A ripening-related transcription factor affects the phenylpropanoid synthesis pathway and the biosynthesis of eugenol in the strawberry fruit.
Abstract
Eugenol is a volatile phenylpropanoid that contributes to flower and ripe fruit scent. In ripe strawberry (Fragaria × ananassa) fruit receptacles, eugenol is biosynthesized by eugenol synthase (FaEGS2). However, the transcriptional regulation of this process is still unknown. We have identified and functionally characterized an R2R3 MYB transcription factor (EMISSION OF BENZENOID II [FaEOBII]) that seems to be the orthologous gene of PhEOBII from Petunia hybrida, which contributes to the regulation of eugenol biosynthesis in petals. The expression of FaEOBII was ripening related and fruit receptacle specific, although high expression values were also found in petals. This expression pattern of FaEOBII correlated with eugenol content in both fruit receptacle and petals. The expression of FaEOBII was repressed by auxins and activated by abscisic acid, in parallel to the ripening process. In ripe strawberry receptacles, where the expression of FaEOBII was silenced, the expression of CINNAMYL ALCOHOL DEHYDROGENASE1 and FaEGS2, two structural genes involved in eugenol production, was down-regulated. A subsequent decrease in eugenol content in ripe receptacles was also observed, confirming the involvement of FaEOBII in eugenol metabolism. Additionally, the expression of FaEOBII was under the control of FaMYB10, another R2R3 MYB transcription factor that regulates the early and late biosynthetic genes from the flavonoid/phenylpropanoid pathway. In parallel, the amount of eugenol in FaMYB10-silenced receptacles was also diminished. Taken together, these data indicate that FaEOBII plays a regulating role in the volatile phenylpropanoid pathway gene expression that gives rise to eugenol production in ripe strawberry receptacles.
The octoploid cultivated strawberry (Fragaria × ananassa) is one of the most economically important, nonclimacteric, soft fruits, in which volatile compounds influence fruit flavor and aroma. Both characteristics contribute to the fruit organoleptic traits and are crucial factors to determine fruit quality.
At present, extensive surveys on the components that contribute to strawberry flavor have been performed. In these studies, more than 360 volatiles have been identified (Latrasse, 1991; Nijssen, 1996; Zabetakis and Holden, 1997), but only 15 to 20 of them in wild varieties of strawberry are believed to be essential for sensory quality, together with nonvolatile sugars and organic acids (Schieberle and Hofmann, 1997). In contrast, in cultivated varieties of strawberry, only about six odor-active compounds have been identified as contributors to fruit flavor (Raab et al., 2006; Ulrich et al., 2007). Strawberry aroma is the result of the combined perception of fruity (ethyl butanoate, ethyl hexanoate, and methyl 2-methylbutanoat), green (Z-3-hexenal), sweaty (butanoic acid and 2-methylbutanoic acid), peach-like (γ-decalactone), and caramel-like [4-hydroxy-2,5-dimethyl-3(2H)-furanone and 2,5-dimethyl-4-methoxy-3(2H)-furanone] flavor notes (Pyysalo et al., 1979; Larsen and Poll, 1992). Several wild strawberry studies have identified some of the key components of strawberry flavor, including acetates of hydroxycinnamoyl alcohols and phenylpropenes, such as eugenol and isoeugenol, which are phenylpropanoid derivatives (Pyysalo et al., 1979; Ulrich et al., 1995, 2007). These volatile compounds, together with volatile benzenoids, such as phenylacetaldehyde, phenylethylalcohol, benzaldehyde, benzyl alcohol, benzyl benzoate, methyl benzoate, benzylacetate, methyl salicylate, and vanillin, are also floral scent components (Schuurink et al., 2006).
To date, few transcription factors (TFs) that regulate the expression of the volatile benzenoid/phenylpropanoid pathway structural genes have been identified. In petunia (Petunia hybrida), the function of EMISSION OF BENZENOID II (PhEOBII) and ODORANT1 (ODO1), two R2R3-MYB TFs, have been described as regulators of the volatile and nonvolatile phenylpropanoid biosynthesis pathway (Verdonk et al., 2005; Spitzer-Rimon et al., 2010). PhEOBII binds and activates the promoter of ODO1 through the union to a MYB-binding site (MBS), indicating a hierarchical link between both TFs (Van Moerkercke et al., 2011). Previous work carried out on protoplast extracted from Arabidopsis (Arabidopsis thaliana) leaves showed binding of PhEOBII to the petunia ISOEUGENOL SYNTHASE (IGS) and to the tobacco (Nicotiana tabacum) Phe ammonia lyase B promoters (Spitzer-Rimon et al., 2010). Reduction of EOBII transcript levels in petunia petals by virus-induced gene silencing leads to a down-regulation of the expression of genes involved in the shikimate and phenylpropanoid pathways, such as CHORISMATE SYNTHASE (CS), CHORISMATE MUTASE (CM), PHENYLALANINE AMMONIA LYASE (PAL2), CONIFERYL ALCOHOL ACETYLTRANSFERASE (CFAT), IGS, BENZOYL-COENZYME A:BENZYL ALCOHOL/PHENYLETHANOL BENZOYLTRANSFERASE (BPBT), and ODO1, whereas no changes were detected in the expression levels of CHALCONE ISOMERASE (CHI) or FLAVANONE 3-HYDROXYLASE (FHT). However, overexpression of EOBII did not produce an increase in the expression levels of ODO1, BPBT, CS, CM, and FHT. This suggests that these genes could be regulated by the synergistic action of EOBII and other TFs. On the other hand, PAL, CFAT, and IGS transcripts were up-regulated, indicating a direct positive regulatory role of the expression of these genes by EOBII (Spitzer-Rimon et al., 2010). Besides, ODO1 was demonstrated to activate the promoter of 5-ENOL-PYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE (EPSPS), hence regulating the floral shikimate pathway that produces benzenoid/pheylpropanoid volatiles (Verdonk et al., 2005). Additionally, ODO1 silencing resulted in a severe decrease of volatile production in petunia flowers, such as phenylacetaldehyde, phenylethylalcohol, methyl benzoate, benzyl acetate, benzyl benzoate, vanillin, and isoeugenol, but it did not affect the production of Phe-derived flavonols and anthocyanins (Verdonk et al., 2005). Ectopic expression of PhODO1 in tomato (Solanum lycopersicum) increased the expression of only a subset of phenylpropanoid-associated genes. Changes in the expression of genes related to flavonoid synthesis were not detected in transgenic fruits. These data were consistent with the metabolomic changes observed (Dal Cin et al., 2011).
Recently, a third TF (EOBI) has been identified in petunia as a regulator of floral scent production through direct modulation of the expression of ODO1 and structural scent-related genes. EOBI is an R2R3-MYB TF that is required for the proper expression of structural and regulatory genes related to floral phenylpropanoid scent production (Spitzer-Rimon et al., 2012). EOBI silencing down-regulated several genes from the shikimate and phenylpropanoid pathways (i.e. EPSPS, CS, CM, PAL, IGS, EGS, and ODO1, among others), which clearly led to reduced levels of floral volatiles, such as benzylbenzoate, phenylethylbenzoate, benzyl alcohol, methylbenzoate, methylsalicylate, isoeugenol, and eugenol (Spitzer-Rimon et al., 2012). However, an EOBI orthologous gene has not been detected in strawberry yet.
Eugenol and isoeugenol are volatile compounds produced by plants as floral attractants of pollinators as well as defense compounds (Koeduka et al., 2006; Pasay et al., 2010). Fruits can also synthesize volatile phenylpropenes that contribute to their aroma (Jordán et al., 2001; Aubert and Pitrat, 2006; Ortiz-Serrano and Gil, 2010). Eugenol production by ripe strawberry fruits has been reported previously (Pyysalo et al., 1979; Zorrilla-Fontanesi et al., 2012). Interestingly, the amount of this volatile is lower in cultivated fruits (F. ananassa) than in wild diploid Fragaria vesca (Pyysalo et al., 1979). By contrast, isoeugenol biosynthesis in strawberry fruits is extremely low in comparison with eugenol levels (Hoffmann et al., 2011).
The last step of the phenylpropene eugenol biosynthesis is catalyzed by EUGENOL SYNTHASE (EGS), an NADPH-dependent reductase belonging to the PIP (for Pinoresinol-lariciresinol reductase, Isoflavone reductase, Phenylcoumaran benzylic ether reductase) family (Min et al., 2003). To date, only a small number of plant EGSs, such as ObEGS1 (from Ocimum basilicum; Koeduka et al., 2006), CbEGS1 and CbEGS2 (from Clarkia breweri), and PhEGS1 (from petunia; Koeduka et al., 2008), have been functionally characterized. These enzymes can use coniferyl acetate as a substrate for eugenol in vitro production. Overexpression of a coniferyl alcohol acetyltransferase (PhCFAT) and PhEGS from petunia in aspen (Populus spp.) increased eugenol production in leaves (Koeduka et al., 2013).
More recently, two different genes (FaEGS1 and FaEGS2) coding two strawberry eugenol synthases have been functionally characterized (Aragüez et al., 2013). FaEGS1 was predominantly expressed in green achenes, whereas the expression of FaEGS2 was specific to ripe fruit receptacles (Aragüez et al., 2013). The expression pattern of both genes correlated with eugenol content of both achenes and fruit receptacles. FaEGS1 as well as FaEGS2 enzymes can use in vitro coniferyl acetate as a substrate for eugenol production (Aragüez et al., 2013). Although eugenol production in strawberry fruit has been clarified, the regulation of the volatile benzenoid/phenylpropanoid structural pathway that renders eugenol in strawberry fruit receptacles has not been elucidated.
Previously performed transcriptomic studies in our research group allowed us to identify a wide group of genes whose expression increased throughout strawberry fruit ripening. One of these genes, FaEOBII (gene28435, 46.063 up), showed a significant sequence homology with the R2R3-MYB, such as the TF PhEOBII. Nevertheless, we have not observed the up-regulation of any orthologous ODO1 or EOBI genes when comparing the transcriptomes of mature red-ripe and immature green receptacles. Although a putative orthologous gene of EOBI from petunia has not been detected in the strawberry genome, putative orthologous genes of ODO1 from petunia have been identified in the strawberry genome (gene32268, gene21264, gene12564, and gene30520). However, none of them present an expression pattern associated with the ripeninng process of the strawberry receptacles (gene21264, 1.559 down; gene12564, 1.468 down; and gene30520, 1.024 up), but the expression of gene32268 (FaODO1), by both microarray studies (13.875 down) and quantitative real-time (qRT)-PCR (23.912 down), was down-regulated during the ripening process.
Here, we present the functional characterization of the R2R3-MYB TF FaEOBII, whose expression is fruit receptacle specific and that is related to eugenol production. This regulatory role is executed through the activation of CINNAMYL ALCOHOL DEHYDROGENASE (FaCAD1) and FaEGS2 gene expression.
RESULTS
Sequence Analysis of the FaEOBII Gene and Protein
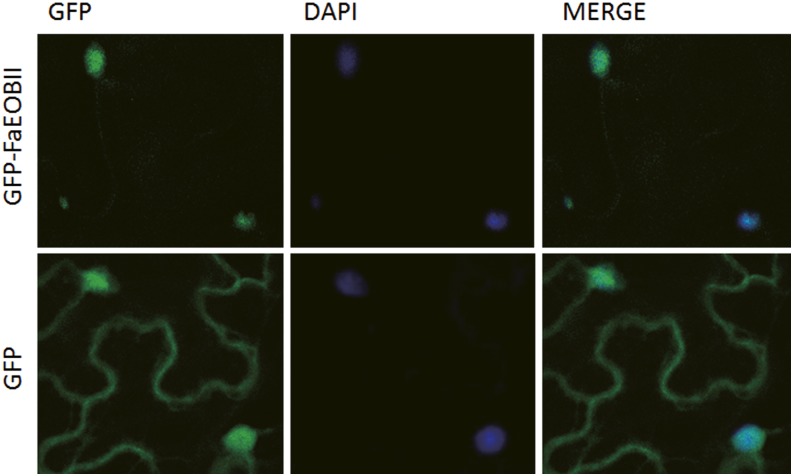
The full-length complementary DNA (cDNA) sequence of the FaEOBII gene contained an open reading frame of 624 bp that encodes a polypeptide of 207 amino acid residues and has a predicted molecular mass of 23.26 kD. WoLF PSORT predicted the nuclear location of this protein (Supplemental Fig. S1). To confirm this hypothesis, we have determined the subcellular location of the FaEOBII protein in vivo. To that end, an N-terminal translational fusion protein construct between FaEOBII and GFP was transiently expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter in Nicotiana benthamiana leaves. As expected, confocal imaging of the fusion protein revealed colocation with the nucleus marker 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) in the parenchyma cells of the abaxial epidermis of tobacco leaves, thus confirming the nuclear location of the FaEOBII protein (Fig. 1). The analysis of the predicted FaEOBII protein revealed the R2 and R3 repeat signatures at its N terminus that are characteristic of the R2R3 DNA-binding MYB proteins (Kranz et al., 1998; Supplemental Fig. S1), which are important for interaction with regulatory sequences in promoters (Kranz et al., 1998; Dubos et al., 2010). We also identified a W/Y-MDDIW transactivation motif (Li et al., 2006) at its C terminus, with three particular amino acids (Trp-193, Asp-197, and Trp-199) conserved in all the sequences analyzed (Supplemental Fig. S2). Based on these features outside the DNA-binding R2R3 domain, FaEOBII can be classified into subgroup 19 of the R2R3-MYB TFs (Dubos et al., 2010; Supplemental Fig. S3; Supplemental Tables S1 and S2). Phylogenetically, FaEOBII shares high amino acid homology with members of this subgroup of MYB TFs from other plants, such as PsMYB26 (Pisum sativum), NlxNsMYB305 (Nicotiana langsdorffii × Nicotiana sanderae), AmMYB305 (Antirrhinum major), AmMYB340 (A. major), AtMYB24 (Arabidopsis), AtMYB21 (Arabidopsis), and PhEOBII (petunia). Functional characterization of all of these TFs has demonstrated that they are involved in the regulation of the volatile benzenoid phenylpropanoid metabolic pathway (Sablowski et al., 1994; Moyano et al., 1996; Uimari and Strommer, 1997; Shin et al., 2002; Li et al., 2006; Liu et al., 2009; Spitzer-Rimon et al., 2010). All of these bioinformatic analyses suggested that FaEOBII could be an R2R3 MYB TF potentially involved in the regulation of those metabolic pathways related to volatile phenylpropanoid production in ripe receptacles.
Figure 1.
Nuclear location of the FaEOBII protein in plant cells. Fluorescence signal was detected using a confocal microscope from GFP-FaEOBII (top row) and GFP (bottom row) expression under the control of the 35S promoter in N. benthamiana leaf epidermis cells. The MERGE column shows merged views of the GFP and DAPI images.
The Expression of FaEOBII Is Highly Expressed in Ripe Fruit Receptacles and Petals and Correlates with Eugenol Production in Both Tissues
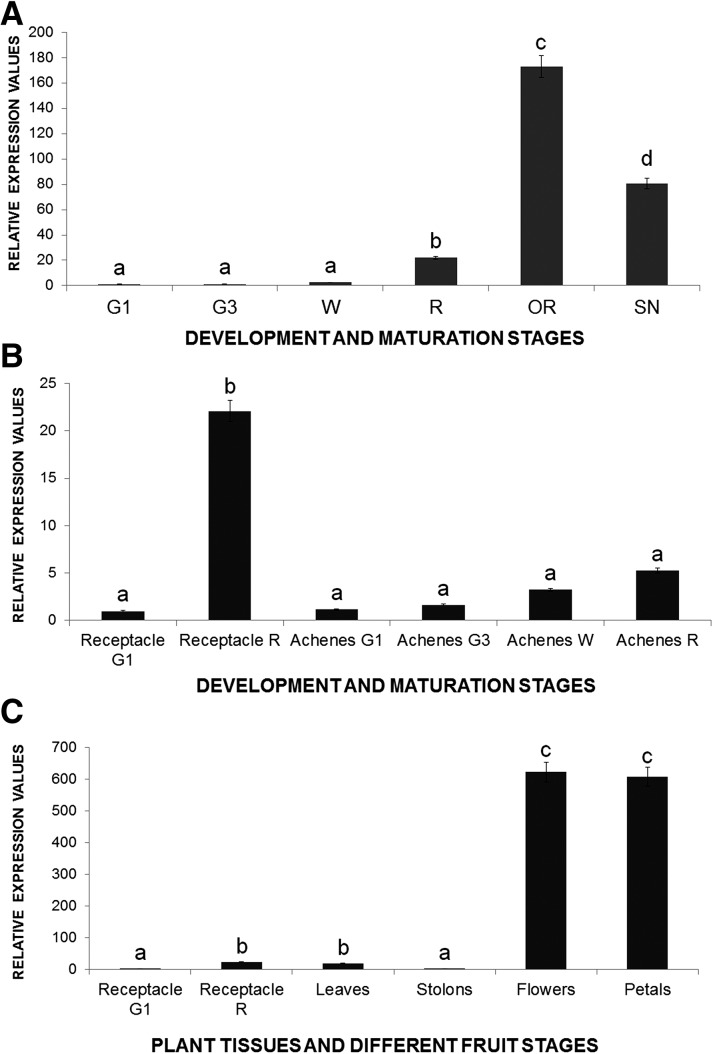
FaEOBII was expressed at very low levels during fruit receptacle elongation at early developmental stages, experiencing an increase in receptacle ripening stages, with a maximum peak taking place in the overripe stage. This maximum expression was followed by a decrease in expression in the senescent stage (Fig. 2A). On the other hand, when compared with those expression values obtained in fruit receptacle at the red stage, FaEOBII was weakly expressed in fruit achenes in all developmental and ripening stages studied (Fig. 2B). This low expression was also observed in other vegetative tissues analyzed, such as leaves and stolons. However, in flowers and specifically in petals, the amount of FaEOBII transcript was higher than in ripe fruits (Fig. 2C). These results showed that FaEOBII expression was not only up-regulated in ripe fruit receptacles but also in petals.
Figure 2.
Analysis by qRT-PCR of FaEOBII expression in developing fruit receptacles (A), achenes (B), and plant tissues (C) of F. ananassa ‘Camarosa.’ G1, Small-sized green fruit; G3, full-sized green fruit; W, white stage; R, ripe stage; OR, overripe stage; SN, senescent stage. Quantification is based on cycle threshold (Ct) values. Relative expression values were calculated relative to receptacles in the G1 stage in all cases, which was assigned an arbitrary value equal to unity. Values are means ± sd of five independent experiments. Statistical significance was determined by one-way ANOVA. Letters indicate significant differences (P < 0.05, Scheffe’s post hoc test).
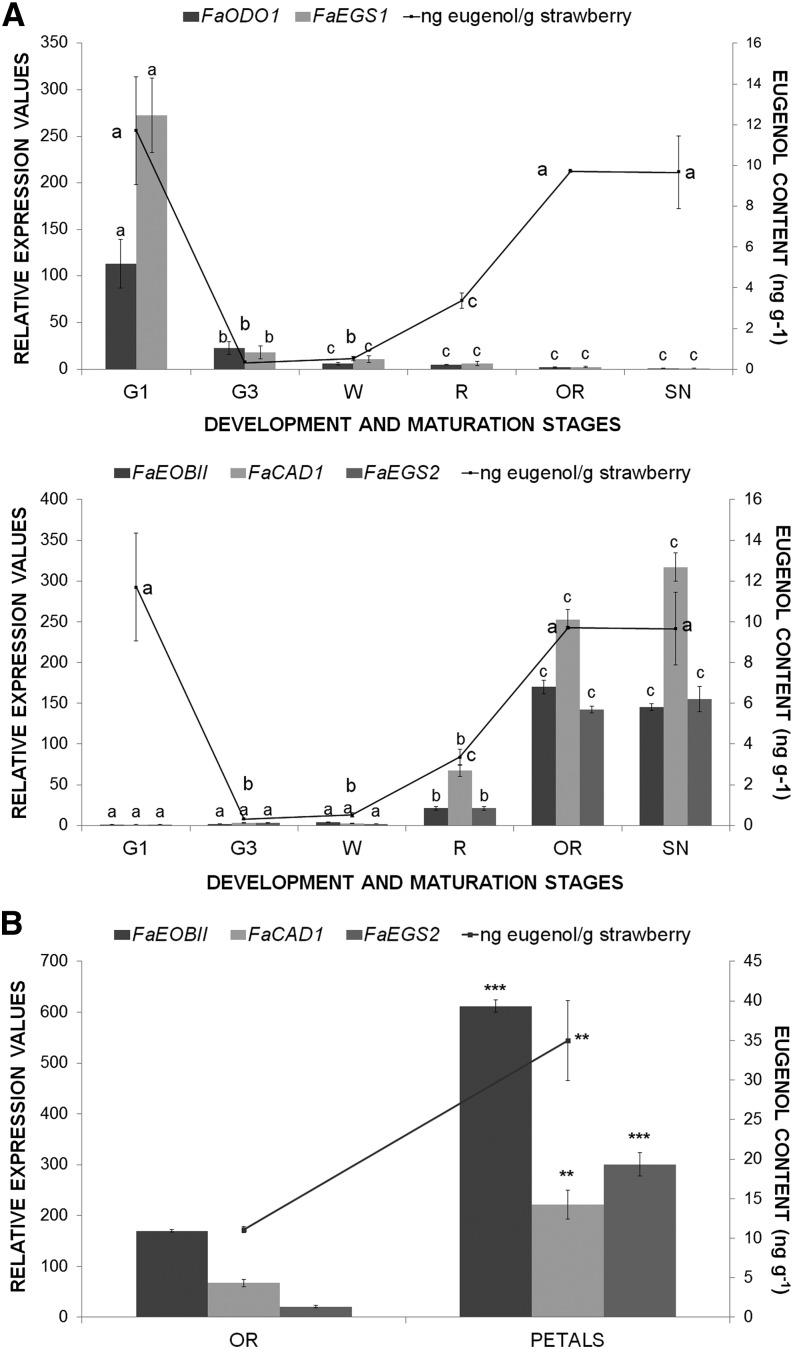
It was shown that throughout the strawberry fruit-ripening process, eugenol accumulates in ripe receptacles (Pyysalo et al., 1979; Zorrilla-Fontanesi et al., 2012; Aragüez et al., 2013). The orthologous gene of FaEOBII in petunia (PhEOBII) has been shown to be involved in eugenol production (Spitzer-Rimon et al., 2010). Thus, we analyzed eugenol production in strawberry fruit receptacles during ripening (Fig. 3A) as well as in those plant tissues where a higher FaEOBII gene expression was observed (Fig. 3B). A clear connection between high FaEOBII gene expression during the ripening stages and high eugenol content was found. In fact, the highest levels of FaEOBII gene expression and of eugenol content were observed in red overripe receptacles and in petals (Fig. 3B). Recently, eugenol production in unripe strawberry fruits has been reported to be mainly restricted to achenes at the green stage (Aragüez et al., 2013) as well as coincident with the expression pattern of FaEGS1 in achenes (Aragüez et al., 2013). Our results indicate that there is also eugenol production in young unripe green receptacles that is coincident with the FaEGS1 gene expression but not with the expression of FaEOBII (Fig. 3A), discounting a regulatory role of FaEOBII in FaEGS1 gene expression. However, in green unripe fruit receptacles, we detected the presence of an ODO1 orthologous gene (Fig. 3A) that could regulate the expression of FaEGS1 in a similar way to what is described for PhODO1 (Verdonk et al., 2005). FaCAD1 encodes a cinnamyl alcohol dehydrogenase that produces cynnamyl alcohols that are, in turn, the substrate for the production of volatile phenylpropanoids such as eugenol. This enzyme works upstream of the FaEGS2 enzyme in the metabolic pathway that renders eugenol. Thus, for this reason, FaCAD1 and FaEGS2 genes have been included in the expression studies. Figure 3A showed a clear correlation in the expression profile of FaEOBII, FaCAD1, and FaEGS2 during the ripening process of strawberry fruit receptacles as well as a concomitant increase in the eugenol content during maturation stages.
Figure 3.
Analysis of spatiotemporal changes in gene expression and content of eugenol. A, Bars represent the developmental expression of strawberry FaODO1, FaEGS1, FaEOBII, FaCAD1, and FaEGS2 genes in receptacles of F. ananassa ‘Camarosa’ obtained by qRT-PCR. B, Bars represent relative expression values of FaEOBII, FaCAD, and FaEGS2 genes in overripe receptacles versus petals. Stages are as defined in Figure 2. Lines represent gas chromatography (GC)-mass spectrometry (MS) quantification of eugenol. Values are given in nanograms of eugenol per gram of strawberry. One-way ANOVA was performed on log-transformed data, and letters indicate significant differences (P < 0.05, lsd post hoc test).
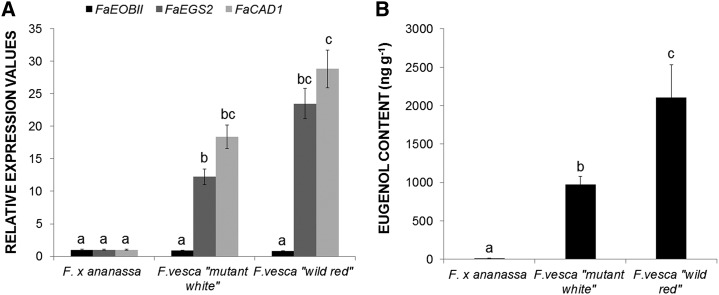
Interestingly, in strawberry fruit receptacles, the expression pattern of FaEOBII was quite similar to that observed for FaEGS2. Similarly, eugenol content, as well as FaCAD1 and FaEGS2 gene expression, were higher in F. vesca than in F. ananassa ripe fruits. In contrast, the expression of FaEOBII was unchanged between both Fragaria spp. (Fig. 4). Using the PLACE server (http://www.dna.affrc.go.jp/PLACE/), several MBSII boxes recognized by the FaEOBII TF were subsequently located in the EGS2 promoter sequences (Supplemental Fig. S4). In view of this bioinformatic analysis of promoter sequences, the FvEGS2 promoter contains more MBSII boxes than the FaEGS2 promoter.
Figure 4.
Analysis of gene expression changes and eugenol content. A, Analysis by qRT-PCR of EOBII, EGS2, and CAD1 gene expression in red ripe receptacles of the cultivated strawberry F. ananassa ‘Camarosa’ and in the wild strawberries F. vesca Mutant White and F. vesca Wild Red of cv Reina de los Valles. Quantification is based on Ct values as described in “Materials and Methods.” The increase in the mRNA value was relative to the F. ananassa Ct value of each experiment, which was assigned an arbitrary value equal to unity. Mean values ± sd of six independent experiments are shown. Statistical significance was determined by one-way ANOVA. Letters indicate significant differences (P < 0.05, Scheffe’s post hoc test). B, GC-MS quantification of eugenol in strawberry fruit varieties. Values are given in nanograms of eugenol per gram of strawberry. One-way ANOVA was performed on log-transformed data, and letters indicate significant differences (P < 0.05, lsd post hoc test).
The Expression of FaEOBII Is Hormonally Regulated throughout Receptacle Fruit Growth and Ripening
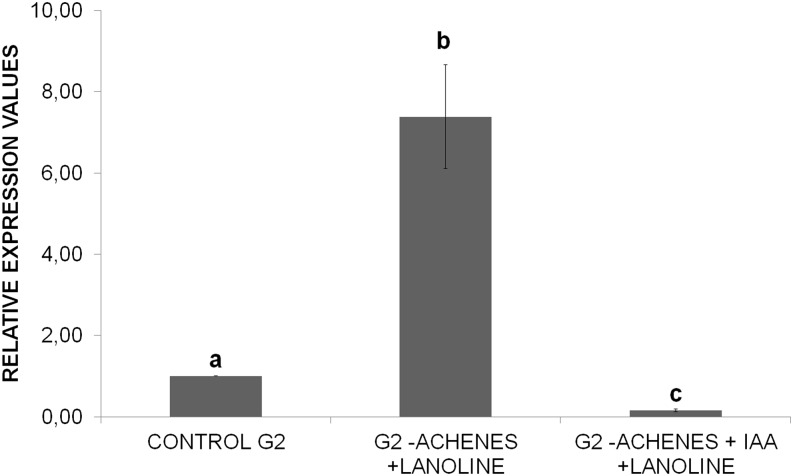
It has been proposed that the abscisic acid (ABA)-auxin ratio in strawberry fruit receptacles constitutes the signal that triggers the fruit-ripening process (Perkins-Veazie, 1995; Jiang and Joyce, 2003). According to this proposal, the auxins that are produced by achenes and released to the fruit receptacle would repress the expression of a vast majority of the ripening-related genes (Medina-Escobar et al., 1997; Manning, 1998; Moyano et al., 1998; Blanco-Portales et al., 2002, 2004; Raab et al., 2006; Medina-Puche et al., 2014). To determine whether FaEOBII gene expression was under auxin control, we determined the changes in FaEOBII gene expression in unripe fruit receptacles after removing the achenes from the fruit surface. This treatment reduces the endogenous content of auxins in the fruit receptacle. After 5 d of treatment, the FaEOBII expression level was clearly increased when compared with what had been observed in untreated control fruits. This increase in the amount of FaEOBII transcript in deachened fruits was abolished by the external application of auxin (indole-3-acetic acid [IAA]), thus indicating that this gene expression is under the negative control of auxins (Fig. 5).
Figure 5.
Analysis of the effects of removing achenes from G2 developing fruits of F. ananassa ‘Camarosa’ on FaEOBII expression by qRT-PCR. Quantification is based on Ct values as described in “Materials and Methods.” Increase in the mRNA value was relative to the receptacle G2 Ct value of each experiment, which was assigned an arbitrary value equal to unity. Values are means ± sd of five independent experiments. CONTROL G2, Middle-sized green fruit receptacle; G2-ACHENES + LANOLINE, G2 fruit receptacles without achenes for 5 d covered by a lanolin paste; G2-ACHENES + IAA + LANOLINE, G2 fruit receptacle without achenes for 5 d treated with the synthetic auxin IAA as a lanolin paste with 1 mm IAA. Statistical significance was determined by one-way ANOVA. Letters indicate significant differences (P < 0.05, Scheffe’s post hoc test).
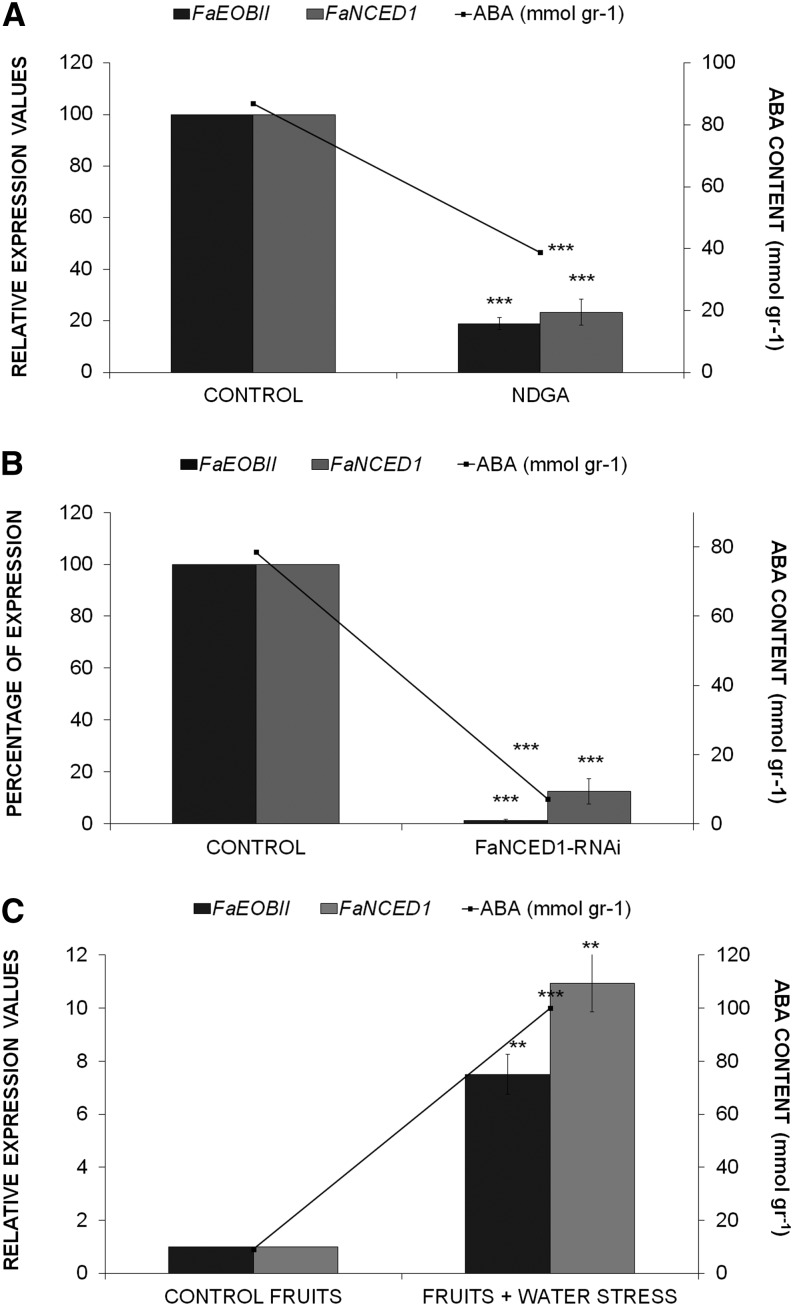
Recent reports have suggested that the phytohormone ABA may be involved in the positive regulation of anthocyanin production by promoting strawberry fruit ripening (Chai et al., 2011; Jia et al., 2011; Medina-Puche et al., 2014). To ascertain if this phytohormone regulates FaEOBII gene expression, we modulated the internal receptacle ABA content in three different experimental conditions: (1) by injecting into the interior of fruit receptacle nordihydroguaiaretic acid (NDGA), an inhibitor of the 9-CIS-EPOXYCAROTENOID DIOXYGENASE (NCED) activity, the key enzyme responsible for ABA biosynthesis (Fig. 6A); (2) by transiently silencing FaNCED1 expression through in vivo agroinfiltration (Fig. 6B); and (3) by depleting water from plants, as water stress is known to increase ABA content in plants (Fig. 6C). Under all conditions, a clear variation in the expression of FaEOBII parallel to the endogenous receptacle ABA content was observed (Fig. 6; Supplemental Fig. S5). These results strongly suggest that ABA can positively regulate the expression of the FaEOBII gene.
Figure 6.
Analysis by qRT-PCR of FaEOBII and FaNCED1 expression (bars) and quantification of ABA concentration (lines). A, CONTROL, Green-white fruits injected with water; NDGA, green-white fruits injected with NDGA (100 μm). B, CONTROL, Control fruits agroinfiltrated with the empty pFRN vector; FaNCED1-RNAi, transgenic strawberry fruits agroinfiltrated with the pFRN-FaNCED1 construct. C, CONTROL FRUITS, Fruits with the pedicels immersed in Murashige and Skoog medium with Suc; FRUITS + WATER STRESS, fruits with their pedicels kept in the air. Mean values ± sd of five independent experiments are shown. Statistical significance with respect to the reference sample (CONTROL) was determined by Student’s t test: **, P < 0.01; and ***, P < 0.001.
Taken together, these results indicate that FaEOBII expression is clearly regulated by both ABA content and auxin content of the fruit receptacle, which provides a putative molecular explanation for both the proposal of Perkins-Veazie (1995) and the findings described by Chai et al. (2011), Jia et al. (2011), and Medina-Puche et al. (2014).
FaEOBII Can Transactivate the Promoters of Genes Involved in Volatile Phenylpropene Production in Strawberry Fruit Receptacles
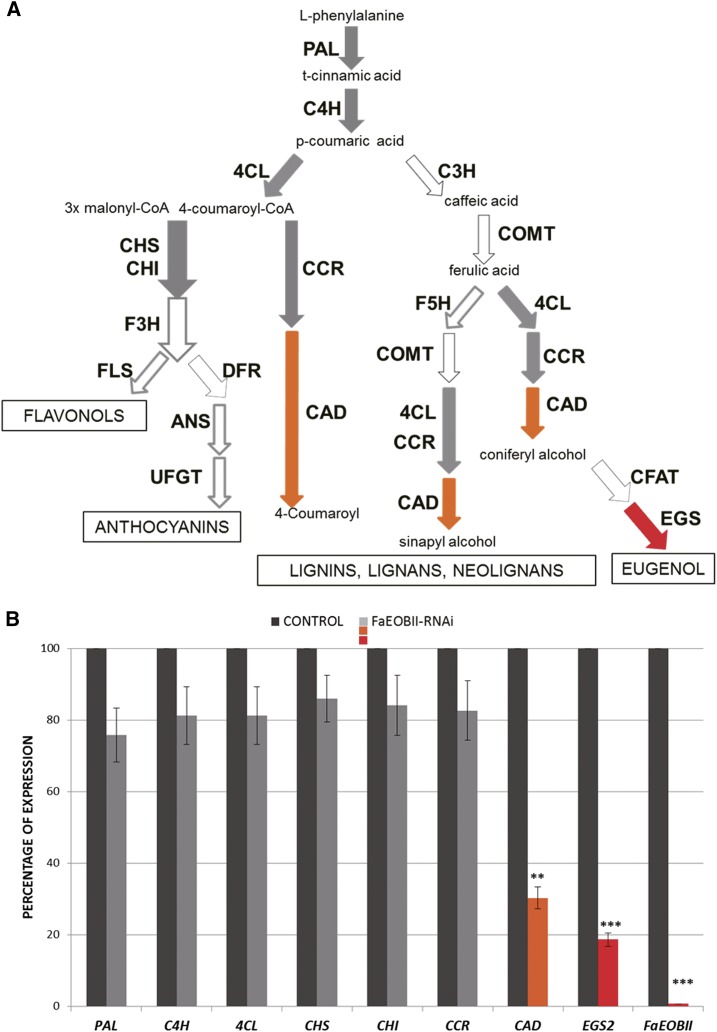
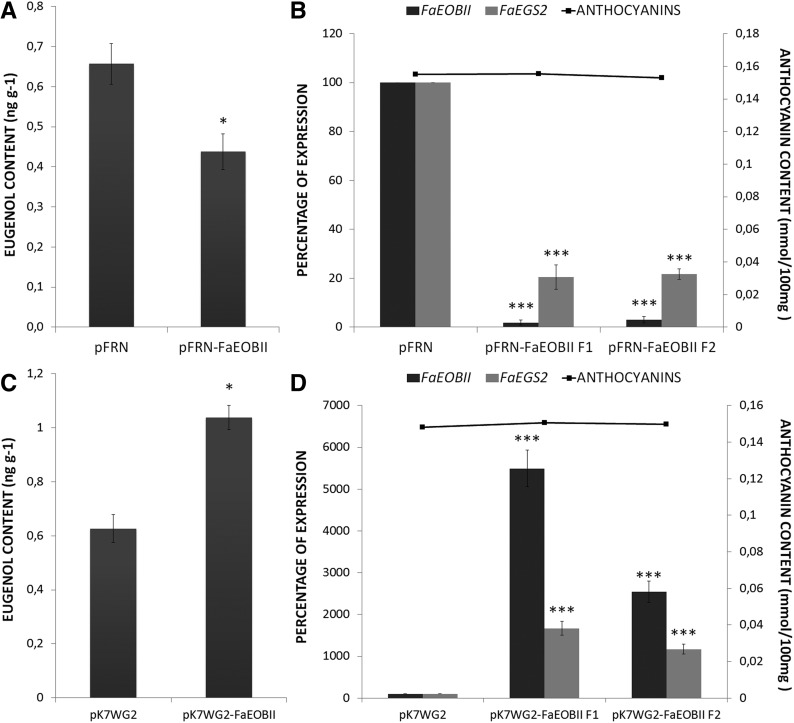
The potential regulatory role played by FaEOBII in the expression of the structural genes belonging to the volatile phenylpropanoid biosynthesis pathway in ripe strawberry receptacles was assessed through transcriptomic studies. For this purpose, the expression of FaEOBII was transiently down-regulated in fruit receptacles by an RNA interference (RNAi) approach. The silencing degree was measured in fruit receptacle by qRT-PCR 14 d after agroinjection. Analyzed transgenic fruits showed an important reduction in FaEOBII expression ranging between 88.3% and 99.5%, whereas no changes in FaEOBII expression were found in control fruits (Figs. 7B and 8B). Fruit receptacles that exhibited a silencing percentage above 99% were used for transcriptomics analysis. A custom-made oligonucleotide-based microarray platform representing 34,496 putative genes from the Fragaria spp. genome (FraGenomics35K) was used. Transcriptome analyses were carried out to compare gene expression profiles of transgenic transiently silenced FaEOBII fruits against control fruits that were agroinfiltrated with the empty pFRN vector. The work focused on the most clearly up- and down-regulated genes by choosing a 2-fold cutoff and a corrected P value of less than 0.01. This microarray analysis revealed that only 10 transcripts were down-regulated, whereas no transcript was up-regulated in transiently FaEOBII-silenced fruits (Table I; Supplemental Tables S3 and S4). Two of them, FaCAD1 (gene20700) and FaEGS2 (gene25260), encode enzymes and were involved in eugenol biosynthesis (Fig. 7A). This down-regulation was also corroborated by qRT-PCR (Fig. 7B; Supplemental Table S4). For this reason, eugenol content in FaEOBII-silenced fruits was analyzed. The results showed a significant decrease of eugenol content (Fig. 8A) in FaEOBII-silenced fruits, whereas anthocyanin content remained unchanged (Fig. 8B).
Figure 7.
FaEOBII silencing effect on the expression of the structural genes belonging to the volatile phenylpropanoid biosynthesis pathway. A, Schematic diagram of the phenylpropanoid pathway in plants. White arrows indicate nonanalyzed genes. Gray arrows represent genes with no significant transcriptomic changes when FaEOBII is silenced. Orange and red arrows represent genes with significant transcriptomic changes in FaEOBII-silenced fruits. C4H, Cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; CHS, chalcone synthase; F3H, flavanone 3-hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol-4-reductase; ANS, anthocyanidin synthase (also called LDOX, leucoanthocyanidin dioxygenase); UFGT, UDP-flavonoid glucosyltransferase; C3H, p-coumarate 3-hydroxylase; COMT, caffeic O-methyltransferase; F5H, ferulic acid 5-hydroxylase; CCR, cynnamoyl CoA reductase; CAD, cynnamyl alcohol dehydrogenase. B, Analysis by qRT-PCR of the expression of genes involved in the phenylpropanoid pathway in FaEOBII-silenced fruits agroinfiltrated with the pFRN-FaEOBII construct, compared with control fruits agroinfiltrated with the empty pFRN vector only. The silencing level is expressed as a percentage. Statistical significance with respect to the reference sample (CONTROL) was determined by Student’s t test: **, P < 0.01; and ***, P < 0.001.
Figure 8.
GC-MS quantification of eugenol, qRT-PCR analysis, and anthocyanin measurements in FaEOBII transitory transgenic fruit receptacles. A and C, GC-MS quantification of eugenol in strawberry fruit receptacles with FaEOBII expression silenced (pFRN-FaEOBII) with respect to control fruits (pFRN; A) and FaEOBII expression increased (pK7WG2-FaEOBII) with respect to control fruits (pK7WG2; C). Values are given in nanograms per gram. Statistical significance with respect to the reference sample (pFRN or pK7WG2) was determined by Student’s t test: *, P < 0.05. B and D, Bars represent the analysis by qRT-PCR of FaEOBII and FaEGS2 expression in transgenic strawberry fruits agroinfiltrated with the pFRN-FaEOBII construct and in control fruits agroinfiltrated with the empty pFRN vector (B) and with the pK7WG2-FaEOBII construct and in control fruits agroinfiltrated with the empty pK7WG2 vector (D). The silencing and overexpression levels are expressed as percentages. Quantification is based on Ct values as described in “Materials and Methods.” The values refer to the mRNA Ct value of control samples, which were assigned an arbitrary value equal to unity. Mean values ± sd of three independent experiments are shown. Lines represent anthocyanin content quantified on fruits analyzed. Statistical significance with respect to the reference sample (pFRN or pK7WG2) was determined by Student’s t test: ***, P < 0.001.
Table I. Microarray data from a transcriptomic comparison between transgenic transiently silenced FaEOBII fruits and control fruits agroinfiltrated with the empty pFRN vector.
Gene identifiers and corresponding annotations are as reported in the F. vesca genome database (http://www.strawberrygenome.org/; Shulaev et al., 2011).
| Gene Identifier | RNAi-FaEOBII | P | Annotation | Species | E Value | Accession No. |
|---|---|---|---|---|---|---|
| gene28435 | 6.549 down | 0.00035 | MYB TF emission of benzenoids II (FaEOBII) | F. ananassa | 0.0 | KM099230 |
| gene25260 | 4.019 down | 0.00437 | Eugenol synthase2 (FaEGS2) | F. ananassa | 4E-148 | KF562266.1 |
| gene20700 | 3.047 down | 0.00501 | Cinnamyl alcohol dehydrogenase (FaCAD1) | F. ananassa | 0.0 | U63534.1 |
| gene20643 | 2.991 down | 0.00343 | β-Glucosidase3 | F. ananassa | 0.0 | JX244263.1 |
| gene32526 | 2.465 down | 0.00262 | DNA-binding with one finger zinc finger protein | Medicago truncatula | 6E-49 | XM_003602109.1 |
| gene25718 | 2.208 down | 0.00167 | Hexokinase | Eriobotrya japonica | 3E-179 | JF414121.2 |
| gene21312 | 2.207 down | 0.002 | Gland development-related protein90-like | Gossypium hirsutum | 2E-21 | EU373080.1 |
| gene12176 | 2.202 down | 0.00174 | Vernalization insensitive | Theobroma cacao | 0.0 | XM_007018178.1 |
| gene18910 | 2.150 down | 0.0011 | β-Hydroxyisobutyryl-CoA hydrolase1 | Arabidopsis | 5E-104 | NM_125991.3 |
| gene06729 | 2.083 down | 0.00487 | Cytochrome P450 | Populus trichocarpa | 0.0 | XM_002307093.1 |
| gene30534 | 2.076 down | 0.00498 | Nonspecific lipid-transfer protein | M. truncatula | 2E-22 | XM_003620358.1 |
In parallel, the expression of FaEOBII was transiently up-regulated in fruit receptacles by overexpression under the control of the CaMV 35S promoter. The increased expression was measured by qRT-PCR, and the results showed a statistically significant increase in both eugenol content (Fig. 8C) and FaEOBII and FaEGS2 gene expression (Fig. 8D) in the FaEOBII overexpression construct receptacles, whereas no changes were detected in the anthocyanin content (Fig. 8D).
FaEOBII Can Activate the Promoter of FvCAD1 That Encodes a Key Enzyme of Eugenol Metabolism
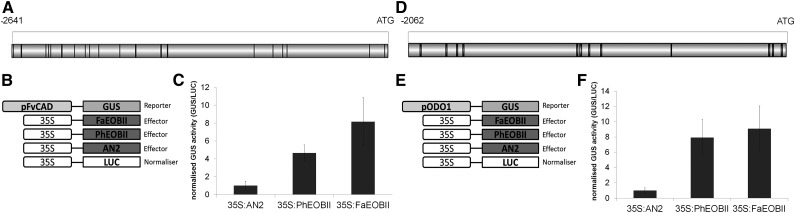
In order to determine the in vivo ability to transactivate the promoters of FaCAD1 and FaEGS2, we conducted transitory assays on N. benthamiana leaves. After several efforts to clone the FaEGS2 promoter, we did not succeed. For this reason, in its place we used the FvCAD1 gene promoter (Fig. 9A) for this assay. For this purpose, we performed transient transactivation assays of the FvCAD1 promoter from F. vesca with the FaEOBII TF in N. benthamiana leaves. The pCAD1:GUS reporter construct was expressed together with the effector construct 35S:FaEOBII. As a control, the same reporter construct was separately coinfiltrated with the R2R3-MYB TFs EOBII (PhEOBII) and ANTHOCYANIN2 (AN2) effector constructs from petunia (Fig. 9B). An 8-fold increase in reporter gene activation was observed when coinfiltration with FaEOBII was compared with control infiltration (AN2). Lower induction values (up to 4.6-fold) were detected with PhEOBII as effector (Fig. 9C). These results indicate that FaEOBII activates the FvCAD1 promoter in N. benthamiana leaves.
Figure 9.
Transactivation of the CAD1 promoter of F. vesca and the ODO1 promoter of petunia ‘Mitchell’ by EOBII in N. benthamiana leaves. A and D, Schematic diagram of pFvCAD1 and pODO1 promoters using the PLACE online database (www.dna.affrc.go.jp) and DOG 2.0.1 for Windows (Illustrator of Protein Domain Structures). The bars at the top indicate the lengths of the promoter fragments relative to the ATG codon. The locations of the MBSII boxes are shown using black lines. B and E, Constructs used for the transactivation assays. C and F, Normalized GUS activity after coinfiltration with A. tumefaciens harboring any of the effector constructs with the pFvCAD1:GUS or the pODO1:GUS reporter construct. Coinfiltration with the CaMV 35S:AN2 effector was used to show EOBII specificity. Coinfiltration with 35S:LUC enabled normalization (averages ± se).
As a positive control in this experiment, the transactivation of the PhODO1 promoter (Van Moerkercke et al., 2011) by FaEOBII was carried out. The results obtained show the accurate completion of the experiment (Fig. 9, D–F).
FaEOBII Binds to MYB-Binding Sequences in Vitro
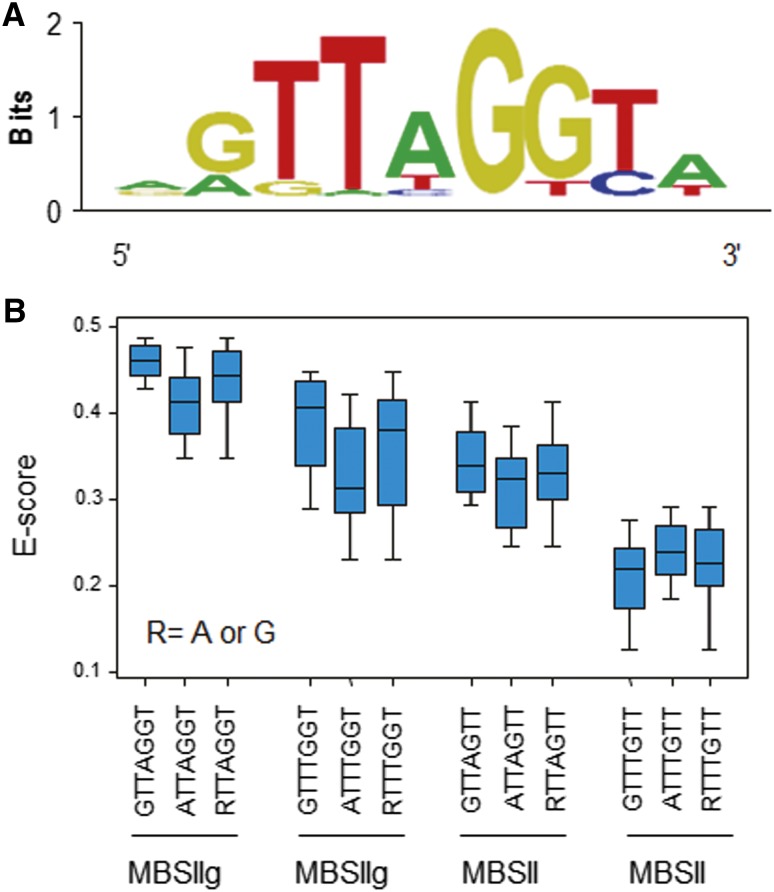
To analyze whether FaEOBII could bind to MYB-binding DNA sequences, we determined the DNA-binding specificity of FaEOBII using the PBM11 microarray, which contains all possible 11-mer sequences (Godoy et al., 2011). After array incubation and data processing (Berger and Bulyk, 2009; Godoy et al., 2011), we obtained the list of the top-scoring 8-mers, shorted in relation to their enrichment scores. The GTTAGGT sequence was obtained with the highest enrichment score, thus indicating that this motif is recognized with the highest affinity by FaEOBII (Fig. 10A). This motif can be interpreted as an MBSIIg box (GKTWGGTR; R, A, or G; K, G, or T; W, A, or T; Prouse and Campbell, 2012), similar to those obtained for other plant R2R3 MYBs (Franco-Zorrilla et al., 2014). Given that some R2R3 MYBs also recognize MBSII elements, we evaluated binding to DNA motifs in which the G nucleotide in the sixth position in the GTTAGGT sequence was replaced by a T residue. In this case, the highest affinity was observed for MBSIIg motifs, in particular RTTAGGT sequences (Fig. 10B). By contrast, binding to their corresponding MBSII elements was very low, thus indicating that FaEOBII does not efficiently recognize these elements (Fig. 10B).
Figure 10.
Identification of FaEOBII-binding motifs in vitro. A, Position weight matrix representation of the top-scoring 9-mer obtained in the seed-and-wobble algorithm. B, Box plot of enrichment scores (E-scores) containing all possible 7 mers indicated representing the MBSIIg and MBSII elements. Blue boxes represent quartiles 25% to 75%, and the black line represents the median of the distribution (quartile 50%). Bars indicate quartiles 1% to 25% (above) and 75% to 100% (below).
FaEOBII Expression Is under the Regulation of the FaMYB10 TF
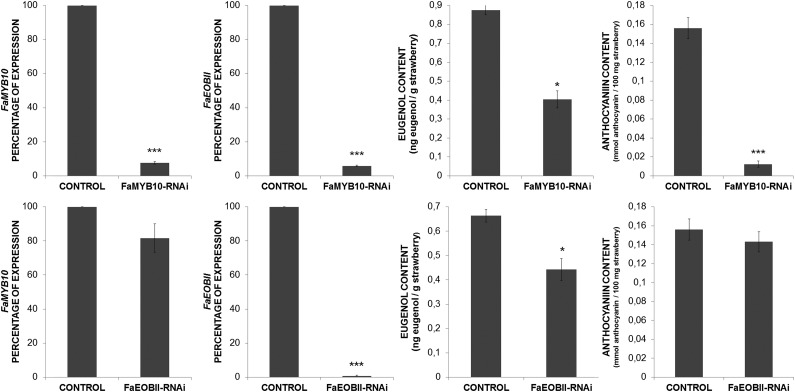
In petunia, a flower-specific gene regulatory network has been described that constitutes the phenylpropanoid volatile biosynthesis pathway that is controlled by three R2R3 MYB TFs, ODO1, EOBII, and EOBI (Verdonk et al., 2005; Spitzer-Rimon et al., 2010, 2012; Van Moerkercke et al., 2011). Recently, in strawberry, it was demonstrated that the transitory silencing of FaMYB10 gene expression led to a down-regulation of structural genes involved in the flavonoid/phenylpropanoid metabolic pathway (Medina-Puche et al., 2014) as well as FaEOBII expression. We have corroborated these microarray results through qRT-PCR experiments. Thus, we have observed that silencing of FaMYB10 expression resulted in a concomitant down-regulation of FaEOBII expression as well as a reduction of both anthocyanin and eugenol contents in the strawberry fruit receptacles. However, after silencing of FaEOBII expression in fruit receptacles, neither the amount of FaMYB10 transcripts nor the anthocyanins was reduced, although the concentration of eugenol decreased significantly (Fig. 11). These results clearly indicate that FaMYB10 regulates FaEOBII expression but not the contrary, and they support even more the involvement of FaEOBII in the regulation of eugenol biosynthesis in red-ripe receptacles.
Figure 11.
qRT-PCR analysis of FaMYB10 and FaEOBII transcript levels and eugenol and anthocyanin content determination in transgenic agroinfiltrated fruits injected with the corresponding RNAi construct. Statistical significance with respect to the control simple was determined by Student’s t test: *, P < 0.05; and ***, P < 0.001 (n = 10).
DISCUSSION
The transcriptional regulatory network of the strawberry fruit-ripening process is poorly known. So far, the functional roles of only a low number of TFs that regulate the expression of the ripening-related genes and related metabolic pathways that determine the organoleptic characteristics of the strawberry ripe fruit have been clarified. Thus, by using a heterologous system, it was proposed that the R2R3 MYB TF FaMYB1 plays a repressor role in the expression of phenylpropanoid and anthocyanin structural genes in ripe fruit receptacles (Aharoni et al., 2001). However, transient silencing of FaMYB1 gene expression in strawberry receptacles did not change the anthocyanin content in transgenic ripe receptacles (Medina-Puche et al., 2014). Similarly, it was demonstrated that the fruit-specific and ripening-related FaMYB10 gene, which encodes an R2R3 MYB TF, regulates the expression of early and late structural genes of the flavonoid/phenylpropanoid metabolic pathway (Medina-Puche et al., 2014). Silencing the expression of this TF, in fruit receptacles, reduced the expression of genes involved in anthocyanin and phenylpropanoid volatile production as well as the expression of a putative MYB TF (FaEOBII; Medina-Puche et al., 2014). This suggests a direct connection between FaMYB10, FaEOBII, and the flavonoid/phenylpropanoid metabolism in ripe fruits. In this article, we present the functional characterization of the role played by FaEOBII throughout the strawberry fruit-ripening process.
FaEOBII Regulates the Expression of Two Structural Genes That Play a Key Role in Eugenol Biosynthesis in Fruit Receptacles
Transcriptomic and qRT-PCR assays in fruit receptacles where the expression of FaEOBII was silenced by RNAi showed a decrease in the expression of a small subset of ripening-related genes (Fig. 7B; Table I; Supplemental Tables S3 and S4). Among them, we found two genes, FaCAD1 (gene20700, coding a cinnamyl alcohol dehydrogenase; Blanco-Portales et al., 2002) and FaEGS2 (gene25260, coding an eugenol synthase; Aragüez et al., 2013), that encode for two key enzymes of the metabolic pathway that produces eugenol. FaCAD1 is an enzyme that catalyzes the production of monolignols that are precursors of phenylpropenes, such as eugenol. Afterward, these monolignol alcohols are acetylated in the hydroxyl group by a coniferyl alcohol acyl transferase. FaEGS2 catalyzes the reductive cleavage of the acetate moiety, rendering the eugenol (Koeduka et al., 2006; Dexter et al., 2007; Aragüez et al., 2013). In contrast, in petunia, PhEOBII regulates the expression of a different set of genes involved in the shikimate and phenylpropanoid pathways, such as CS, CM, PAL2, CFAT, IGS, BPBT, and ODO1 (Spitzer-Rimon et al., 2010). Thus, our results indicate that, in contrast to what is described in petunia, in strawberry receptacle, it seems that FaEOBII directly regulates the expression of both FaCAD1 and FaEGS2 genes. This possibility was also supported by the fact that FaEOBII can transactivate in vivo the FvCAD1 promoter (Fig. 9) and can also specifically bind to MBSII boxes, MYB cis-regulatory sequences, which are present in the promoters of those genes regulated by R2R3 MYB TFs (Fig. 10). Additionally, the statistically significant decrease of eugenol (Fig. 8A) in strawberry fruit receptacles where the expression of FaEOBII was silenced (Fig. 8B) confirms the direct connection between the functional activity of this TF and eugenol biosynthesis. However, as stated previously, the lack of a strong correlation between the percentage of gene silencing and eugenol content reduction could be due to the fact that the amount of transcript (arbitrary units of intensity [a.u.i.]) corresponding to FaEGS2 in ripening receptacle transcriptome is very high (FaEGS2-gene25260, a.u.i. = 10,732.21). The amount of eugenol quantified in pFRN-FaEOBII fruits (Fig. 8A) could be explained by two reasons. First, the expression of FaEGS2 was not reduced sufficiently (Fig. 8B) or the residual FaEGS2 transcript in the fruit could be translated into a sufficient amount of enzyme capable of catalyzing the production of eugenol. Second, the half-life of the enzyme may be high enough so that the detected eugenol was indeed catalyzed by FaEGS2 enzyme in the fruit prior to the silencing of FaEOBII.
Furthermore, to confirm the regulatory role of FaEOBII in the biosynthesis of eugenol, qRT-PCR analysis was carried out in fruit receptacles overexpressing the gene FaEOBII. The results showed not only an increase in the expression of FaEOBII and FaEGS2 genes (Fig. 8D) but also a higher eugenol content with respect to control fruits (Fig. 8C). Although the increase of eugenol is significant in fruits overexpressing FaEOBII with respect to control fruits (Fig. 8C), it is difficult to detect higher increases. It is necessary to consider that, in control fruits, the expression of FaEOBII and FaEGS2 in the transcriptome of red-ripened receptacles is already very high (FaEOBII-gene28435, a.u.i. = 3,198.538; FaEGS2-gene25260, a.u.i. = 10,732.21), as is eugenol content, so the production system of eugenol may be saturated in fruits overexpressing FaEOBII and greater increases of eugenol may not be detected.
FvEGS2 and FaEGS2 Have Different Expression Patterns That Correlate with MBSII cis-Regulatory Elements in Their Promoters
Gene expression analyses of EGS2 and EOBII were performed by qRT-PCR in ripe F. vesca and F. ananassa fruits. Significant differences in EGS2 expression were observed between the two species, up to more than 20-fold in F. vesca than in F. ananassa, whereas EOBII expression values were constant in both species (Fig. 4A). Bioinformatic analysis of promoter sequences located along the EGS2 promoter identified several MBSII boxes recognized by FaEOBII, containing more MBSII boxes of the FvEGS2 promoter than the FaEGS2 promoter (Supplemental Fig. S4). These results suggest that higher expression levels of EGS2 and eugenol content in F. vesca than in F. ananassa (Fig. 4) could be the result of a higher degree of FvEGS2 promoter activation by FaEOBII due to the presence of a greater number of MBS regulatory boxes (Supplemental Fig. S4). Nevertheless, results in F. vesca may also indicate the presence of other TFs that regulate, synergistically with EOBII, EGS2 expression.
FaEOBII Expression Is Ripening Related and Correlates with Eugenol Production in Different Strawberry Plant Tissues
Eugenol is the main phenylpropanoid volatile compound that contributes to strawberry aroma (Pyysalo et al., 1979; Ulrich et al., 1995, 2007; Zorrilla-Fontanesi et al., 2012). In ripe strawberry fruits, eugenol is biosynthesized by the action of FaEGS2, whose coding gene has a ripening-related expression pattern and presents its maximum expression at the red-ripe stage (Aragüez et al., 2013). Besides, the expression of FaEGS2 was specific to ripe fruit receptacles (Aragüez et al., 2013).
The expression pattern of the FaEOBII gene was ripening related and mainly confined to fruit receptacles, although a high expression level in petals was found (Fig. 2). Interestingly, the expression pattern of FaEOBII was similar to that of FaCAD1 and FaEGS2 (Fig. 3A) and other ripening-related genes that are responsible for the organoleptic properties of the strawberry fruit (Medina-Escobar et al., 1997; Moyano et al., 1998; Trainotti et al., 1999; Blanco-Portales et al., 2002, 2004; Benítez-Burraco et al., 2003; Raab et al., 2006; Griesser et al., 2008; Cumplido-Laso et al., 2012; Molina-Hidalgo et al., 2013; Medina-Puche et al., 2014). Besides, the maximum FaEOBII gene expression as well as FaCAD1 and FaEGS2 expression were found in those tissues, such as red-ripened receptacles or petals, where eugenol content was highest (Fig. 3B). This indicates that, as in the case of PhEOBII in petunia petals (Spitzer-Rimon et al., 2010; Van Moerkercke et al., 2011), FaEOBII could regulate the expression of those structural genes that are responsible for the production of eugenol in both ripe fruit receptacles and petals.
In Ripe Receptacles, Eugenol Production Is Regulated by the Interaction between FaMYB10 and FaEOBII
Recently, the key role played by FaMYB10 in the regulation of those genes that are part of the flavonoid/phenylpropanoid metabolism has been functionally characterized (Medina-Puche et al., 2014). In that work, after silencing FaMYB10 expression, a down-regulation of the expression of FaCAD1, FaEGS2, and FaEOBII was observed (Medina-Puche et al., 2014). We have confirmed this result by qRT-PCR, and we also showed that, after FaMYB10 silencing, there is a concomitant reduction in the receptacle eugenol content (Fig. 11). These results, together with the fact that both TFs share the same expression pattern, support the fact that the expression of FaEOBII is under the control of FaMYB10 during the fruit-ripening process. This regulatory network is quite different from the one observed in petunia petals (Spitzer-Rimon et al., 2010; Van Moerkercke et al., 2011). In petunia, the regulation of the production of hydroxycinnamoyl alcohols, phenylpropenes such as eugenol and isoeugenol, as well as other volatile benzenoid compounds is carried out in a concerted manner by three R2R3 MYB TFs named PhEOBII, ODO1, and EOBI (Verdonk et al., 2003, 2005; Schuurink et al., 2006; Colquhoun et al., 2010b, 2011; Spitzer-Rimon et al., 2010, 2012; Colquhoun and Clark, 2011; Van Moerkercke et al., 2011). PhEOBII directly regulates the expression of ODO1 and EOBI, which in turn regulate the expression of several structural genes corresponding to the biosynthesis of volatile phenylpropanoids. On the contrary, in ripe strawberry receptacles, FaMYB10 would activate the expression of FaEOBII, which could subsequently trigger both the expression of FaCAD1 and FaEGS2 genes and eugenol production. Thus, although the involvement of the EOBII TF in the regulation of eugenol production has been maintained in both petunia and strawberry, the regulatory loops involved are clearly different. This may be because of the evolutionary distance between petunia (an asterid) and F. ananassa (a rosid).
FaEOBII Expression Is Hormonally Regulated in a Similar Way to That of Other Strawberry Ripening-Related TFs
As in the case of FaMYB10, FaEOBII gene expression was regulated in an antagonist manner by auxins and ABA, the two most important hormones that control strawberry developmental and ripening processes (Perkins-Veazie, 1995; Chai et al., 2011; Jia et al., 2011; Medina-Puche et al., 2014). Thus, the decline in the inner contents of auxins in green unripe receptacles gave rise to an increased FaEOBII expression (Fig. 5). This expression model has been described for other receptacle-specific and ripening-related genes (Moyano et al., 1998; Blanco-Portales et al., 2002; Raab et al., 2006; Cumplido-Laso et al., 2012; Molina-Hidalgo et al., 2013). On the contrary, when ABA content in the receptacle was diminished by transient silencing of FaNCED1 expression or inhibition of the corresponding enzyme activity by the inhibitor NDGA, a decrease in FaEOBII expression was observed. However, when the concentration of ABA was increased in fruit receptacles as a consequence of subjecting the fruits to water stress, an increase in FaEOBII gene expression was observed (Fig. 6). In both climacteric and nonclimacteric fruits, water deficiency generated a rise in ABA content, which accelerated ripening and was accompanied by changes in both metabolites and gene expression (Castellarin et al., 2007; Gong et al., 2010). This dual regulation has also been observed for other genes whose expression is also up-regulated throughout the strawberry fruit-ripening process (Cumplido-Laso et al., 2012; Daminato et al., 2013; Molina-Hidalgo et al., 2013) and molecularly supported both the proposal of Perkins-Veazie (1995) and the findings described by Chai et al. (2011), Jia et al. (2011), and Medina-Puche et al. (2014). This hormonally regulated expression pattern was also similar to that observed for the FaMYB10 gene (Medina-Puche et al., 2014), and it strengthens the relationship between FaMYB10 and FaEOBII. In this sense, we cannot discount that the hormonal regulation of FaEOBII could be indirect as a result of the regulation of FaEOBII expression by FaMYB10.
CONCLUSION
In summary, this work presents novel functional data indicating that FaEOBII is a hormonal and ripening-related TF that regulates, by a novel regulatory network, key structural genes of the phenylpropanoid pathway that are related to eugenol biosynthesis in the strawberry fruit receptacle.
MATERIALS AND METHODS
Plant Material
Strawberry plants (Fragaria × ananassa ‘Camarosa,’ an octoploid cultivar, and Fragaria vesca, a diploid wild strawberry) were grown under field conditions in Huelva, in southwestern Spain. F. ananassa fruits were harvested at different developmental stages: small-sized green fruits (2–3 g), full-sized green fruits (4–7 g), white fruits (5–8 g), full-ripe red fruits (6–10 g), overripe fruits (6–10 g), and senescent fruits (6–10 g). Selected overripe fruits were harder and dark redder than red ones, while senescent fruits were softer. Other tissues were harvested as well, such as stolons, roots, crowns, flowers, petals, and expanding leaves. All tissues were immediately frozen in liquid nitrogen and stored at −80°C. Nicotiana benthamiana and strawberry (F. ananassa ‘Elsanta’) plants used for agroinfiltration were grown in plant chambers at 25°C, 10,000 lux, and 80% humidity and afterward were held in a greenhouse.
Auxin Treatment
Achenes of two sets of 50 full-sized green fruits (G3) each were carefully removed from their receptacles. One set of deachened G3 fruits was treated with IAA in lanolin paste (1 mL) containing 1 mm IAA in 1% (w/v) dimethyl sulfoxide. The other set of G3 deachened fruits (control group) was treated with the same paste but without IAA. Auxin treatments, sample collection, and analysis were performed according to Medina-Puche et al. (2014).
Water Stress Treatment
Fruits at the green to white stages from F. ananassa ‘Elsanta’ were used in this experiment. For water stress treatments, fruit pedicels were kept outdoors, whereas those of control fruits were immersed in Murashige and Skoog medium with Suc (renewed every 2 d). The stress treatment condition and sample analysis followed the instructions of Medina-Puche et al. (2014).
NDGA Treatment
NDGA is an ideal inhibitor of the NCED enzyme and was used to block ABA biosynthesis (Creelman et al., 1992). Strawberry fruits (‘Elsanta’) at mature green stages were used in this study. The treatment was performed in accordance with Medina-Puche et al. (2014). These samples were used for ABA content measurement and relative expression of the FaEOBII gene.
Preparation of Deuterated ABA and ABA Extraction Procedure
Deuterated ABA was used as an internal standard. Both the deuterated ABA preparation and ABA extraction from strawberry samples followed the instructions of Medina-Puche et al. (2014).
HPLC-Mass Spectrometry Conditions
In order to determine the ABA amount in strawberry fruits, we used an HPLC-mass spectrometry system (Varian 1200L Triple Quadrupole) with a column (150- × 2.1-mm-i.d. Phenomenex C18 with a 3-µm particle). The conditions and procedure used for the analysis were those described by Medina-Puche et al. (2014).
Solid-Phase Microextraction of Eugenol
Eugenol was analyzed in F. ananassa ‘Elsanta’ fruits infiltrated with Agrobacterium tumefaciens carrying pFRN, pFRN-FaEOBII, pK7WG2.0, or pK7WG2-FaEOBII and F. ananassa ‘Camarosa’ and F. vesca fruits at different developmental stages. Ten strawberry fruits were harvested per sample, and three samples were measured on each occasion. Fruits were ground to a fine powder under liquid nitrogen. Saturated NaCl (300 µL) was added to 1 g, 200 mg, or 10 mg of powdered F. ananassa, F. vesca white fruits, or F. vesca red fruits, respectively. β-Caryophyllene (3 ng) was added as an internal standard. Solid-phase microextraction was performed on a 7890A GC system (Agilent; http://www.home.agilent.com/) coupled to a 7200A QTOF-MS device (Agilent). Samples were incubated for 10 min at 50°C, after which volatiles were extracted on a 100-μm PDMS fiber (Supelco; www.sigmaaldrich.com/Supelco) for 30 min at 50°C. Extracted compounds were desorbed for 1 min at 250°C in the injector port in splitless injection mode. Volatiles were separated on an HP5MS column (5% phenyl methyl silox, 30 m × 250 μm × 0.25 μm), with a constant helium flow of 1.2 mL min−1. Oven temperature was set to 40°C for 3 min, ramped at 5°C min−1 to 250°C, and held at 250°C for 5 min. Twenty mass spectra were recorded every second (mass-to-charge ratio 30–500) at an ionization energy of 70 eV. Eugenol was measured in commercial strawberry, and no eugenol was detected above the detection limits of the system. Therefore, a calibration curve of eugenol (Acros Organics) carried out in extracts of these strawberries lacking eugenol was used to identify and quantify it. Statistical significance was tested with one-way ANOVA and an lsd post hoc test or with Student’s t test using SPSS software.
Total Anthocyanidin Analysis
Anthocyanidin extraction was performed following the instructions of Medina-Puche et al. (2014). The anthocyanidin presence was measured at 515 nm. The amount of anthocyanins was calculated by using Emolar = 36,000 L mol–1 cm–1 (Woodward, 1972; Bustamante et al., 2009). Three replicates were performed for each analyzed treatment.
RNA Isolation
Total RNA was isolated from independent pools of strawberry fruits at different growth and ripening stages and from vegetative tissues in accordance with Asif et al. (2000). Achenes were always removed from fruits, and only receptacle RNA was extracted and purified. The total RNA obtained was treated with DNase I (RNase free; Invitrogen), following the manufacturer’s instructions, and further purified using the RNeasy Mini Kit (Qiagen). RNA samples were considered DNA free when no amplicons corresponding to the analyzed genes were observed using RNA as template in a standard PCR.
Microarray Generation and Analysis
We compared the cDNA sequences contained in Kevin Folta’s libraries (Shulaev et al., 2011) with our own previously obtained cDNA libraries (Bombarely et al., 2010). The percentage identity between the cDNA sequences from F. ananassa and those from F. vesca was always over 98.6% and similar to that found previously (Bombarely et al., 2010). Therefore, we decided to generate a custom-made oligonucleotide-based (60-mer length) microarray platform containing a total of 34,187 singletons corresponding to those sequences published in the Strawberry Genome Project (https://strawberry.plantandfood.co.nz/).
For each sequence, four different oligonucleotides were printed per block, and four blocks were printed per data set. Samples of total RNA were DNase I treated and afterward purified with Qiagen columns, in accordance with the manufacturer’s conditions. Sample labeling (Cy3), hybridization, and data normalization were performed by NimbleGen Systems, in accordance with the procedures described in its expression analysis section (http://www.nimblegen.com/). Briefly, 10 μg of total receptacle RNA was processed using the Roche cDNA Synthesis System, and cDNAs were purified using High Pure PCR Product Purification following recommendations. Three biological replicates were performed, obtaining three independent cDNA populations. Each cDNA sample (1 μg) was random primer labeled with Cy3-nonamers using NimbleGen One-Color DNA Labeling Kits, in accordance with Roche NimbleGen’s standard protocol. Using random assignment, each Cy3-labeled cDNA sample was applied to custom-made strawberry 12 × 135K array formats (each slide containing 12 independent arrays, each with 140,856 probes covering 34,187 genes, four probes per target gene). The array was then hybridized for 16 h at 42°C, washed, dried, and scanned at a 2-μm resolution using a NimbleGen MS 200 Microarray Scanner (Roche NimbleGen). NimbleScan version 2.6 software was used to extract fluorescence intensity signals from scanned images and to perform robust multiarray analyses to generate raw gene expression values.
Differential expression analysis was performed using the Array Star software (DNASTAR). Moderated Student’s t test and false discovery rate (Benjamini and Hochberg, 1995) for multiple testing corrections were used at P < 0.01 to statistically identify significant differences.
Validation of Microarray Data and Expression Analysis by qRT-PCR
Expression analyses of FaEOBII (KM099230) and the genes herein analyzed and studied in all physiological conditions were performed by qRT-PCR using the iCycler system (Bio-Rad), as described previously (Benítez-Burraco et al., 2003). First strand cDNA was obtained from 2 μg of total RNA using the iScript kit (Bio-Rad) following the manufacturer’s instructions. Supplemental Table S5 depicts the primer sequences used for quantitative amplification. Each reaction was performed at least in triplicate, and the corresponding Ct values were normalized using the Ct value corresponding to an interspacer 26S-18S strawberry RNA gene (Benítez-Burraco et al., 2003; Raab et al., 2006; Encinas-Villarejo et al., 2009; Cumplido-Laso et al., 2012; Molina-Hidalgo et al., 2013). All these values were then used to determine the relative increase or decrease of FaEOBII expression in the samples as compared with those in the control, in accordance with Pedersen (2001).
The interspacer 26S-18S gene (primers 5′-ACCGTTGATTCGCACAATTGGTCATCG-3′ and 5′-TACTGCGGGTCGGCAATCGGACG-3′) was selected as a control gene owing to its constitutive expression throughout all of the different tested experimental conditions (Supplemental Table S6). The efficiency of each particular qRT-PCR and the melting curves of the products were also analyzed to ensure the existence of a single amplification peak corresponding to a unique molecular species (melting temperature was 87°C for the FaEOBII gene and 91.5°C for the interspacer 26S-18S gene).
Cloning Full-Length cDNA of FaEOBII and Sequence Analysis
We used the resources of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and the European Bioinformatics Institute server (http://www.ebi.ac.uk/) for the in silico study of the FaEOBII gene sequence. To compare the nucleotide FaEOBII sequence against databases, we used the National Center for Biotechnology Information BLASTN program as an alignment tool. Multiple sequence alignment and phylogenetic tree construction were performed with the European Bioinformatics Institute ClustalW2 program or the MegAlign program (from the Lasergene suite) as well as the FigTree program (http://tree.bio.ed.ac.uk/software/figtree/). Prediction of domains and functional sites was performed with the InterProScan database (version 4.8; www.ebi.ac.uk/Tools/pfa/iprscan/), and prediction of cellular location was performed with the WoLF PSORT program (http://www.genscript.com/psort/wolf_psort.html). BLASTN has also been used to determine gene location in the F. vesca genome in the Genome Database for Rosaceae data bank (http://www.rosaceae.org).
Generation of FaEOBII-RNAi and FaEOBII Overexpression Constructs and Transfection of Strawberry Fruit by Agroinfiltration
The FaEOBII gene was cloned into pFRN binary vector (Supplemental Fig. S6; courtesy of Marten Denekamp, Department of Molecular Cell Biology, University of Utrecht) using Gateway technology (Invitrogen). A 448-bp conserved region from the FaEOBII cDNA was PCR amplified and used in the silencing construct, using forward 5′-AGGGCCGTGGACCATGGAAGAGG-3′ and reverse 5′-CGCCGTCGGGGCGCATAGAGG-3′ primers. The resulting fragment was cloned into pCR8/GW/TOPO (Invitrogen) and subsequently transferred to the Gateway pFRN vector using LR Clonase (Invitrogen). The RNAi construct generated (pFRN-FaEOBII) was tested by sequencing prior to strawberry plant transformation.
The 624-bp coding sequence of strawberry FaEOBII was amplified from F. ananassa cDNA using the oligonucleotides attB1-EOBII-fw (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAACAATGGATAAAAAACCATGCAATTCATC-3′) and attB2-EOBII-rv (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTACTCTCCATTTAGTAACTGCATAGACC-3′), cloned into the pDONR221 (Invitrogen) plasmid by the BP Gateway reaction (Invitrogen), and subsequently transferred to the Gateway pK7WG2.0 vector using LR Clonase (Invitrogen). The overexpression construct generated (OE-FaEOBII) was tested by sequencing prior to strawberry plant transformation.
The A. tumefaciens strain AGL0 (Lazo et al., 1991) containing the pFRN-FaEOBII or empty pFRN vector and pK7WG2-FaEOBII or empty pK7WG2 vector was used to inject entire strawberry fruits (F. ananassa ‘Elsanta’), according to Hoffmann et al. (2006). Overexpression of FaEOBII was carried out by the coinfiltration of A. tumefaciens harboring the silencing suppressor pTBSV-p19 (Voinnet et al., 2003). A total of 15 to 25 strawberry plants and 30 to 40 agroinjected fruits were inoculated and analyzed, respectively. The same protocol was followed to generate the FaNCED1-RNAi and FaMYB10-RNAi constructs and to silence FaNCED1 and FaMYB10 expression in strawberry fruits. Forward and reverse primers used for FaNCED1 were 5′-CGGTGCCGCCAGCTACGCAT-3′ and 5′-GTGGGGGCCGCCAGAGGGAT-3′, while those for FaMYB10 were 5′-AGATGACTAGATGATTGCTTGCCG-3′ and 5′-TGCCGGACGATTGCCAGGAAG-3′, respectively (Medina-Puche et al., 2014).
In all cases, the silencing or overexpression percentages were determined by comparing the amounts of FaEOBII, FaNCED1, or FaMYB10 transcripts in RNAi and overexpression agroinjected fruits against those observed in empty pFRN or pK7WG2 vector agroinfiltrated fruits, respectively.
Subcellular Location Analysis
The 624-bp coding sequence of strawberry FaEOBII was amplified from F. ananassa cDNA using the oligonucleotides attB1-EOBII-fw (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAACAATGGATAAAAAACCATGCAATTCATC-3′) and attB2-EOBII-rv (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTACTCTCCATTTAGTAACTGCATAGACC-3′) and cloned into the pDONR221 (Invitrogen) plasmid by the BP Gateway reaction (Invitrogen). FaEOBII was fused in frame with GFP at the N-terminal position by LR Gateway recombination (Invitrogen) in the destination vector pK7WGF2 (Karimi et al., 2002), resulting in the 35S::GFP::FaEOBII fusion. The generated GFP construct (pK7WGF2-FaEOBII) was tested through sequencing prior to N. benthamiana leaf transformation.
Agroinfiltration experiments were performed on N. benthamiana leaves. The A. tumefaciens strain GV3101 (Weigel and Glazebrook, 2006) containing pK7WGF2-FaEOBII, the empty pK7WGF2, and the silencing suppressor pTBSV-p19 (Voinnet et al., 2003) was grown at 28°C in Luria-Bertani medium with appropriate antibiotics. Cells were harvested and resuspended in an infiltration buffer (10 mm MgSO4, 10 mm MES, and 1 mm acetosyringone). This step was repeated once, and the concentration of bacterial suspension was measured by spectroscopy (optical density at 600 nm) and adjusted to a final concentration of 0.5 to 0.8. Then, the cells carrying the pK7WGF2-FaEOBII and the empty pK7WGF2 constructs were mixed with cells containing the p19 vector pairwise and left at room temperature for 2 to 4 h in darkness. Bacterial suspensions were syringe infiltrated through the abaxial surface of the leaves, and material was collected after 4 d. To locate nuclei precisely, N. benthamiana leaf tissues were incubated with DAPI. Leaves were observed by epifluorescence microscopy (LSM 5 EXCITER; Carl Zeiss), with excitation/emission settings of 488/500 to 515 nm for GFP and 405/449 to 461 nm for DAPI.
Protein Purification and Determination of FaEOBII DNA-Binding Specificity
Translational fusion of FaEOBII to Maltose Binding Protein was obtained by cloning its corresponding cDNA into the pMAL-c2 vector (New England Biolabs) using Gateway technology. Donor template was obtained through PCR amplification of FaEOBII with specific oligonucleotides attB1-EOBII-fw (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAACAATGGATAAAAAACCATGCAATTCATC-3′) and attB2-EOBII-rv (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTACTCTCCATTTAGTAACTGCATAGACC-3′). The recombinant insert was verified by sequencing, and the plasmid was introduced into Escherichia coli BL-21. The expression and purification of recombinant protein were also described for the pMAL purification system (New England Biolabs). FaEOBII DNA-binding specificity was determined using protein-binding microarrays (PBM11) using soluble protein extracts from an induced E. coli culture, as described by Godoy et al. (2011).
Isolation and in Silico Analysis of the CAD Promoter
The promoter region of CAD was identified from the strawberry genome database (http://www.strawberry.org; Shulaev et al., 2011), and PLACE (www.dna.affrc.go.jp) was used to detect cis-acting regulatory DNA elements in the CAD promoter (Higo et al., 1999). For further analysis, a 2,638-bp fragment of the promoter region, including the 5′ untranslated region, was isolated from genomic DNA using primers pCAD-up (5′-ACTCCGACGTTTGTTGAGAGA-3′) and pCAD-low (5′-TCTCGATCTGTGGCAGATTCTT-3′) and following the manufacturer’s instructions for the Phire Plant Direct PCR Kit (Thermo Scientific). PCR products were purified and cloned into the pCR8/GW/TOPO entry vector (Invitrogen) by TOPOTA Cloning (Invitrogen). pCAD1 was fused with GUS by LR Gateway recombination (Invitrogen) in destination vector pKGWFS7 (Karimi et al., 2002), resulting in pCAD1::GUS fusion. The generated GUS construct (pKGWFS7-pCAD1) was sequence verified prior to N. benthamiana leaf transformation.
Transient Transactivation Assay in N. benthamiana Leaves
A. tumefaciens GV3101 (pMP90) cultures harboring reporter or effector constructs were grown overnight and diluted in infiltration buffer (50 mm MES, pH 5.8, 0.5% [w/v] Glc, 2 mm Na3PO4, and 100 μm acetosyringone) to an optical density at 600 nm of 0.3 prior to infiltration in N. benthamiana leaves. Combinations of reporter and effector constructs were coinfiltrated in leaves of glasshouse-grown N. benthamiana plants and incubated for 48 h. Infiltration regions were marked, and five leaf discs were pooled per sample. Six leaves on two plants were infiltrated for each effector/reporter combination (n = 6), and the experiment was repeated twice. To enable normalization, leaves were coinfiltrated with a CaMV 35S:LUCIFERASE (LUC)-harboring A. tumefaciens.
Tissues were homogenized in liquid N2, and proteins were extracted from 50 mg of tissue in 150 μL of cold luciferase cell culture lysis reagent (25 mm Tris phosphate, pH 7.8, 2 mm dithiothreitol, 2 m 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 1% [v/v] Triton X-100, and 10% [v/v] glycerol [Promega; http://www.promega.com]) supplemented with a complete protease inhibitor (Roche; http://www.roche.com). Samples were vortexed for 30 s and centrifuged at 4°C for 30 min at 13,000g. GUS activity of crude extracts was determined spectrophotometrically using 1 mm 4-methylumbelliferyl-d-glucuronide in cell culture lysis reagent supplemented with 10 mm β-mercaptoethanol as substrate after a 30-min incubation period at 37°C. Measurements were performed in a FluoroCount Microplate Fluorometer (Packard BioScience; http://www.packardinstrument.com) using a 360-nm excitation filter and a 460-nm emission filter. For LUC measurements, 20 μL of extract was analyzed in 80 μL of luciferase assay buffer (20 mm Tricine, 2.67 mm MgSO4, 0.1 mm EDTA, 33 mm dithiothreitol, 270 μm CoA, 530 μm ATP, and 470 μm d-luciferin, pH 7.8; Leeuwen et al., 2000) and measured in a FluoroCount Microplate Fluorometer (Packard BioScience) using a 560-nm emission filter (Van Moerkercke et al., 2011).
For promoter analysis in N. benthamiana leaves and the transactivation assays, the pODO1:GUS reporter construct was used as a control and pCAD1:GUS was used as a test. GUS and LUC activity were determined as described by Van Moerkercke et al. (2011). For the transactivation assays, 35S:FaEOBII was used as the effector construct. As a control, the previously described constructs 35S:PhEOBII, 35S:AN2, and 35S:LUC (Verdonk et al., 2005; Van Moerkercke et al., 2011) were used.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FaEOBII (KM099230), FaMYB10 (EU155162), PhEOBII (EU360893), FvEOBII (XP004304632), FaEGS1 (KF562264), FaEGS2 (KF562266), FaCAD1 (U63534), and FaNCED1 (HQ290318).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Screenshots corresponding to the results of bioinformatic predictions.
Supplemental Figure S2. Sequence alignment of R2R3-MYB proteins.
Supplemental Figure S3. Phylogenetic tree of 206 MYB TFs.
Supplemental Figure S4. Schematic diagram of the EGS2 promoter from F. ananassa and F. vesca.
Supplemental Figure S5. Overview of the phenotypes of fruits analyzed in studies of gene expression regulated by ABA.
Supplemental Figure S6. Structure of the binary vector pFRN.
Supplemental Table S1. Subgroup classification of 61 plant MYB sequences examined in this study and additional information.
Supplemental Table S2. MYB TFs from strawberry.
Supplemental Table S3. Total microarray data from a transcriptomic comparison between transgenic transiently silenced FaEOBII fruits and control fruits.
Supplemental Table S4. Microarray and qRT-PCR results of the differentially regulated genes after silencing FaEOBII.
Supplemental Table S5. Primer sequences used for qRT-PCR.
Supplemental Table S6. Gene expression values of different housekeeping genes obtained by qRT-PCR analysis.
Supplementary Material
Glossary
- TF
transcription factor
- qRT
quantitative real-time
- cDNA
complementary DNA
- CaMV
cauliflower mosaic virus
- DAPI
4′,6-diamidino-2-phenylindole dihydrochloride
- ABA
abscisic acid
- IAA
indole-3-acetic acid
- NDGA
nordihydroguaiaretic acid
- RNAi
RNA interference
- a.u.i.
arbitrary units of intensity
- Ct
cycle threshold
- GC
gas chromatography
- MS
mass spectrometry
Footnotes
This work was supported by the Spanish Ministerio de Ciencia e Innovación (grant nos. BIO2010–19322, BIO2010–21739, CSD2007–00057, and EUI2008–03666), by the Spanish Ministerio de Educación y Ciencia within the framework of the Formación del Profesorado Universitario program (Ph.D. fellowship to L.M.-P.), and by the Campus de Excelencia Internacional Agroalimentario 3 from the University of Córdoba (postdoctoral contract to R.B.-P.).
References
- Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JN, O’Connell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28: 319–332 [DOI] [PubMed] [Google Scholar]
- Aragüez I, Osorio S, Hoffmann T, Rambla JL, Medina-Escobar N, Granell A, Botella MÁ, Schwab W, Valpuesta V (2013) Eugenol production in achenes and receptacles of strawberry fruits is catalyzed by synthases exhibiting distinct kinetics. Plant Physiol 163: 946–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M, Dhawan P, Nath P (2000) A simple procedure for the isolation of high quality RNA from ripening banana fruit. Plant Mol Biol Rep 18: 109–115 [Google Scholar]
- Aubert C, Pitrat M (2006) Volatile compounds in the skin and pulp of Queen Anne’s pocket melon. J Agric Food Chem 54: 8177–8182 [DOI] [PubMed] [Google Scholar]
- Benítez-Burraco A, Blanco-Portales R, Redondo-Nevado J, Bellido ML, Moyano E, Caballero JL, Muñoz-Blanco J (2003) Cloning and characterization of two ripening-related strawberry (Fragaria × ananassa cv. Chandler) pectate lyase genes. J Exp Bot 54: 633–645 [DOI] [PubMed] [Google Scholar]
- Benjamini YH, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Berger MF, Bulyk ML (2009) Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat Protoc 4: 393–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Portales R, López-Raéz JA, Bellido ML, Moyano E, Dorado G, González-Reyes JA, Caballero JL, Muñoz-Blanco J (2004) A strawberry fruit-specific and ripening-related gene codes for a HyPRP protein involved in polyphenol anchoring. Plant Mol Biol 55: 763–780 [DOI] [PubMed] [Google Scholar]
- Blanco-Portales R, Medina-Escobar N, López-Ráez JA, González-Reyes JA, Villalba JM, Moyano E, Caballero JL, Muñoz-Blanco J (2002) Cloning, expression and immunolocalization pattern of a cinnamyl alcohol dehydrogenase gene from strawberry (Fragaria × ananassa cv. Chandler). J Exp Bot 53: 1723–1734 [DOI] [PubMed] [Google Scholar]
- Bombarely A, Merchante C, Csukasi F, Cruz-Rus E, Caballero JL, Medina-Escobar N, Blanco-Portales R, Botella MA, Muñoz-Blanco J, Sánchez-Sevilla JF, et al. (2010) Generation and analysis of ESTs from strawberry (Fragaria × ananassa) fruits and evaluation of their utility in genetic and molecular studies. BMC Genomics 11: 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CA, Civello PM, Martínez GA (2009) Cloning of the promoter region of β-xylosidase (FaXyl1) gene and effect of plant growth regulators on the expression of FaXyl1 in strawberry fruit. Plant Sci 177: 49–56 [Google Scholar]
- Castellarin SD, Matthews MA, Di Gaspero G, Gambetta GA (2007) Water deficits accelerate ripening and induce changes in gene expression regulating flavonoid biosynthesis in grape berries. Planta 227: 101–112 [DOI] [PubMed] [Google Scholar]
- Chai YM, Jia HF, Li CL, Dong QH, Shen YY (2011) FaPYR1 is involved in strawberry fruit ripening. J Exp Bot 62: 5079–5089 [DOI] [PubMed] [Google Scholar]
- Colquhoun TA, Clark DG (2011) Unraveling the regulation of floral fragrance biosynthesis. Plant Signal Behav 6: 378–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun TA, Kim JY, Wedde AE, Levin LA, Schmitt KC, Schuurink RC, Clark DG (2011) PhMYB4 fine-tunes the floral volatile signature of Petunia × hybrida through PhC4H. J Exp Bot 62: 1133–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun TA, Schimmel BCJ, Kim JY, Reinhardt D, Cline K, Clark DG (2010a) A petunia chorismate mutase specialized for the production of floral volatiles. Plant J 61: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun TA, Verdonk JC, Schimmel BCJ, Tieman DM, Underwood BA, Clark DG (2010b) Petunia floral volatile benzenoid/phenylpropanoid genes are regulated in a similar manner. Phytochemistry 71: 158–167 [DOI] [PubMed] [Google Scholar]
- Creelman RA, Bell E, Mullet JE (1992) Involvement of a lipoxygenase-like enzyme in abscisic acid biosynthesis. Plant Physiol 99: 1258–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumplido-Laso G, Medina-Puche L, Moyano E, Hoffmann T, Sinz Q, Ring L, Studart-Wittkowski C, Caballero JL, Schwab W, Muñoz-Blanco J, et al. (2012) The fruit ripening-related gene FaAAT2 encodes an acyl transferase involved in strawberry aroma biogenesis. J Exp Bot 63: 4275–4290 [DOI] [PubMed] [Google Scholar]
- Dal Cin V, Tieman DM, Tohge T, McQuinn R, de Vos RC, Osorio S, Schmelz EA, Taylor MG, Smits-Kroon MT, Schuurink RC, et al. (2011) Identification of genes in the phenylalanine metabolic pathway by ectopic expression of a MYB transcription factor in tomato fruit. Plant Cell 23: 2738–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daminato M, Guzzo F, Casadoro G (2013) A SHATTERPROOF-like gene controls ripening in non-climacteric strawberries, and auxin and abscisic acid antagonistically affect its expression. J Exp Bot 64: 3775–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter R, Qualley A, Kish CM, Ma CJ, Koeduka T, Nagegowda DA, Dudareva N, Pichersky E, Clark D (2007) Characterization of a petunia acetyltransferase involved in the biosynthesis of the floral volatile isoeugenol. Plant J 49: 265–275 [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Encinas-Villarejo S, Maldonado AM, Amil-Ruiz F, de los Santos B, Romero F, Pliego-Alfaro F, Muñoz-Blanco J, Caballero JL (2009) Evidence for a positive regulatory role of strawberry (Fragaria × ananassa) Fa WRKY1 and Arabidopsis At WRKY75 proteins in resistance. J Exp Bot 60: 3043–3065 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R (2014) DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA 111: 2367–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy M, Franco-Zorrilla JM, Pérez-Pérez J, Oliveros JC, Lorenzo O, Solano R (2011) Improved protein-binding microarrays for the identification of DNA-binding specificities of transcription factors. Plant J 66: 700–711 [DOI] [PubMed] [Google Scholar]
- Gong P, Zhang J, Li H, Yang C, Zhang C, Zhang X, Khurram Z, Zhang Y, Wang T, Fei Z, et al. (2010) Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J Exp Bot 61: 3563–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesser M, Vitzthum F, Fink B, Bellido ML, Raasch C, Munoz-Blanco J, Schwab W (2008) Multi-substrate flavonol O-glucosyltransferases from strawberry (Fragaria × ananassa) achene and receptacle. J Exp Bot 59: 2611–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T, Kalinowski G, Schwab W (2006) RNAi-induced silencing of gene expression in strawberry fruit (Fragaria × ananassa) by agroinfiltration: a rapid assay for gene function analysis. Plant J 48: 818–826 [DOI] [PubMed] [Google Scholar]
- Hoffmann T, Kurtzer R, Skowranek K, Kiessling P, Fridman E, Pichersky E, Schwab W (2011) Metabolic engineering in strawberry fruit uncovers a dormant biosynthetic pathway. Metab Eng 13: 527–531 [DOI] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY (2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol 157: 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Joyce D (2003) ABA effects on ethylene production, PAL activity, anthocyanin and phenolic contents of strawberry fruit. Plant Growth Regul 39: 171–174 [Google Scholar]
- Jordán MJ, Tandon K, Shaw PE, Goodner KL (2001) Aromatic profile of aqueous banana essence and banana fruit by gas chromatography-mass spectrometry (GC-MS) and gas chromatography-olfactometry (GC-O). J Agric Food Chem 49: 4813–4817 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Koeduka T, Fridman E, Gang DR, Vassão DG, Jackson BL, Kish CM, Orlova I, Spassova SM, Lewis NG, Noel JP, et al. (2006) Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc Natl Acad Sci USA 103: 10128–10133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeduka T, Louie GV, Orlova I, Kish CM, Ibdah M, Wilkerson CG, Bowman ME, Baiga TJ, Noel JP, Dudareva N, Pichersky E (2008) The multiple phenylpropene synthases in both Clarkia breweri and Petunia hybrida represent two distinct protein lineages. Plant Journal 54: 362–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeduka T, Suzuki S, Iijima Y, Ohnishi T, Suzuki H, Watanabe B, Shibata D, Umezawa T, Pichersky E, Hiratake J (2013) Enhancement of production of eugenol and its glycosides in transgenic aspen plants via genetic engineering. Biochem Biophys Res Commun 436: 73–78 [DOI] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, et al. (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16: 263–276 [DOI] [PubMed] [Google Scholar]
- Larsen M, Poll L (1992) Odour thresholds of some important aroma compounds in strawberries. Z Lebensm Unters Forsch 195: 120–123 [Google Scholar]
- Latrasse A. (1991) Volatile compounds in food and beverages. In Maarse H, ed, Fruits III. Marcel-Dekker, New York, pp 329–387 [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (N Y) 9: 963–967 [DOI] [PubMed] [Google Scholar]
- Leeuwen W, Hagendoorn MM, Ruttink T, Poecke R, Plas LW, Krol A (2000) The use of the luciferase reporter system for in planta gene expression studies. Plant Mol Biol Rep 18: 143–144 [Google Scholar]
- Li J, Yang X, Wang Y, Li X, Gao Z, Pei M, Chen Z, Qu LJ, Gu H (2006) Two groups of MYB transcription factors share a motif which enhances trans-activation activity. Biochem Biophys Res Commun 341: 1155–1163 [DOI] [PubMed] [Google Scholar]
- Liu G, Ren G, Guirgis A, Thornburg RW (2009) The MYB305 transcription factor regulates expression of nectarin genes in the ornamental tobacco floral nectary. Plant Cell 21: 2672–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K. (1998) Isolation of a set of ripening-related genes from strawberry: their identification and possible relationship to fruit quality traits. Planta 205: 622–631 [DOI] [PubMed] [Google Scholar]
- Medina-Escobar N, Cárdenas J, Moyano E, Caballero JL, Muñoz-Blanco J (1997) Cloning, molecular characterization and expression pattern of a strawberry ripening-specific cDNA with sequence homology to pectate lyase from higher plants. Plant Mol Biol 34: 867–877 [DOI] [PubMed] [Google Scholar]
- Medina-Puche L, Cumplido-Laso G, Amil-Ruiz F, Hoffmann T, Ring L, Rodríguez-Franco A, Caballero JL, Schwab W, Muñoz-Blanco J, Blanco-Portales R (2014) MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria × ananassa fruits. J Exp Bot 65: 401–417 [DOI] [PubMed] [Google Scholar]
- Min T, Kasahara H, Bedgar DL, Youn B, Lawrence PK, Gang DR, Halls SC, Park H, Hilsenbeck JL, Davin LB, et al. (2003) Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases and their relationship to isoflavone reductases. J Biol Chem 278: 50714–50723 [DOI] [PubMed] [Google Scholar]
- Molina-Hidalgo FJ, Franco AR, Villatoro C, Medina-Puche L, Mercado JA, Hidalgo MA, Monfort A, Caballero JL, Muñoz-Blanco J, Blanco-Portales R (2013) The strawberry (Fragaria × ananassa) fruit-specific rhamnogalacturonate lyase 1 (FaRGLyase1) gene encodes an enzyme involved in the degradation of cell-wall middle lamellae. J Exp Bot 64: 1471–1483 [DOI] [PubMed] [Google Scholar]
- Moyano E, Martínez-Garcia JF, Martin C (1996) Apparent redundancy in myb gene function provides gearing for the control of flavonoid biosynthesis in Antirrhinum flowers. Plant Cell 8: 1519–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyano E, Portero-Robles I, Medina-Escobar N, Valpuesta V, Muñoz-Blanco J, Caballero JL (1998) A fruit-specific putative dihydroflavonol 4-reductase gene is differentially expressed in strawberry during the ripening process. Plant Physiol 117: 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijssen LM. (1996) Volatile Compounds in Food: Qualitative and Quantitative Data. TNO Nutrition and Food Research Institute, Zeist, The Netherlands [Google Scholar]
- Ortiz-Serrano P, Gil JV (2010) Quantitative comparison of free and bound volatiles of two commercial tomato cultivars (Solanum lycopersicum L.) during ripening. J Agric Food Chem 58: 1106–1114 [DOI] [PubMed] [Google Scholar]
- Pasay C, Mounsey K, Stevenson G, Davis R, Arlian L, Morgan M, Vyszenski-Moher D, Andrews K, McCarthy J (2010) Acaricidal activity of eugenol based compounds against scabies mites. PLoS ONE 5: e12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen SB. (2001) Multiplex relative gene expression analysis by real-time RT-PCR using the iCycler iQ detection system. BioRadiations 107: 10–11 [Google Scholar]
- Perkins-Veazie P. (1995) Growth and ripening of strawberry fruit. Hortic Rev (Am Soc Hortic Sci) 17: 267–297 [Google Scholar]
- Prouse MB, Campbell MM (2012) The interaction between MYB proteins and their target DNA binding sites. Biochim Biophys Acta 1819: 67–77 [DOI] [PubMed] [Google Scholar]
- Pyysalo T, Honkanen E, Hirvi T (1979) Volatiles of wild strawberries, Fragaria vesca L., compared to those of cultivated berries, Fragaria × ananassa cv Senga Sengana. J Agric Food Chem 27: 19–22 [Google Scholar]
- Raab T, López-Ráez JA, Klein D, Caballero JL, Moyano E, Schwab W, Muñoz-Blanco J (2006) FaQR, required for the biosynthesis of the strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone, encodes an enone oxidoreductase. Plant Cell 18: 1023–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RW, Moyano E, Culianez-Macia FA, Schuch W, Martin C, Bevan M (1994) A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J 13: 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieberle P, Hofmann T (1997) Evaluation of the character impact odorants in fresh strawberry juice by quantitative measurements and sensory studies on model mixtures. J Agric Food Chem 45: 227–232 [Google Scholar]
- Schuurink RC, Haring MA, Clark DG (2006) Regulation of volatile benzenoid biosynthesis in petunia flowers. Trends Plant Sci 11: 20–25 [DOI] [PubMed] [Google Scholar]
- Shin B, Choi G, Yi H, Yang S, Cho I, Kim J, Lee S, Paek NC, Kim JH, Song PS, et al. (2002) AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. Plant J 30: 23–32 [DOI] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer-Rimon B, Farhi M, Albo B, Cna’ani A, Ben Zvi MM, Masci T, Edelbaum O, Yu Y, Shklarman E, Ovadis M, et al. (2012) The R2R3-MYB-like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent-related genes in petunia. Plant Cell 24: 5089–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer-Rimon B, Marhevka E, Barkai O, Marton I, Edelbaum O, Masci T, Prathapani N-K, Shklarman E, Ovadis M, Vainstein A (2010) EOBII, a gene encoding a flower-specific regulator of phenylpropanoid volatiles’ biosynthesis in petunia. Plant Cell 22: 1961–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainotti L, Spolaore S, Pavanello A, Baldan B, Casadoro G (1999) A novel E-type endo-β-1,4-glucanase with a putative cellulose-binding domain is highly expressed in ripening strawberry fruits. Plant Mol Biol 40: 323–332 [DOI] [PubMed] [Google Scholar]
- Uimari A, Strommer J (1997) Myb26: a MYB-like protein of pea flowers with affinity for promoters of phenylpropanoid genes. Plant J 12: 1273–1284 [DOI] [PubMed] [Google Scholar]
- Ulrich D, Komes D, Olbricht K, Hoberg E (1995) Analysis of strawberry flavour-quantification of the volatile components of varieties of cultivated and wild strawberries. Z Lebensm Unters Forsch 200: 217–220 [Google Scholar]
- Ulrich D, Komes D, Olbricht K, Hoberg E (2007) Diversity of aroma patterns in wild and cultivated Fragaria accessions. Genet Resour Crop Evol 54: 1185–1196 [Google Scholar]
- Van Moerkercke A, Haring MA, Schuurink RC (2011) The transcription factor EMISSION OF BENZENOIDS II activates the MYB ODORANT1 promoter at a MYB binding site specific for fragrant petunias. Plant J 67: 917–928 [DOI] [PubMed] [Google Scholar]
- Verdonk JC, Haring MA, van Tunen AJ, Schuurink RC (2005) ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell 17: 1612–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdonk JC, Ric de Vos CH, Verhoeven HA, Haring MA, van Tunen AJ, Schuurink RC (2003) Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochemistry 62: 997–1008 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2006) In planta transformation of Arabidopsis. Cold Spring Harb Protoc 2006: 107–120 [Google Scholar]
- Woodward JR. (1972) Physical and chemical changes in developing strawberry fruits. J Sci Food Agric 23: 465–473 [DOI] [PubMed] [Google Scholar]
- Zabetakis I, Holden MA (1997) Strawberry flavour: analysis and biosynthesis. J Sci Food Agric 74: 421–434 [Google Scholar]
- Zorrilla-Fontanesi Y, Rambla JL, Cabeza A, Medina JJ, Sánchez-Sevilla JF, Valpuesta V, Botella MA, Granell A, Amaya I (2012) Genetic analysis of strawberry fruit aroma and identification of O-methyltransferase FaOMT as the locus controlling natural variation in mesifurane content. Plant Physiol 159: 851–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.