006). GJB2 is a transmembrane protein that belongs to the connexin family. Connexins have a role in many physiological processes and embryonic development including the microvasculature. Defects in GJB2 lead to the most common form of congenital deafness (Apps et al, 2007). Thus, most GJB2 studies have centred on this pathology. Nevertheless, a recent study reported the expression of GJB2 in lymphatic endothelium in the mouse embryo, and that the deletion of GJB2 in mice disrupted the development of lymphatic vessels and was embryonic lethal (Dicke et al, 2011). PROM2 is a multi-pass membrane protein and belongs to the prominin family of pentaspan membrane glycoproteins. PROM2 is at present comparatively uncharacterised.
Low shear stress and turbulent flow are mechanical factors that regulate endothelial gene expression (Wasserman and Topper, 2004). Around 600 genes are regulated by shear stress in the endothelial cell (Ando and Yamamoto, 2009; Mura et al, 2012). Like the previously described TEMs CLEC14A and Robo4, STEAP1, PCDH7 and BIRC5 are all upregulated in the endothelium exposed to reduced shear stress (Bicknell et al, unpublished data). Reduced blood flow and shear stress may account for their expression on tumour vessels.
To conclude, our work not only enhances our knowledge of proteins that are differentially expressed on the lung tumour endothelium but has also identified several promising biomarkers/targets for future investigation. Cell-surface expression of some of these targets, for example STEAP1, will facilitate the pre-clinical validation with antibodies. Further work is needed to characterise their functions and their roles in the endothelial biology and angiogenesis in the lung.
Acknowledgments
This project was funded by Knowledge Transfer Partnership KTP007696 awarded by the Technology Strategy Board to RB (University of Birmingham) and DC(AstraZeneca plc). We thank the staff of the Birmingham Heartlands Hospital for help in rapidly obtaining fresh tissue and the University of Birmingham Technology Hub for help with sequencing and bioinformatics analysis.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Allport JR, Weissleder R. Murine Lewis lung carcinoma-derived endothelium expresses markers of endothelial activation and requires tumor-specific extracellular matrix in vitro. Neoplasia. 2003;5:205–217. doi: 10.1016/S1476-5586(03)80053-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves PM, Faure O, Graff-Dubois S, Cornet S, Bolonakis I, Gross DA, Miconnet I, Chouaib S, Fizazi K, Soria JC, Lemonnier FA, Kosmatopoulos K. STEAP, a prostate tumor antigen, is a target of human CD8+ T cells. Cancer Immunol Immunother. 2006;55:1515–1523. doi: 10.1007/s00262-006-0165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S.2010. HTSeq: Analysing high-throughput sequencing data with Python [Online]. Available at http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html [Accessed]. [DOI] [PMC free article] [PubMed]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando J, Yamamoto K. Vascular mechanobiology: endothelial cell responses to fluid shear stress. Circ J. 2009;73:1983–1992. doi: 10.1253/circj.cj-09-0583. [DOI] [PubMed] [Google Scholar]
- Apps SA, Rankin WA, Kurmis AP. Connexin 26 mutations in autosomal recessive deafness disorders: a review. Int J Audiol. 2007;46:75–81. doi: 10.1080/14992020600582190. [DOI] [PubMed] [Google Scholar]
- Armstrong LJ, Heath VL, Sanderson S, Kaur S, Beesley JF, Herbert JM, Legg JA, Poulsom R, Bicknell R. ECSM2, an endothelial specific filamin a binding protein that mediates chemotaxis. Arterioscler Thromb Vasc Biol. 2008;28:1640–1646. doi: 10.1161/ATVBAHA.108.162511. [DOI] [PubMed] [Google Scholar]
- Bach PB, Mirkin JN, Oliver TK, Azzoli CG, Berry DA, Brawley OW, Byers T, Colditz GA, Gould MK, Jett JR, Sabichi AL, Smith-BINDMAN R, Wood DE, Qaseem A, Detterbeck FC. Benefits and harms of CT screening for lung cancer: a systematic review. JAMA. 2012;307:2418–2429. doi: 10.1001/jama.2012.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluk P, Hashizume H, Mcdonald DM. Cellular abnormalities of blood vessels as targets in cancer. Curr Opin Genet Dev. 2005;15:102–111. doi: 10.1016/j.gde.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Bergethon K, Shaw AT, Ou SH, Katayama R, Lovly CM, Mcdonald NT, Massion PP, Siwak-Tapp C, Gonzalez A, Fang R, Mark EJ, Batten JM, Chen H, Wilner KD, Kwak EL, Clark JW, Carbone DP, Ji H, Engelman JA, Mino-Kenudson M, Pao W, Iafrate AJ. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhati R, Patterson C, Livasy CA, Fan C, Ketelsen D, Hu Z, Reynolds E, Tanner C, Moore DT, Gabrielli F, Perou CM, Klauber-Demore N. Molecular characterization of human breast tumor vascular cells. Am J Pathol. 2008;172:1381–1390. doi: 10.2353/ajpath.2008.070988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier C, Sharma S, Wigler M. Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc Natl Acad Sci USA. 1987;84:9270–9274. doi: 10.1073/pnas.84.24.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins CJ, Emond MR, Biswas S, Jontes JD. Differential expression, alternative splicing, and adhesive properties of the zebrafish delta1-protocadherins. Neuroscience. 2011;199:523–534. doi: 10.1016/j.neuroscience.2011.09.061. [DOI] [PubMed] [Google Scholar]
- Dachs GU, Chaplin DJ. Microenvironmental control of gene expression: implications for tumor angiogenesis, progression, and metastasis. Semin Radiat Oncol. 1998;8:208–216. doi: 10.1016/s1053-4296(98)80046-5. [DOI] [PubMed] [Google Scholar]
- Dicke N, Pielensticker N, Degen J, Hecker J, Tress O, Bald T, Gellhaus A, Winterhager E, Willecke K. Peripheral lymphangiogenesis in mice depends on ectodermal connexin-26 (Gjb2) J Cell Sci. 2011;124:2806–2815. doi: 10.1242/jcs.084186. [DOI] [PubMed] [Google Scholar]
- Favre CJ, Mancuso M, Maas K, Mclean JW, Baluk P, Mcdonald DM. Expression of genes involved in vascular development and angiogenesis in endothelial cells of adult lung. Am J Physiol Heart Circ Physiol. 2003;285:H1917–H1938. doi: 10.1152/ajpheart.00983.2002. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin H, Bray F, Forman D, Mathers C, Parkin D. GLOBOCAN 2008 v1.2, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10. International Agency for Research on Cancer: Lyon, France; 2010. [Google Scholar]
- Garcia-Hernandez Mde L, Gray A, Hubby B, Kast WM. In vivo effects of vaccination with six-transmembrane epithelial antigen of the prostate: a candidate antigen for treating prostate cancer. Cancer Res. 2007;67:1344–1351. doi: 10.1158/0008-5472.CAN-06-2996. [DOI] [PubMed] [Google Scholar]
- Goffin JR, Anderson IC, Supko JG, Eder JP, Jr, Shapiro GI, Lynch TJ, Shipp M, Johnson BE, Skarin AT. Phase I trial of the matrix metalloproteinase inhibitor marimastat combined with carboplatin and paclitaxel in patients with advanced non-small cell lung cancer. Clin Cancer Res. 2005;11:3417–3424. doi: 10.1158/1078-0432.CCR-04-2144. [DOI] [PubMed] [Google Scholar]
- Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, Shepherd FA. Non-small-cell lung cancer. Lancet. 2011;378:1727–1740. doi: 10.1016/S0140-6736(10)62101-0. [DOI] [PubMed] [Google Scholar]
- Gomes IM, Maia CJ, Santos CR. STEAP proteins: from structure to applications in cancer therapy. Mol Cancer Res. 2012;10:573–587. doi: 10.1158/1541-7786.MCR-11-0281. [DOI] [PubMed] [Google Scholar]
- Guduric-Fuchs J, O'connor A, Cullen A, Harwood L, Medina RJ, O'neill CL, Stitt AW, Curtis TM, Simpson DA. Deep sequencing reveals predominant expression of miR-21 amongst the small noncoding RNAs in retinal microvascular endothelial cells. J Cell Biochem. 2012;113 (6:2098–2111. doi: 10.1002/jcb.24084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo CB, Wang S, Deng C, Zhang DL, Wang FL, Jin XQ. Relationship between matrix metalloproteinase 2 and lung cancer progression. Mol. Diagn Ther. 2007;11:183–192. doi: 10.1007/BF03256240. [DOI] [PubMed] [Google Scholar]
- Heath EI, O'reilly S, Humphrey R, Sundaresan P, Donehower RC, Sartorius S, Kennedy MJ, Armstrong DK, Carducci MA, Sorensen JM, Kumor K, Kennedy S, Grochow LB. Phase I trial of the matrix metalloproteinase inhibitor BAY12-9566 in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2001;48:269–274. doi: 10.1007/s002800100330. [DOI] [PubMed] [Google Scholar]
- Herbert JM, Stekel D, Sanderson S, Heath VL, Bicknell R. A novel method of differential gene expression analysis using multiple cDNA libraries applied to the identification of tumour endothelial genes. BMC Genomics. 2008;9:153. doi: 10.1186/1471-2164-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M, Yang E, Matcuk G, Deng D, Sampas N, Tsalenko A, Tabibiazar R, Zhang Y, Chen M, Talbi S, Ho YD, Wang J, Tsao PS, Ben-Dor A, Yakhini Z, Bruhn L, Quertermous T. Identification of endothelial cell genes by combined database mining and microarray analysis. Physiol Genomics. 2003;13:249–262. doi: 10.1152/physiolgenomics.00186.2002. [DOI] [PubMed] [Google Scholar]
- Holthofer H, Virtanen I, Kariniemi AL, Hormia M, Linder E, Miettinen A. Ulex europaeus I lectin as a marker for vascular endothelium in human tissues. Lab Invest. 1982;47:60–66. [PubMed] [Google Scholar]
- Huang YT, Heist RS, Chirieac LR, Lin X, Skaug V, Zienolddiny S, Haugen A, Wu MC, Wang Z, Su L, Asomaning K, Christiani DC. Genome-wide analysis of survival in early-stage non-small-cell lung cancer. J Clin Oncol. 2009;27:2660–2667. doi: 10.1200/JCO.2008.18.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert RS, Vivanco I, Chen E, Rastegar S, Leong K, Mitchell SC, Madraswala R, Zhou Y, Kuo J, Raitano AB, Jakobovits A, Saffran DC, Afar DE. STEAP: a prostate-specific cell-surface antigen highly expressed in human prostate tumors. Proc Natl Acad Sci USA. 1999;96:14523–14528. doi: 10.1073/pnas.96.25.14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huminiecki L, Bicknell R. In silico cloning of novel endothelial-specific genes. Genome Res. 2000;10:1796–1806. doi: 10.1101/gr.150700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CJ, Garbett PK, Nissen B, Schrieber L. Binding of human endothelium to Ulex europaeus I-coated Dynabeads: application to the isolation of microvascular endothelium. J Cell Sci. 1990;96 (Pt 2:257–262. doi: 10.1242/jcs.96.2.257. [DOI] [PubMed] [Google Scholar]
- Janne PA, Meyerson M. ROS1 rearrangements in lung cancer: a new genomic subset of lung adenocarcinoma. J Clin Oncol. 2012;30:878–879. doi: 10.1200/JCO.2011.39.4197. [DOI] [PubMed] [Google Scholar]
- John A, Tuszynski G. The role of matrix metalloproteinases in tumor angiogenesis and tumor metastasis. Pathol Oncol Res. 2001;7:14–23. doi: 10.1007/BF03032599. [DOI] [PubMed] [Google Scholar]
- Johnson DH, Fehrenbacher L, Novotny WF, Herbst RS, Nemunaitis JJ, Jablons DM, Langer CJ, Devore RF, 3rd, Gaudreault J, Damico LA, Holmgren E, Kabbinavar F. Randomized phase II trial comparing bevacizumab plus carboplatin and paclitaxel with carboplatin and paclitaxel alone in previously untreated locally advanced or metastatic non-small-cell lung cancer. J Clin Oncol. 2004;22:2184–2191. doi: 10.1200/JCO.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Jones DT, Lechertier T, Mitter R, Herbert JM, Bicknell R, Jones JL, Li JL, Buffa F, Harris AL, Hodivala-Dilke K. Gene expression analysis in human breast cancer associated blood vessels. PLoS One. 2012;7:e44294. doi: 10.1371/journal.pone.0044294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Chung HS, Sun W, Kim H. Spatiotemporal expression pattern of non-clustered protocadherin family members in the developing rat brain. Neuroscience. 2007;147:996–1021. doi: 10.1016/j.neuroscience.2007.03.052. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovly CM, Carbone DP. Lung cancer in 2010: One size does not fit all. Nat Rev Clin Oncol. 2011;8:68–70. doi: 10.1038/nrclinonc.2010.224. [DOI] [PubMed] [Google Scholar]
- Meyer T. Vascular Disruptive Agents for the Treatment of Cancer. Springer; 2010. [Google Scholar]
- Mura M, Swain RK, Zhuang X, Vorschmitt H, Reynolds G, Durant S, Beesley JF, Herbert JM, Sheldon H, Andre M, Sanderson S, Glen K, Luu NT, Mcgettrick HM, Antczak P, Falciani F, Nash GB, Nagy ZS, Bicknell R. Identification and angiogenic role of the novel tumor endothelial marker CLEC14A. Oncogene. 2012;31:293–305. doi: 10.1038/onc.2011.233. [DOI] [PubMed] [Google Scholar]
- Nair A, Klusmann MJ, Jogeesvaran KH, Grubnic S, Green SJ, Vlahos I. Revisions to the TNM staging of non-small cell lung cancer: rationale, clinicoradiologic implications, and persistent limitations. Radiographics. 2011;31:215–238. doi: 10.1148/rg.311105039. [DOI] [PubMed] [Google Scholar]
- Office for National Statistics . Births and Deaths in England and Wales, 2010 (Statistical Bulletin) Office for National Statistics: London, UK; 2011. [Google Scholar]
- Oh P, Li Y, Yu J, Durr E, Krasinska KM, Carver LA, Testa JE, Schnitzer JE. Subtractive proteomic mapping of the endothelial surface in lung and solid tumours for tissue-specific therapy. Nature. 2004;429:629–635. doi: 10.1038/nature02580. [DOI] [PubMed] [Google Scholar]
- Pallis AG, Syrigos KN. Targeting tumor neovasculature in non-small-cell lung cancer. Crit Rev Oncol Hematol. 2013;86:130–142. doi: 10.1016/j.critrevonc.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Relf M, Lejeune S, Scott PA, Fox S, Smith K, Leek R, Moghaddam A, Whitehouse R, Bicknell R, Harris AL. Expression of the angiogenic factors vascular endothelial cell growth factor, acidic and basic fibroblast growth factor, tumor growth factor beta-1, platelet-derived endothelial cell growth factor, placenta growth factor, and pleiotrophin in human primary breast cancer and its relation to angiogenesis. Cancer Res. 1997;57:963–969. [PubMed] [Google Scholar]
- Safranek J, Pesta M, Holubec L, Kulda V, Dreslerova J, Vrzalova J, Topolcan O, Pesek M, Finek J, Treska V. Expression of MMP-7, MMP-9, TIMP-1 and TIMP-2 mRNA in lung tissue of patients with non-small cell lung cancer (NSCLC) and benign pulmonary disease. Anticancer Res. 2009;29:2513–2517. [PubMed] [Google Scholar]
- Sah NK, Khan Z, Khan GJ, Bisen PS. Structural, functional and therapeutic biology of survivin. Cancer Lett. 2006;244:164–171. doi: 10.1016/j.canlet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Sasaroli D, Gimotty PA, Pathak HB, Hammond R, Kougioumtzidou E, Katsaros D, Buckanovich R, Devarajan K, Sandaltzopoulos R, Godwin AK, Scholler N, Coukos G. Novel surface targets and serum biomarkers from the ovarian cancer vasculature. Cancer Biol Ther. 2011;12:169–180. doi: 10.4161/cbt.12.3.16260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scagliotti G, Brodowicz T, Shepherd FA, Zielinski C, Vansteenkiste J, Manegold C, Simms L, Fossella F, Sugarman K, Belani CP. Treatment-by-histology interaction analyses in three phase III trials show superiority of pemetrexed in nonsquamous non-small cell lung cancer. J Thorac Oncol. 2011;6:64–70. doi: 10.1097/JTO.0b013e3181f7c6d4. [DOI] [PubMed] [Google Scholar]
- Sharma S, Birchmeier C, Nikawa J, O'neill K, Rodgers L, Wigler M. Characterization of the ros1-gene products expressed in human glioblastoma cell lines. Oncogene Res. 1989;5:91–100. [PubMed] [Google Scholar]
- Sottile J. Regulation of angiogenesis by extracellular matrix. Biochim Biophys Acta. 2004;1654:13–22. doi: 10.1016/j.bbcan.2003.07.002. [DOI] [PubMed] [Google Scholar]
- ST Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, Kinzler KW. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beijnum JR, Dings RP, Van Der Linden E, Zwaans BM, Ramaekers FC, Mayo KH, Griffioen AW. Gene expression of tumor angiogenesis dissected: specific targeting of colon cancer angiogenic vasculature. Blood. 2006;108:2339–2348. doi: 10.1182/blood-2006-02-004291. [DOI] [PubMed] [Google Scholar]
- Vasudev NS, Reynolds AR. Anti-angiogenic therapy for cancer: current progress, unresolved questions and future directions. Angiogenesis. 2014;17:471–494. doi: 10.1007/s10456-014-9420-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voellenkle C, Rooij J, Guffanti A, Brini E, Fasanaro P, Isaia E, Croft L, David M, Capogrossi MC, Moles A, Felsani A, Martelli F. Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel microRNAs. RNA. 2012;18:472–484. doi: 10.1261/rna.027615.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang LD, Li B, Sheng ZX. Gefitinib compared with systemic chemotherapy as first-line treatment for chemotherapy-naive patients with advanced non-small cell lung cancer: a meta-analysis of randomised controlled trials. Clin Oncol (R Coll Radiol) 2012;24:396–401. doi: 10.1016/j.clon.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Wasserman SM, Topper JN. Adaptation of the endothelium to fluid flow: in vitro analyses of gene expression and in vivo implications. Vasc Med. 2004;9:35–45. doi: 10.1191/1358863x04vm521ra. [DOI] [PubMed] [Google Scholar]
- Yang D, Holt GE, Velders MP, Kwon ED, Kast WM. Murine six-transmembrane epithelial antigen of the prostate, prostate stem cell antigen, and prostate-specific membrane antigen: prostate-specific cell-surface antigens highly expressed in prostate cancer of transgenic adenocarcinoma mouse prostate mice. Cancer Res. 2001;61:5857–5860. [PubMed] [Google Scholar]
- Yoshida K, Yoshitomo-Nakagawa K, Seki N, Sasaki M, Sugano S. Cloning, expression analysis, and chromosomal localization of BH-protocadherin (PCDH7), a novel member of the cadherin superfamily. Genomics. 1998;49:458–461. doi: 10.1006/geno.1998.5271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.