Abstract
Background:
Tumour cells exclusively express the embryonic M2 isoform of pyruvate kinase (PKM2). PKM2 expression levels have been correlated with the effect of platinum compounds in cancer cell lines and xenograft models. The potential predictive role of PKM2 in patients with metastatic/advanced non-small-cell lung cancer (NSCLC) receiving platinum-based chemotherapy as first-line was investigated.
Methods:
Quantitative real-time PCR was used to assess the expression of PKM2 in tumour samples from 148 and 157 NSCLC patients in the training and the validation set, respectively. All patients received front-line platinum-based chemotherapy. PKM2 mRNA expression was also analysed in a control group of 85 NSCLC patients treated with non-platinum containing regimens.
Results:
In the training set, high PKM2 mRNA levels were associated with decreased progression-free survival (PFS; 4.9 months vs 6.4, P=0.006), overall survival (OS; 10.1 vs 17.0 months, P=0.01) and disease control rate (DCR; 57.7% vs 74.3% P=0.021) compared to patients with low PKM2 levels. In the validation set, high PKM2 mRNA levels were also associated with deceased PFS (3.7 vs 5.9 months, P=0.006), OS (8.3 vs 16.8 months, P=0.003) and DCR (57.7% vs 70.9% P=0.049) compared to those with low PKM2 mRNA levels. There was no correlation between the PKM2 mRNA levels and the PFS (5.6 vs 5.9, P=0.43) or the OS (9.8 vs 10.1, P=0.51) in the control group. Multivariate analysis revealed high PKM2 mRNA expression as an independent predictive factor for the poor patients' outcome.
Conclusions:
PKM2 expression may be a predictive biomarker of platinum sensitivity in advanced NSCLC patients treated with platinum-based chemotherapy.
Keywords: pyruvate kinase M2, glycolysis, NSCLC
Systemic platinum-based chemotherapy remains the mainstay for the treatment of advanced non-small-cell lung cancer (NSCLC) since it improves survival, symptom control and quality of life compared to best supportive care (Schiller et al, 2002). Despite these advances, response to front-line chemotherapy remains poor since patients experience disease progression on an average of 4–6 months from the treatment initiation, and 1-year survival rate is <45% (Fossella et al, 2003).
Cisplatin exerts its cytotoxic effect by reacting with DNA, causing inter- and intra-strand DNA crosslinks that result in the formation of DNA adducts (Ferry et al, 2000). A major limitation of cisplatin efficacy is due to intrinsic or acquired resistance by tumour cells. Multiple mechanisms contribute to cisplatin resistance including decreased drug accumulation due to decreased cellular uptake and increased cellular efflux, increased drug detoxification by thiols and increased DNA repair (Kelland, 2007). In the last decade numerous studies have focused on molecules that are components of the pathways that regulate resistance mechanisms and their potential role as factors that could predict the response to cisplatin treatment.
Metabolic requirements in cancer cells are fundamentally different from those in normal differentiated adult cells since they are characterised by increased glucose uptake and lactate production, regardless of oxygen availability (Levine and Puzio-Kuter, 2010). This altered metabolic phenotype known as ‘aerobic glycolysis' or ‘Warburg effect' is likely preferred by the tumour cells to efficiently convert glucose into the macromolecules needed for tumour growth (Warburg, 1956). One of the most established key regulators of aerobic glycolysis is the embryonic M2 isoform of pyruvate kinase (PKM2), which is preferentially expressed in cancer and in all rapidly proliferating cells (Christofk et al, 2008a). Pyruvate kinase catalyses the last step of glycolysis by the formation of pyruvate and ATP from phosphoenolpyruvate (PEP) and ADP. PKM2 activity is allosterically regulated by binding of fructose 1,6-biphosphate, and also by interactions with tyrosine-phosphorylated proteins in response to growth signals and by post-translational modifications (Christofk et al, 2008b; Lv et al, 2011; Anastasiou et al, 2011, 2012). PKM2 can switch from a highly-active tetrameric form to a low-active dimeric form with low affinity to PEP (Mazurek et al, 2005; Christofk et al, 2008b).
Recently, several studies have reported that PKM2 is more than a regulator of aerobic glycolysis suggesting multiple non-metabolic functions with diverse implications during tumourigenesis (Harris et al, 2011; Luo and Semenza, 2012; Tamada et al, 2012b). PKM2 has been described to act as a transcriptional co-activator and as a protein kinase (Luo et al, 2011; Yang et al, 2011, 2012; Gao et al, 2012), and has also been found to play a role in the control of reactive oxygen species (ROS) concentrations and glutathione antioxidant protection (Anastasiou et al, 2011; Tamada et al, 2012a). The latter appears to imply a possible role of PKM2 to cisplatin resistance.
There are few in vitro studies investigating the tumoural expression of PKM2 and the effect of platinum compounds in cancer cell lines, but the results still remain confusing. Decreased PKM2 protein and activity was found in cisplatin- and oxaliplatin-resistant gastric and colorectal cell lines, respectively (Yoo et al, 2004; Martinez-Balibrea et al, 2009). In contrast, the combination of siRNA targeting the PKM2 and cisplatin increased apoptosis and decreased tumour volume in a lung cancer xenograft model (Guo et al, 2010). In the present study, we investigated the predictive significance of PKM2 mRNA expression in tumours from NSCLC patients treated with front-line platinum-based chemotherapy and provided evidence of its potential role as a predictive biomarker.
Materials and Methods
Patient population
Formalin-fixed, paraffin-embedded (FFPE) tissues from 148 consecutive patients with histologically confirmed stage IIIB (with pleural effusion) and IV NSCLC who were treated with front-line platinum-based chemotherapy were retrospectively collected and analysed (training set). The validation set consisted of 157 NSCLC patients with unresectable stage IIIB (with pleural effusion) or IV from an independent cohort of patients receiving platinum-based chemotherapy doublets in the front-line setting. Furthermore, 85 NSCLC patients who were treated with front-line non-platinum-based doublets were enrolled (control group). The used platinum-based regimens in the three cohorts of patients are presented in Table 1. All the above mentioned patients received front-line treatment in the context of two randomised trials conducted by the Hellenic Oncology Research Group (Georgoulias et al, 2001, 2005). The study was approved by the Ethics and Scientific Committees of the University General Hospital of Heraklion. All patients gave their written informed consent for the use of their tissue for translational research.
Table 1. Characteristics of patients and tumours in the training, validation and control set.
|
Training |
Validation |
Control |
||||
|---|---|---|---|---|---|---|
| Feature | N | % | N | % | N | % |
| 148 | 157 | 85 | ||||
|
Gender | ||||||
| Male | 121 | 82 | 135 | 86 | 78 | 91 |
| Female | 27 | 18 | 22 | 14 | 7 | 8 |
| Median age (range) years | 60 (34–78) |
61 (31–80) |
62 (37–79) |
|||
|
Tumour type | ||||||
| Squamous | 44 | 30 | 44 | 28 | 17 | 20 |
| Non-squamous | 104 | 70 | 113 | 72 | 68 | 80 |
|
ECOG PS | ||||||
| 0–1 | 127 | 86 | 135 | 86 | 80 | 94 |
| 2 | 21 | 14 | 21 | 14 | 5 | 6 |
|
Stage | ||||||
| IIIB (wet) | 38 | 25 | 41 | 26 | 24 | 28 |
| IV | 110 | 75 | 116 | 74 | 61 | 72 |
|
Platinum-based first-line | ||||||
| Platinum+docetaxel | 110 | 74 | 111 | 71 | NA | |
| Platinum+docetaxel+Avastin | 18 | 12 | 22 | 14 | NA | |
| Platinum+gemcitabine | 15 | 10 | 17 | 11 | NA | |
| Platinum+pemetrexed | 5 | 4 | 7 | 4 | NA |
|
|
Non-platinum-based first-line | ||||||
| Docetaxel+gemcitabine | NA |
NA |
85 |
100 |
||
| Post-progression treatment | 100 | 68 | 105 | 67 | 55 | 65 |
|
EGFR mutational status | ||||||
| EGFR wt | 27 | 34 | 10 | |||
| EGFR mut | 4 | 4 | 1 | |||
| EGFR uknown | 117 | 119 | 74 | |||
|
PKM2 mRNA expression | ||||||
| Median (range) | 12.77 (0.34–71.88) |
NA |
12.73 (0.36–70.34) |
|||
| High expression | 74 | 50 | 78 | 49 | 42 | 49 |
| Low expression | 74 | 50 | 79 | 51 | 43 | 51 |
Abbreviations: ECOG PS=Eastern Cooperative Oncology Group Performance Status; NA=not applicable.
Specimen selection and RNA extraction
Formalin-fixed, paraffin-embedded tumour sections were examined by a pathologist (EL) in order to identify the most appropriate tumour areas for dissection. Serial sections of 5 μm were prepared and stained with Nuclear Fast Red (Sigma-Aldrich, St Lious, MO, USA). In the case of samples with <80% tumour cells, an Eppendorf piezoelectric microdissector (Eppendorf, Hamburg, Germany) was used to procure only malignant cells. TRIzol LS (Invitrogen, Carlsbad, CA, USA) was used for RNA extraction, followed by DNase (DNA-free, Ambion, Austin, TX, USA) treatment in order to avoid genomic DNA contamination (Papadaki et al, 2009).
mRNA expression analysis
Reverse transcription and RT–qPCR have been described elsewhere (Papadaki et al, 2009). Briefly, 200 ng of total RNA were used for cDNA synthesis using SuperScript III reverse transcriptase (Invitrogen). Relative cDNA quantification for PKM2 and β-actin and phosphoglycerate kinase 1 (PGK) as internal controls was performed using the ABI Prism 7900 HT Sequence Detection System (AB, Foster City, CA, USA).
The primers and 5′-labelled fluorescent reporter dye (6FAM) probe sets were designed using the Primer Express 2.0 Software (AB) according to the Ref Seq NM_002654 for PKM2 and were as follows: PKM2, 5′-GCCATAATCGTCCTCACCAAGT-3′ (forward), 5′-GCACGTGGGCGGTATCTG-3′ (reverse) and 5′-CAGGTCTGCTCACCAGG-3′ (probe). The primers and probe sequences for both housekeeping genes, β-actin and PGK have been reported elsewhere (Saridaki et al, 2011). Comparative Ct method was used for gene expression analysis using both β-actin and PGK as reference genes and commercial RNA (Stratagene, La Jolla, CA, USA) as calibrators. Final expression values were determined as follows: 2−(ΔCT sample−ΔCT calibrator), where ΔCT values of the calibrator and sample were determined by subtracting the CT value of the target gene from the mean value of both reference genes. In all experiments, only triplicates with a s.d. of the CT value <0.25 were accepted. In addition, genomic DNA contamination was excluded by including non-reverse-transcribed RNA as a control for each sample.
Immunohistochemistry and staining evaluation
Approximately, 4 μm-thick FFPE tissue sections were stained with haematoxylin and eosin and histopathologically verified by a pathologist. The primary antibody used for PKM2 staining was directed against the specific sequence of exon 9 that is unique to PKM2 (rabbit polyclonal Ab, cat. no. 3198, Cell Signaling, Danvers, MA, USA; dilution 1 : 600). Immunostaining was performed using Ultra Vision LP Quanto Detection System HRP Polymer (Thermo Fisher Scientific, Fremont, CA, USA). Sections stained for PKM2 were previously treated in Tris-EDTA buffer (pH 9.0) for 13 min. Staining evaluation was performed by two independent pathologists (EL and AK) blinded to each other's scores and to each patient's clinical information. Staining of PKM2 was scored as the product of the intensity on a scale of 0–5: low and 5.1–8: high expression, as described elsewhere (Yang et al, 2011).
Study design and statistical analysis
Progression-free survival (PFS) and overall survival (OS) were calculated from the start of treatment to the first documented disease progression or death, respectively. Objective responses were recorded according to the RECIST criteria (Therasse et al, 2000). Cutoff points were calculated according to the median value for the mRNA expression. Samples with mRNA expression above or equal to the median were considered as samples with high expression, while those with value below the median as samples with low expression. All the laboratory analyses were performed blinding to the clinical data.
The potential association between baseline characteristics, response and gene expression levels were compared with either the two-sided Fisher's exact test or the chi-square test for categorical variables and the Kruskal–Wallis test for continuous variables. The normality of continuous variables was verified with the Kolmogorov–Smirnov test. The association of risk factors with time-to-event end points was analysed with the log-rank test and the Kaplan–Meier method was used to plot the corresponding time-to-progression and survival curves. A univariate Cox regression analysis, with hazard ratios (HRs) and 95% confidence intervals (CIs), was used to assess the association between each potential prognostic factor and survival and time to progression. These factors were then included in a multivariate Cox proportional hazards regression model with a stepwise procedure (both forward and backward) to evaluate the independent significance of different variables on survival and time to progression. Statistical significance was set at P=0.05.
Results
Patients' characteristics and clinical features
The main clinical characteristics and gene mRNA levels of PKM2 in the three cohorts of patients are summarised in Table 1. Among the three groups analysed, the median age ranged from 60 to 62 years old and patients were predominantly males. All three groups were predominantly constituted from non-squamous type of tumours and most patients had stage IV disease. The EGFR mutation status of the tested patients is also shown in Table 1.
PKM2 mRNA expression and patients' outcome
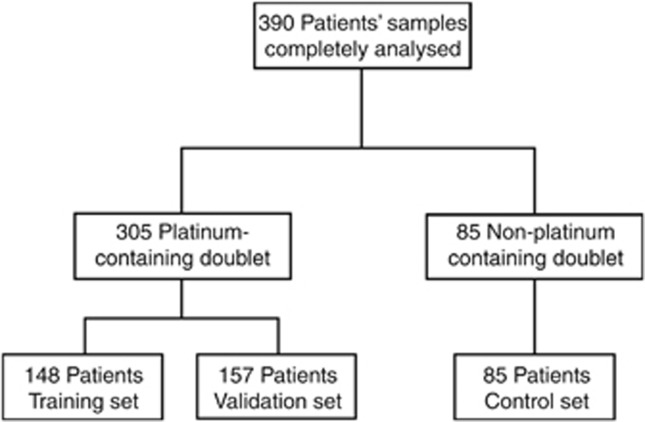
In total, 390 NSCLC patients were treated with front-line platinum and/or non-platinum containing doublets in the context of two randomised trials conducted by the Hellenic Oncology Research Group. Among the 305 patients treated with platinum containing doublets, 148 and 157 of them were analysed as the training and the validation set, respectively. The remaining 85 platinum-naive patients constituted the control set (Figure 1). PKM2 mRNA expression was successfully assessed in all of the samples analysed and the level of expression was associated with PFS and OS.
Figure 1.
Flow chart of NSCLC patients analysed for PKM2 mRNA expression.
The median mRNA expression level was 12.77 (minimum, maximum: 0.34, 71.88) for the training set and the same cutoff was used for the analysis of the validation and control sets. In 30 tumour specimens, which were randomly selected from the training and the validation sets that were stained for PKM2 protein expression by immunohistochemistry, there was no correlation between PKM2 mRNA and protein expression (Spearman's test, P=0.275; data not shown). Moreover, there was no significant correlation between the PKM2 mRNA expression and the patients' age, gender, tumour histology, stage and PS (all P-values >0.05).
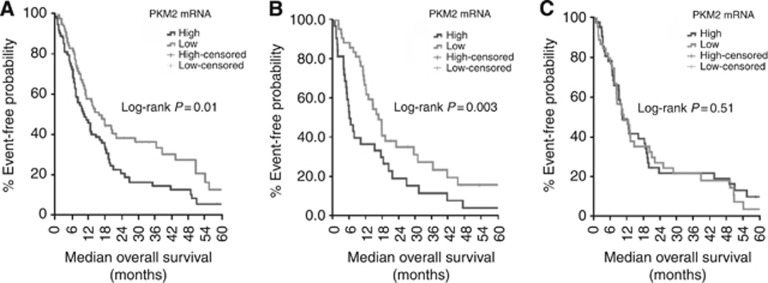
In the training set, patients with high tumoural PKM2 mRNA levels had significantly shorter median PFS (4.9 vs 6.4 months; P=0.006; Figure 2A) compared to patients with low tumoural PKM2 mRNA levels. Similarly, patients with high mRNA expression of PKM2 were significantly associated with decreased median OS (10.1 vs 17.0; P=0.01; Figure 3A) compared to the patients with low expression levels. On the contrary, there was no significant correlation between the PKM2 mRNA levels and the objective response to cisplatin-based chemotherapy (P=0.497; Table 2). However, when the analysis was performed according to the clinical benefit (DCR) a significant correlation was observed between PKM2 mRNA expression levels and DCR (Table 2; 74.3% and 57.7% DCR for patients with low and high PKM2 mRNA expression, respectively; P=0.021).
Figure 2.
(A) PFS according to PKM2 mRNA expression in the training set. (B) PFS according to PKM2 mRNA expression in the validation set. (C) PFS according to PKM2 mRNA expression in the control set. Higher levels of PKM2 mRNA were associated with decreased PFS in the training (A) and in validation set (B) but not in the control group (C).
Figure 3.
(A) OS according to PKM2 mRNA expression in the training set. (B) OS according to PKM2 mRNA expression in the validation set. (C) OS according to PKM2 mRNA expression in the control set. Higher levels of PKM2 mRNA were associated with decreased OS in the training (A) and validation set (B) but not in the control group (C).
Table 2. ORR and DCR according to PKM2 mRNA expression in the training, validation set and whole population.
|
ORR,
N
(%) |
DCR,
N
(%) |
|||||
|---|---|---|---|---|---|---|
| PKM2 mRNA expression | CR+PR | SD+PD | P | CR+PR+SD | PD | P |
|
Training | ||||||
| High | 32 (43.2) | 42 (56.8) | 0.497 | 42 (57.7) | 32 (42.3) | 0.021 |
| Low | 31 (41.9) | 43 (58.1) | 55 (74.3) | 19 (25.7) | ||
|
Validation | ||||||
| High | 25 (32.1) | 53 (67.9) | 0.390 | 45 (57.7) | 33 (42.3) | 0.049 |
| Low | 28 (35.4) | 51 (64.6) | 56 (70.9) | 23 (29.1) | ||
|
Whole population | ||||||
| High | 57 (37.5) | 95 (62.5) | 0.590 | 87 (57.2) | 65 (42.3) | 0.014 |
| Low | 59 (38.6) | 94 (61.4) | 111 (72.5) | 42 (27.5) | ||
Abbreviations: CR=complete response; DCR=disease control rate; ORR=objective response rate; PD=progressive disease; PR=partial response; SD=stable disease.
Results in the validation set were similar to those in the training set. Median PFS was significantly decreased in patients with high mRNA expression of PKM2 (3.7 vs 5.9 months; P=0.006; Figure 2B) in comparison with those with low mRNA levels. Furthermore, patients with high PKM2 mRNA levels had significantly decreased median OS (8.3 vs 16.8; P=0.003; Figure 3B) as compared with those whose tumours had low PKM2 mRNA levels. Similarly to the training set, although there was no significant correlation between PKM2 mRNA levels and objective response rate (P=0.390; Table 2) in the validation set, a marginal correlation with the DCR (70.9% vs 57.7% P=0.049; Table 2) was observed.
Finally, the analysis of the whole group of patients (after combining the training and the validation sets) clearly revealed that high PKM2 mRNA expression was associated with decreased PFS (3.9 vs 6.3 months; P=0.001), OS (9.6 vs 16.8 months; P<0.001) and DCR (57.2% vs 72.5% P=0.014).
The expression values of PKM2 mRNA in the control set were quite similar with that recorded in the training set since the median mRNA expression level for the control set was 12.73 (minimum, maximum: 0.46, 72.17) and no significant difference was observed in comparison with that of the training set (P=0.057). Despite that, and as opposed to the patients treated with platinum-based regimens both in the training and validation sets, there was no difference in terms of PFS (5.6 vs 5.9; P=0.43; Figure 2C) and OS (9.8 vs 10.1; P=0.51; Figure 3C) among patients with low and high PKM2 mRNA expression.
Univariate and multivariate analysis
Univariate analysis in the whole group of patients enrolled in both the training and validation sets revealed that patients with high tumoural PKM2 mRNA expression levels (HR: 1.89, 95% CI: 1.48–2.27; P=0.003), PS of 2 (HR: 2.64, 95% CI: 1.89–3.17; P=0.001) as well as stage IV (HR: 1.77, 95% CI: 1.11–2.31; P=0.03) were significantly associated with decreased PFS, whereas age >70 years (P=0.77), gender (P=0.51), histology (P=0.61) and tumour differentiation (P=0.14) did not show any significant correlation with the PFS (Table 3). Similarly, high PKM2 mRNA expression (HR: 1.93, 95% CI: 1.54–2.46; P=0.002) and PS of 2 (HR: 2.87, 95% CI: 1.92–3.44; P<0.001) were significantly associated with decreased OS. In contrast, age >70 years (P=0.23), gender (P=0.48), stage IV (P=0.17), histology (P=0.94) and tumour differentiation (P=0.31) were not significantly associated with decreased OS (Table 3).
Table 3. Univariate analysis for PFS and OS.
| Hazard ratio | 95% CI | P-value | |
|---|---|---|---|
|
PFS | |||
| PKM2 expression (high vs low) | 1.89 | 1.48–2.27 | 0.003 |
| PS (2 vs 0–1) | 2.64 | 1.89–3.17 | 0.001 |
| Age (>70 vs ⩽70 years) | 1.24 | 0.87–1.62 | 0.77 |
| Gender (male vs female) | 1.19 | 0.84–1.41 | 0.51 |
| Stage (IV vs IIIB) | 1.77 | 1.11–2.31 | 0.03 |
| Histology (squamous vs non-squamous) | 1.32 | 0.85–1.54 | 0.61 |
| Tumour differentiation (low vs well-moderate) | 1.41 | 0.90–1.77 | 0.14 |
|
OS | |||
| PKM2 expression (high vs low) | 1.93 | 1.54–2.46 | 0.002 |
| PS (2 vs 0–1) | 2.87 | 1.92–3.44 | <0.001 |
| Age (>70 vs ⩽70 years) | 1.35 | 0.91–1.88 | 0.23 |
| Gender (male vs female) | 1.16 | 0.81–1.33 | 0.48 |
| Stage (IV vs IIIB) | 1.45 | 0.96–1.85 | 0.17 |
| Histology (squamous vs non-squamous) | 1.17 | 0.89–1.28 | 0.94 |
| Tumour differentiation (low vs well-moderate) | 1.32 | 0.87–1.32 | 0.31 |
Abbreviations: CI=confidence interval; OS=overall survival; PFS=progression-free survival; PS=performance status.
Multivariate Cox regression analysis revealed that high tumoural PKM2 mRNA expression (HR: 1.81, 95% CI: 1.40–2.38; P=0.002) as well as PS of 2 (HR: 3.97, 95% CI: 2.77–4.16; P<0.001) emerged as independent predictive factors for decreased PFS (Table 4). Similarly, high PKM2 mRNA levels (HR: 1.97, 95% CI: 1.45–2.46; P=0.001) and PS of 2 (HR: 4.01, 95% CI: 3.56–5.06; P<0.001; Table 4) were revealed as independent predictive factors for shorter OS.
Table 4. Multivariate analysis for PFS and OS.
| Hazard ratio | 95% CI | P-value | |
|---|---|---|---|
|
PFS | |||
| PKM2 expression (high vs low) | 1.81 | 1.40–2.38 | 0.002 |
| PS (2 vs 0–1) | 3.97 | 2.77–4.16 | <0.001 |
| Stage (IV vs IIIB) | 1.34 | 0.96–1.88 | 0.13 |
|
OS | |||
| PKM2 expression (high vs low) | 1.97 | 1.45–2.46 | 0.001 |
| PS (2 vs 0–1) | 4.01 | 3.56–5.06 | <0.001 |
Abbreviations: CI=confidence interval; OS=overall survival; PFS=progression-free survival; PS=performance status.
Discussion
In the current study, we investigated the role of tumoural PKM2 mRNA expression levels as a predictive factor in the outcome of metastatic NSCLC patients treated with front-line platinum-based chemotherapy. In a training set of 148 patients, those with tumours having low mRNA levels of PKM2 presented significantly higher PFS (P=0.006) and OS (P=0.01) and DCR (P=0.021) as well. Our results were confirmed in an independent cohort of 157 patients who have also been treated with cisplatin-based chemotherapy in the first-line setting. Patients with low PKM2 mRNA levels attained statistically significant increase of PFS (P=0.006) and OS (P=0.003) and higher DCR (P=0.049) as well. Unlike the results in the training and validation set, in the control group of 85 patients, who did not received platinum-based chemotherapy, PKM2 mRNA levels were not correlated with PFS (P=0.43) and OS (P=0.51). This observation clearly suggests that the predictive value of PKM2 mRNA levels is mainly related to the platinum compounds. This association could not be attributed to a possible effect of tyrosine kinase inhibitors administered to our patients since the number of patients who received this anti-EGFR treatment was very low. Furthermore, multivariate analysis revealed that high PKM2 mRNA expression was an independent predictive factor for shorter PFS and decreased OS in both the training and validation sets.
PKM2 is one of the four isoforms of pyruvate kinase, the enzyme that catalyses the formation of pyruvate and ATP from PEP and ADP (Mazurek, 2010). The four isoforms of pyruvate kinase are encoded by two genes that are expressed in a cell- and tissue-specific manner. The L and R isoenzymes, derived from the PKLR gene are expressed in the liver and red blood cells, respectively (Noguchi et al, 1987). The PKM gene encodes the M1- and M2-type isoenzymes (Noguchi et al, 1986). It consists of 12 exons, of which 9 and 10 are alternatively spliced in a mutually exclusive manner to give rise to the PKM1 and PKM2 isoforms, respectively (Noguchi et al, 1986). Alternative splicing in PKM gene is regulated by heterogeneous nuclear ribonucleoproteins hnRNP, under the control of c-Myc (Clower et al, 2010; David et al, 2010). From the four isoforms of pyruvate kinase, cancer cells exclusively express the M2 isoform (Christofk et al, 2008a). PKM2 but not PKM1 is necessary for aerobic glycolysis since the replacement of PKM2 by PKM1 reduced the capacity of tumour cell lines to develop into a tumour (Christofk et al, 2008a). PKM2 is negatively regulated in response to growth factors by binding to tyrosine-phosphorylated proteins (Christofk et al, 2008b; Hitosugi et al, 2009). Phosphorylation of PKM2 results in the formation of its inactive dimeric form that enables the diversion of glycolytic intermediates into anabolic pathways (Hitosugi et al, 2009; Mazurek, 2010). Also, PKM2 enzymatic activity can be modulated by a variety of post-translational modifications such as acetylation, sumoylation, ubiquitination and oxidation (Anastasiou et al, 2011; Lv et al, 2011; Luo and Semenza, 2012; Yang and Lu, 2013). The lack of significant correlation between PKM2 mRNA and protein expression observed in the current study could be explained on the basis of post-translational modifications mentioned above. It is obvious that IHC and mRNA expression analysis in a larger cohort of patients would confirm this observation.
The role of PKM2 to modulate the cytotoxicity of cisplatin and its derivatives is as yet not fully explored. Proteomic analysis showed that PKM2 is downregulated in oxaliplatin-resistant cell lines, while high mRNA expression was associated with higher response rate in oxaliplatin-treated colorectal cancer patients (Martinez-Balibrea et al, 2009). In the same line of evidence it was shown that PKM2 protein and activity were lower in cisplatin-resistant human gastric carcinoma cell lines (Yoo et al, 2004). In contrast, our results are in the opposite direction, since low PKM2 mRNA levels were associated with better outcome of NSCLC patients both in the training and validation set. This evidence is in agreement with previous results from our laboratory on SCLC patients treated with platinum-based chemotherapy, since patients with low expression levels of PKM2 attained significantly better PFS and OS (Karachaliou et al, 2013). Accordingly, results from previous studies showed that inhibition of PKM2 mRNA expression by siRNA targeting in combination with chemotherapeutic agents, significantly increased apoptosis and decreased tumour volume in xenograft models (Guo et al, 2010).
Several mechanisms of resistance to platinum compounds, either intrinsically or acquired, have been described. Decreased membrane transport, increased cytoplasmic detoxification, increased DNA repair activity and increased tolerance to DNA damage are the major mechanisms that can contribute to cisplatin resistance (Siddik, 2003; Kelland, 2007). Excision repair complementation group 1 (ERCC1) and breast cancer susceptibility gene 1 (BRCA1) that participate in DNA-repair pathways have been considered as potential predictive factors, since their level of expression seems to be correlated and influence cisplatin efficacy in a variety of solid tumours (Postel-Vinay et al, 2012). It is also well established that platinum analogues interact with sulfur-containing thiomolecules such as glutathione and metallothionein leading to its inactivation and subsequent failure from binding to DNA (Siddik, 2003; Yang et al, 2006; Kelland, 2007). Also, studies in preclinical models and tumour samples suggest that elevated levels of glutathione or glutathione-related enzymes are associated with limited cisplatin efficacy (Godwin et al, 1992; Yang et al, 2005, 2006). Furthermore, glutathione as an antioxidant offers protection from ROS that are known to accumulate and induce apoptosis after radiotherapy and chemotherapeutic drug treatment (Bragado et al, 2007; Choi et al, 2007). Recent studies suggest that PKM2 might play a role in the control of glutathione and ROS concentrations, implying a possible role to cisplatin resistance. In human lung cancer cells, increased concentrations of ROS can inhibit PKM2 activity through oxidation of cysteine 358 (Cys358; Anastasiou et al, 2011). This resulted in the diversion of glucose intermediates into pentose phosphate pathway (PPP), which produces nicotinamide adenine dinucleotide phosphate (NADPH). Nicotinamide adenine dinucleotide phosphate provides reducing equivalents for the reduction of oxidised GSH (GSSG) to reduced GSH, thereby increasing ROS detoxification (Anastasiou et al, 2011). The above regulation mechanism of ROS concentrations by PKM2 seems to be specific since an oxidation-resistant mutant form of PKM2 failed to confer antioxidant response (Anastasiou et al, 2011). Additionally, PKM2 has been described to regulate ROS accumulation by interacting with the cell-surface marker of stem cells, CD44 (Tamada et al, 2012a, 2012b). This interaction promotes glycolysis and increases the flux to PPP resulting to the production of NADPH and to the subsequent increase of reduced GSH and decrease ROS accumulation (Tamada et al, 2012a). The role of CD44 to inhibit ROS accumulation in cancer cells has also been addressed earlier (Ishimoto et al, 2011). It is questionable if the observed association between tumoural PKM2 mRNA expression and the poor patient's outcome in our study could be explained on the basis of the resistance mechanisms described above. Preliminary data from our laboratory strengthens this possibility since they demonstrate a strong positive correlation between PKM2 and CD44 expression in ovarian tumour specimens from patients treated with cisplatin in front line (data not shown).
Tumour cells have multiple ways to regulate PKM2 both in transcriptional and post-transcriptional level, in order to ensure anabolic metabolism and survival under hypoxic conditions, PKM2 acting in a positive feedback loop interacts with HIF1α through the prolyl hydroxylase 3 and promotes the activation of HIF1α regulated genes (Luo et al, 2011). This positive feedback loop maintains expression of PKM2 and other glycolytic enzymes in high levels. Accordingly, the above mechanism seems to be biologically relevant since preliminary data from our laboratory have demonstrated that there is a strong positive correlation between PKM2 and HIF1α mRNA expression in NSCLC tumour samples (data not shown). A strong positive correlation also exist between PKM2 and c-Myc mRNA expression (data not shown). Therefore, as argued by our data, measuring PKM2 mRNA levels by RT–PCR in clinical samples seem to be reasonable.
Although our results are retrospectively originated, in the best of our knowledge this study is the first one providing evidence for the predictive significance of a biomarker by validating the results in an independent cohort of patients. Also, unpublished data from our laboratory have evaluated the predictive significance of PKM2 in other tumour types treated with cisplatin or its derivatives. However, this evidence has to be interpreted with caution and any clinical relevance of the tumoural PKM2 mRNA expression should be validated prospectively. Furthermore, it is an interesting issue to investigate whether PKM2 mRNA are influenced by chemotherapy by using a rebiopsy in the metastatic tumour. In addition, it remains a challenge that has to be answered using in vitro models, to elucidate at which level of PKM2 regulation, either transcriptional or post-transcriptional, could modulate anticancer-drug cytotoxicity.
Acknowledgments
This work was partially supported by a research grant from the Hellenic Society for Medical Oncology (HeSMO) and the Cretan Association for Biomedical Research (CABR).
The authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, Thomas CJ, Vander Heiden MG, Cantley LC. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334 (6060:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, Yang H, Mattaini KR, Metallo CM, Fiske BP, Courtney KD, Malstrom S, Khan TM, Kung C, Skoumbourdis AP, Veith H, Southall N, Walsh MJ, Brimacombe KR, Leister W, Lunt SY, Johnson ZR, Yen KE, Kunii K, Davidson SM, Christofk HR, Austin CP, Inglese J, Harris MH, Asara JM, Stephanopoulos G, Salituro FG, Jin S, Dang L, Auld DS, Park HW, Cantley LC, Thomas CJ, Vander Heiden MG. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8 (10:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragado P, Armesilla A, Silva A, Porras A. Apoptosis by cisplatin requires p53 mediated p38alpha MAPK activation through ROS generation. Apoptosis. 2007;12 (9:1733–1742. doi: 10.1007/s10495-007-0082-8. [DOI] [PubMed] [Google Scholar]
- Choi KJ, Piao YJ, Lim MJ, Kim JH, Ha J, Choe W, Kim SS. Overexpressed cyclophilin A in cancer cells renders resistance to hypoxia- and cisplatin-induced cell death. Cancer Res. 2007;67 (8:3654–3662. doi: 10.1158/0008-5472.CAN-06-1759. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452 (7184:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452 (7184:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci USA. 2010;107 (5:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463 (21:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry KV, Hamilton TC, Johnson SW. Increased nucleotide excision repair in cisplatin-resistant ovarian cancer cells: role of ERCC1-XPF. Biochem Pharmacol. 2000;60 (9:1305–1313. doi: 10.1016/s0006-2952(00)00441-x. [DOI] [PubMed] [Google Scholar]
- Fossella F, Pereira JR, von Pawel J, Pluzanska A, Gorbounova V, Kaukel E, Mattson KV, Ramlau R, Szczesna A, Fidias P, Millward M, Belani CP. Randomized, multinational, phase III study of docetaxel plus platinum combinations versus vinorelbine plus cisplatin for advanced non-small-cell lung cancer: the TAX 326 study group. J Clin Oncol. 2003;21 (16:3016–3024. doi: 10.1200/JCO.2003.12.046. [DOI] [PubMed] [Google Scholar]
- Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45 (5:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgoulias V, Ardavanis A, Tsiafaki X, Agelidou A, Mixalopoulou P, Anagnostopoulou O, Ziotopoulos P, Toubis M, Syrigos K, Samaras N, Polyzos A, Christou A, Kakolyris S, Kouroussis C, Androulakis N, Samonis G, Chatzidaki D. Vinorelbine plus cisplatin versus docetaxel plus gemcitabine in advanced non-small-cell lung cancer: a phase III randomized trial. J Clin Oncol. 2005;23 (13:2937–2945. doi: 10.1200/JCO.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Georgoulias V, Papadakis E, Alexopoulos A, Tsiafaki X, Rapti A, Veslemes M, Palamidas P, Vlachonikolis I. Platinum-based and non-platinum-based chemotherapy in advanced non-small-cell lung cancer: a randomised multicentre trial. Lancet. 2001;357 (9267:1478–1484. doi: 10.1016/S0140-6736(00)04644-4. [DOI] [PubMed] [Google Scholar]
- Godwin AK, Meister A, O'Dwyer PJ, Huang CS, Hamilton TC, Anderson ME. High resistance to cisplatin in human ovarian cancer cell lines is associated with marked increase of glutathione synthesis. Proc Natl Acad Sci USA. 1992;89 (7:3070–3074. doi: 10.1073/pnas.89.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Zhang Y, Chen T, Wang Y, Xue J, Zhang Y, Xiao W, Mo X, Lu Y. Efficacy of RNAi targeting of pyruvate kinase M2 combined with cisplatin in a lung cancer model. J Cancer Res Clin Oncol. 2010;137 (1:65–72. doi: 10.1007/s00432-010-0860-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris I, McCracken S, Mak TW. PKM2: a gatekeeper between growth and survival. Cell Res. 2011;22 (3:447–449. doi: 10.1038/cr.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, Xie J, Gu TL, Polakiewicz RD, Roesel JL, Boggon TJ, Khuri FR, Gilliland DG, Cantley LC, Kaufman J, Chen J. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2 (97:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H, Oshima M, Ikeda T, Asaba R, Yagi H, Masuko T, Shimizu T, Ishikawa T, Kai K, Takahashi E, Imamura Y, Baba Y, Ohmura M, Suematsu M, Baba H, Saya H. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell. 2011;19 (3:387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- Karachaliou N, Papadaki C, Lagoudaki E, Trypaki M, Sfakianaki M, Koutsopoulos A, Mavroudis D, Stathopoulos E, Georgoulias V, Souglakos J. Predictive value of BRCA1, ERCC1, ATP7B, PKM2, TOPOI, TOPOmicron-IIA, TOPOIIB and C-MYC genes in patients with small cell lung cancer (SCLC) who received first line therapy with cisplatin and etoposide. PLoS One. 2013;8 (9:e74611. doi: 10.1371/journal.pone.0074611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7 (8:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330 (6009:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145 (5:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Semenza GL. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol Metab. 2012;23 (11:560–566. doi: 10.1016/j.tem.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H, Zha Z, Liu Y, Li Z, Xu Y, Wang G, Huang Y, Xiong Y, Guan KL, Lei QY. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42 (6:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balibrea E, Plasencia C, Gines A, Martinez-Cardus A, Musulen E, Aguilera R, Manzano JL, Neamati N, Abad A. A proteomic approach links decreased pyruvate kinase M2 expression to oxaliplatin resistance in patients with colorectal cancer and in human cell lines. Mol Cancer Ther. 2009;8 (4:771–778. doi: 10.1158/1535-7163.MCT-08-0882. [DOI] [PubMed] [Google Scholar]
- Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2010;43 (7:969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15 (4:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261 (29:13807–13812. [PubMed] [Google Scholar]
- Noguchi T, Yamada K, Inoue H, Matsuda T, Tanaka T. The L- and R-type isozymes of rat pyruvate kinase are produced from a single gene by use of different promoters. J Biol Chem. 1987;262 (29:14366–14371. [PubMed] [Google Scholar]
- Papadaki C, Mavroudis D, Trypaki M, Koutsopoulos A, Stathopoulos E, Hatzidaki D, Tsakalaki E, Georgoulias V, Souglakos J. Tumoral expression of TXR1 and TSP1 predicts overall survival of patients with lung adenocarcinoma treated with first-line docetaxel-gemcitabine regimen. Clin Cancer Res. 2009;15 (11:3827–3833. doi: 10.1158/1078-0432.CCR-08-3027. [DOI] [PubMed] [Google Scholar]
- Postel-Vinay S, Vanhecke E, Olaussen KA, Lord CJ, Ashworth A, Soria JC. The potential of exploiting DNA-repair defects for optimizing lung cancer treatment. Nat Rev Clin Oncol. 2012;9 (3:144–155. doi: 10.1038/nrclinonc.2012.3. [DOI] [PubMed] [Google Scholar]
- Saridaki Z, Tzardi M, Papadaki C, Sfakianaki M, Pega F, Kalikaki A, Tsakalaki E, Trypaki M, Messaritakis I, Stathopoulos E, Mavroudis D, Georgoulias V, Souglakos J. Impact of KRAS, BRAF, PIK3CA mutations, PTEN, AREG, EREG expression and skin rash in ⩾ 2 line cetuximab-based therapy of colorectal cancer patients. PLoS One. 2011;6 (1:e15980. doi: 10.1371/journal.pone.0015980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346 (2:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22 (47:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- Tamada M, Nagano O, Tateyama S, Ohmura M, Yae T, Ishimoto T, Sugihara E, Onishi N, Yamamoto T, Yanagawa H, Suematsu M, Saya H. Modulation of glucose metabolism by CD44 contributes to antioxidant status and drug resistance in cancer cells. Cancer Res. 2012;72 (6:1438–1448. doi: 10.1158/0008-5472.CAN-11-3024. [DOI] [PubMed] [Google Scholar]
- Tamada M, Suematsu M, Saya H. Pyruvate kinase M2: multiple faces for conferring benefits on cancer cells. Clin Cancer Res. 2012;18 (20:5554–5561. doi: 10.1158/1078-0432.CCR-12-0859. [DOI] [PubMed] [Google Scholar]
- Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92 (3:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123 (3191:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Yang W, Lu Z. Regulation and function of pyruvate kinase M2 in cancer. Cancer Lett. 2013;339:153–158. doi: 10.1016/j.canlet.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Ebbert JO, Sun Z, Weinshilboum RM. Role of the glutathione metabolic pathway in lung cancer treatment and prognosis: a review. J Clin Oncol. 2006;24 (11:1761–1769. doi: 10.1200/JCO.2005.02.7110. [DOI] [PubMed] [Google Scholar]
- Yang W, Toffa SE, Lohn JW, Seifalian AM, Winslet MC. Malignant ascites increases the antioxidant ability of human ovarian (SKOV-3) and gastric adenocarcinoma (KATO-III) cells. Gynecol Oncol. 2005;96 (2:430–438. doi: 10.1016/j.ygyno.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480 (7375:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14 (12:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BC, Ku JL, Hong SH, Shin YK, Park SY, Kim HK, Park JG. Decreased pyruvate kinase M2 activity linked to cisplatin resistance in human gastric carcinoma cell lines. Int J Cancer. 2004;108 (4:532–539. doi: 10.1002/ijc.11604. [DOI] [PubMed] [Google Scholar]